Abstract
Large-scale, quantitative analysis of gene expression can be accomplished by microarray or RNA-seq analysis. While these methods are applicable to genome-wide analysis, it is often desirable to quantify expression of a more limited set of genes in thousands or even tens of thousands of biological samples. For example, some studies may need to monitor a sizable panel of key genes under many different experimental conditions, during development, or treated with a large library of small molecules, for which current genome-wide methods are either inefficient or cost-prohibitive. This unit presents a method that permits quantitative profiling of several hundred selected genes in a large number of samples by coupling RNA-mediated oligonucleotide Annealing, Selection, and Ligation with Next-Gen sequencing (RASL-seq). The method even allows direct analysis of RNA levels in cell lysates and is also adaptable to full automation, making it ideal for large-scale analysis of multiple biological pathways or regulatory gene networks in the context of systematic genetic or chemical genetic perturbations.
Keywords: Gene Expression, RNA-mediated oligonucleotide Annealing, Selection, Ligation with multiplex sequencing (RASL-seq), Bar-Coding Strategies, High Throughput Screening (HTS)
INTRODUCTION
While microarray and RNA-seq have been widely used to profile the transcriptome in cells or tissues, it remains a major challenge to use these genome-scale methods to conduct “two-dimensional” analysis (i.e. gene expression profiling under a large array of experimental conditions). Such experimental capability would facilitate pathway dissection and elucidation of gene networks in different genetic backgrounds, in response to internal and external cues, or through genome-wide RNAi screens. Here we describe an experimental approach to obtain these objectives by taking advantage of the increasing power of high throughput sequencing, a method we referred to as RASL-seq, which is designed to follow a large panel of specific genes (hundreds) under thousands or even tens of thousands conditions.
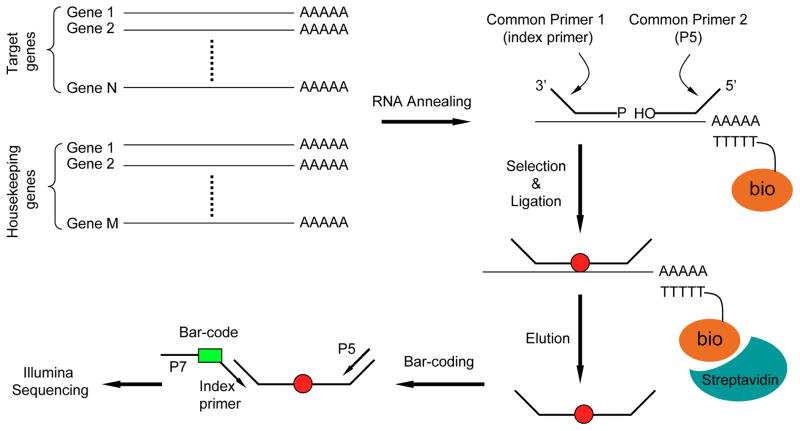
As diagramed in Figure 1, a pair of oligonucleotides is designed to target a specific splice junction region in each transcript from a selected panel of genes (Yeakley et al., 2002). Each oligonucleotide also carries a universal primer sequence and one of the primers also contains a 5′ phosphate. A pool of such probes is annealed to total RNA, and corresponding mRNAs are captured using oligo-dT biotin beads. After washing away free oligonucleotides, the aligned pairs of probes correctly annealed to their complementary templates are ligated by T4 ligase, thus converting the pairs of single probes to single PCR amplicons. The products from each sample (conditions) are indexed by performing a limited PCR amplification using a set of bar-coded primers in conjunction with a common primer. The products are pooled, purified, quantified, and subjected to high throughput sequencing.
Figure 1.
Overview of the RASL-seq technology. All steps, including annealing, selection, ligation and elution, can be carried out manually or on a customized Biomek FX robot. One of the targeting oligos (the upstream one) contains a 5′ phosphate. Each oligo contains a specific universal primer as indicated. After incorporating bar-codes during PCR, the products are pooled, purified, and sequenced on Illumina sequencer (GAII or HiSeq 2000). The first sequencing run reads the ligated region from the P5 primer and the second identifies the bar-coded region from the index primer.
A first sequencing reaction reads the ligated region of 40nt (which is the entire length of the ligated products) and a second sequence reaction decodes the bar-code region. The rationale behind this bar-coding strategy is to introduce the bar-code to each sample during post-ligation PCR. Because the positional information for individual clusters on the Illumina flowcell is recorded in the computer during the first round of sequencing, the bar-code information from the second round of sequencing can be directly linked to the sequences in the ligated region.
In our hands, we are able to index 1536 samples (which corresponds to four 384-well plates), each with a unique bar-code primer (we have synthesized a collection of 1536 bar-coded primers for this purpose), and pool all samples for sequencing in each Solexa/Illumina lane. This level of multiplexing thus permits profiling up to five hundred genes following genome-wide RNAi treatment on a pair of 8-lane Illumina flowcells that can be simultaneously processed in a single HiSeq2000 run (1536×2×8=24,576). It is optional to perform the assay with individual isolated RNA or directly on cell lysates. The entire procedure can be manually performed or implemented on a custom robot.
STRATEGIC PLANNING
Probe design
We normally design three probe pairs for each gene according to exon-exon junctions near the 3′ end of target transcript. Avoid extreme G/C content in probe design, which can also be aided by keeping all designed probes in a chosen Tm range. In any case, all probes need to be experimentally tested as described below. Considering the cost, we normally do not purify synthesized oligos because of the relatively short length.
As illustrated in Figure 1, the probe designed to hybridize on the 3′ side of the exon-exon juction has the configuration in which the 3′ portion of the probe corresponds to the common index primer and the 5′ portion to a specific 20nt target sequence complementary to an exonic sequence in a chosen transcript. This probe needs to contain a phosphate at its 5′ end.
The probe designed to hybridize on the 5′ side of the exon-exon junction has the configuration in which the 3′ portion contains a 20nt target complementary to the exonic sequence adjacent to the corresponding 3′ probe and the 5′ portion corresponds to the common P5 primer.
If a transcript lacks convenient splice junctions (e.g. intronless genes), select a number of relatively unique 40nt regions to design targeting oligos. Because of the polyA selection step in our assay, interference of genomic DNA is minimized. In addition, the oligo annealing step takes place at 65°C followed by gradual cooling, which is insufficient to denature genome DNA for efficient annealing.
Pool synthetic oligo probes according to the final concentration defined in the binding cocktail. Note that we will find out whether individual oligos work as expected from the test run described below. This avoids tedious validation for the quality of individual oligos.
Conduct a preliminary profiling using the pooled probes on 1 μg total RNA from the cell type to be analyzed, ideally under two different experimental conditions where the expected fold-changes have been pre-determined.
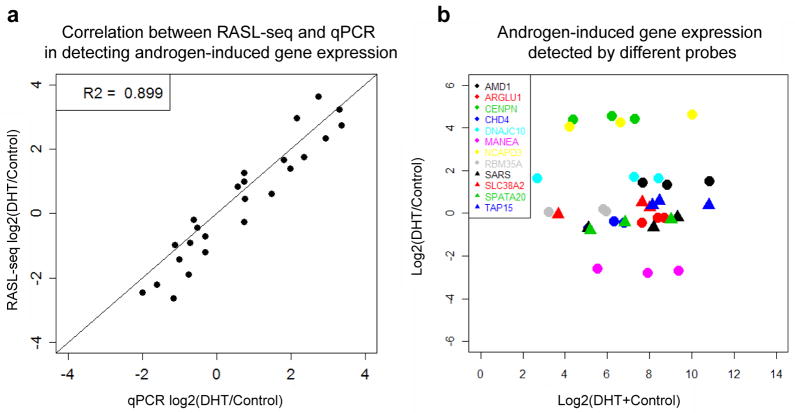
Select one pair of probes per transcript based on two criteria. First, the pair gives the expected fold-difference according to prior profiling experiments or qPCR, and second, the count per probe is between 100 and 5000 after considering the final levels of multiplexing (for example, if 100 samples to be multiplexed to monitor a 100-gene signature, the total counts in a single sample should be between 1×104 and 5×105). As shown in Figure 2, we found that the ligation efficiency of different probe pairs that target the same transcripts can significantly vary, but the recorded fold-changes are relatively similar. By choosing probe pairs that produce sequence reads in a similar range, one can select high efficiency probes to target low abundance transcripts and low efficiency probe to target high abundance transcripts, thus balancing the readouts among transcripts of different abundance.
We also detected a small number of probe pairs that produce very high reads, which might be due to their ligation templated by other non-specific sequences. Discard these pairs to prevent them contributing too many sequences in the final data set.
Figure 2.
Reproducibility of RASL-seq and similar fold-differences recorded by different oligonucleotides targeting different regions in the same transcripts. (A) Validation of androgen-induced gene expression by RT-qPCR on LNCaP cells. (B) Similar fold-differences despite distinct targeting efficiencies with different probe sets against the same transcripts.
BASIC PROTOCOL
Materials
Oligonucleotide probes, which are designed based on the rationale and principle described below. Note that each upstream oligonucleotide contains a 5′ phosphate, which can be incorporated during chemical synthesis or using T4 polynucleotide kinase.
Biotinylated oligo (dT)25. This can be chemically synthesized or directly purchased from various vendors (IDT or Life Technologies).
Streptavidin-coated magnetic beads (Fisher Scientific, cat. # 09-981-134, alternatively, oligo dT-coated plates can also be used), wash the beads with washing buffer twice before use.
MELT™ Total Nucleic Acid Isolation System for cell lysis (Ambion, cat. # AM1983).
2X binding cocktail: 40 mM Tris-HCl, pH 7.6, 1 M NaCl, 2 mM EDTA, 0.2% SDS, 5 nM each oligo probe (this provides a final concentration of 0.1 pmoles per oligo per reaction), 100 nM biotinylated oligo(dT)25, and 5 μl of streptavidin-coated magnetic beads.
Washing buffer: 20 mM Tris-HCl, pH 7.6, 0.1 M NaCl, 1 mM EDTA, 0.1% Tween-80.
T4 DNA ligase (Fermentas, cat # EL0011).
Amplitaq Gold DNA polymerase (Life Technology, cat. # N8080241).
10 mM dNTPs.
NucleoSpin® Extract II Kit for PCR purification (Clontech, cat. # 740609.50).
-
PCR primers:
The bar-coding primer: 5′-CAAGCAGAAGACGGCATACGAG(XXXXXXX)TAG CTGATGCTACGACCACAGG -3′). Each of these primers contains a 7nt bar-code sequence (barcodes consist of any nucleotide combination, and we require at least two base differences between any two barcodes) between the P7 primer sequence (5′-CAAGCAGAAGACGGCATACGAG-3′) at the 5′ end and the index primer sequence (5′-TAGCTGATGCTACGACCACAGG-3′) at the 3′ end. Each of these primers is used in combination with a P5 common primer (5′-AATGATACGGCGACCACCGAGAT-3′) for PCR. The sequences of P5 and P7 primers correspond to those on the Illumina flowcell for cluster generating (note that the nomenclature of primers is according to the Solexa system at Illumina). Thermal cycler (single or tetrade).
Illumina sequencer (ideally HiSeq2000).
Choice of the starting materials for RASL-seq
-
1
The assay can begin with isolated RNA or directly on cell lysates. If using isolated RNA, dispense ~1 μg total RNA in 20 μl RNase-free H2O in each well of 384-well plates. The minimal amount of RNA can be as low as 10 ng, which corresponds to ~1,000 cells.
-
2
If the assay is to be directly performed on cell lysates, first culture ~3,000 cells in each well of 384-well plates. This applies to all types of cells of different sizes (note that if ES cells are used, avoid having too many cells in each well, which may release too much genome DNA to cause high viscosity of the solution).
-
3
After specific treatments (e.g. RNAi, chemicals, etc), aspirate the medium, leaving ~10 μl medium in each well.
-
4
Add 10 μl of the MELT cocktail to each well and incubate at room temperature for 5 min. The lysate should be largely transparent and the viscosity is minimal. Too many cells in the culture cause high viscosity because of released genomic DNA, which should be avoided.
-
5
Proceed to the next step or keep the plates at −80°C (for any period of time as long as the content does not dry up).
RASL reaction
-
6
Add 20 μl of 2X binding cocktail (containing magnetic beads) to each well in 384-well plates containing total RNA or cell lysate.
-
7
Heat the plates at 65°C for 10 min to denature RNA followed by incubation at 45°C for 60 min to allow probe annealing.
-
8
Transfer the plates to a magnetic stand and aspirate the supernatant, leaving ~10 μl in each well.
-
9
Add 50 μl of washing buffer per well and fully resuspend the beads.
-
10
Wash the beads 5 times (repeating step 8 and 9).
-
11
Wash the beads once with 50 μl of 1X ligation buffer.
-
12
Add 20 μl of 1X ligation buffer containing 5 U (Weiss unit) of T4 DNA ligase and pipette the solution to resuspend the beads.
-
13
Incubate the plates at 37°C for 60 min.
-
14
Wash the beads 3 times with the washing buffer (as in step 8 and 9).
-
15
Resuspend the beads in 30 μl H2O.
-
16
Incubate the plates at 65°C for 5 min to release ligated probes from their templates.
-
17
Transfer the plates to the magnetic stand and then pipette 5 μl of supernatant containing released probes from each well into the corresponding well in a new 384-well PCR plate.
-
18
Store the original plates at −20°C (as backup in case of problems during bar-coding; the ligated DNA should be very stable for years). Note that the original cell culture plates are used throughout the entire RASL procedure.
Bar-coding
-
19
To each well containing 5 μl of ligated probes, add 10 μl of the PCR cocktail containing 1X Amplitag Gold PCR buffer, 0.6 μM P5 primer, 0.6 μM bar-coded primer, 1.5 mM MgCl2, 200 μM dNTPs, and 0.45 U Amplitag Gold DNA polymerase.
-
20
Run PCR by heating the plate at 94°C for 10 min followed by 20 to 25 cycles at 94°C for 30 sec, 58°C for 30 sec and 72°C for 30 sec.
-
21
Examine a number of samples on agarose gel to detect PCR products (correct amplicons should run at the position of 114nt above smaller free primers). Note that the PCR cycle number may be reduced if a more sensitive method is used to monitor PCR products.
-
22
Pool all PCR products from one to four 384-well plates together (depending on the level of multiplexing desired) and purify the PCR products away from free primers with NucleoSpinR Extract II Kit according to manufacturer’s instruction.
Sequencing and analysis
-
23
Use Qubit fluorometer (Life Technologies) to quantify the amount of pooled and purified PCR products to make an estimate for loading onto individual lanes of a Solexa sequencer (note that the estimate can be made with other methods, such real time PCR or using Nanodrop and the amount also depends on the specific seqeuncer to be used). For sequencing on HiSeq 2000, we aim to generate ~200 million clusters per lane.
-
24
Sequence 40nt using the P5 primer to read the ligated probe region.
-
25
Strip-off the sequencing products (which has been described in the Illumina sequencing manual) and then prime the flowcell with the index primer to sequence the bar-code region.
-
26
Map the sequence tags to the sequences that correspond to intended ligation products and segregate tags for individual samples based on their bar-codes.
-
27
Compile tag numbers for each gene in each sample for further analysis.
COMMENTARY
Background information
The RASL procedure and its DNA-based version call DASL (DNA-templated oligonucleotide Annealing, Seleciton, Ligation) are based on the LMA (Ligation Mediated Assay) described by Landdegren and Hood (Landegren et al., 1988) by including the annealing step to offset random ligation. RASL was originally designed to detect mRNA isoforms (Yeakley et al., 2002) and its DNA version is the basis of the GoldenGate kit for large-scale genotyping marketed by Illumina. RASL/DASL have been applied to profile gene expression on formalin-fixed, paraffin embedded biological samples because of their ability to accommodate heavily fragmented RNA (Fan et al., 2004; Li et al., 2006).
The original RASL/DASL assays were all based on quantification on microarrays, which produce analog information. Because the RASL/DASL products are directly compatible with high throughput sequencing, we re-designed the assays by incorporating a bar-coding strategy, thus permitting analysis of a large number of samples. It is important to emphasize that the RASL-seq platform is not designed for genome-wide analysis, due to the limitation of current deep sequencing technologies. However, RASL-seq is ideal for quantitative analysis of a large selected panel of genes important for a biological pathway or disease phenotype. According to the sequencing capacity of HiSeq2000, we are able to monitor 100 to 500 genes (in which we normally include 30 to 50 housekeeping genes as built-in controls) in a highly multiplex fashion.
The RASL-seq procedure described here was designed with two major applications in mind. One is to facilitate large-scale, quantitative analysis of gene expression in various genetic studies, which has been demonstrated in the elucidation of circadian regulation of the plant defense system (Wang et al., 2011). The platform is ideally suited for genome-wide RNAi analysis, which permits analysis of many genes in a regulatory pathway(s) against systematic genetic perturbations in the cells. This offers a tremendous advantage over current RNAi screens based on a single functional readout. The second major application is to use this digital system to pursue pathway-centric chemical screening by utilizing a disease-associated gene signatures to identify small molecules that can specifically intervene with a specific disease pathway(s). We have recently applied this approach to the hormone-refractory prostate cancer problem (Li et al., submitted). This permits for the first time using high throughput sequencing for high throughput chemical screening, which we refer to as the HTS2 platform.
Critical parameters and troubleshooting
It is important to have a test run on the oligo pool. We first take a small aliquot from the master plates containing individually synthesized oligos to make a small pool. Ideally, conduct the test run on RNA from two biological samples where differential expression of individual genes to be targeted has been determined by microarray or RNA-seq. This allows identification of individual oligo pairs that can reproduce the fold differences. In this test run, we usually encounter a few pairs that produce very high reads for unknown reasons. Eliminate those pairs in making the final pool. It may be also desirable to include some spike-in controls to determine whether the expected fold-differences can be detected. In some applications where the known differences cannot be experimentally determined, we use a reference RNA from Strategen to identify oligos that can record the expression of their target genes in the range of 50 to 5000 counts per gene per assay.
Another problem we frequently encountered is a high level of random ligation (i.e. unrelated oligos are ligated to one another). Normally, such random ligation is less than 10% among the total ligated products, which can be ignored because they will not be counted as the expected products during bioinformatics analysis of the data. When this reaches 30% or more, however, it is problematic, which is usually due to low levels of RNA or to RNA degradation that causes failure in polyA selection. If cell lysates are used, make sure that there are sufficient cells in each well. When the cell number is more than 10,000, on the other hand, the high viscosity due to released genomic DNA will cause loss of beads during polyA selection and/or buffer change. In this case, the polyA selection may be carried out on oligo-dT coated plates so that released DNA and cell debris can be removed during the polyA selection step.
We normally conduct RASL reactions in a separate room to avoid contamination from previous PCR reactions. The PCR cycle number should be kept minimal because the main purpose of this step is for bar-coding. We normally run 20 to 25 cycles, which produces sufficient materials for inspection of the products from individual wells on agarose gel. The pooled material is more than enough for sequencing. If no need for such inspection, 10 to 15 cycles should be sufficient, as long as the pooled sample has measurable quantity for estimation before loading onto the flowcell.
Cost-effectiveness
There are two major expenses associated with RASL-seq. The first is the cost for olignucleotide synthesis. Although various optimized oligo design algorithms are available, we found that they cannot fully substitute experimental tests to identify probes with the ability to report fold-differences and read density in a reasonable range. We thus recommend synthesizing 3 sets of probes for each transcript. This will cost ~$30 per gene. However, if the pool will be used for more than 1,000 assays, the average cost will be $3 per assay. If the scale is for 100,000 assays, for example, the oligo cost will become insignificant.
The second cost is the RASL-seq procedure from individual reactions to high throughput sequencing. On the HiSeq2000 platform, we are able to multiplex four 384 plates per lane, and thus each full scale run on two flowcells (each contains 8 lanes) will score 2×8×4×384=24,576 samples. Based on a rough estimate of the sequencing cost at $10,000 per run, the cost per sample is ~$0.4. Besides the sequencing cost, we estimate the reagent cost (enzymes, tips, and chemicals) is ~$1 per sample. Thus, the total cost is similar to conventional large-scale genetic or chemical screenings.
Automation and other technical considerations
The RASL-seq procedure was designed with full automation in mind. Indeed, all steps simply involve addition or aspiration of solution to or from 384 well plates because all biochemical reactions take place on the solid phase (magnetic beads or oligo-dT coated plates). We have now fully implemented the protocol on a Beckman Biomek FX robot, which takes ~5 hrs to complete all procedures before bar-coding. We recommend manual assays on less than ten 384 plates and robotic assays for larger scale applications. In our hands, the results from manual assays are slightly more robust because of efficient removal of free probes that reduces the overall rate of random ligation.
Anticipated Results
The amount of the library obtained following PCR amplification depends on the number of genes selected, their expression levels, the number and type of cells and individual probe efficiency. The decoding rate (the sequence of a ligated product that can be uniquely assigned to a specific sample) is usually >95%, and the rate of random ligation is <10%. If all potential problems are addressed as described above, the assay usually produces a good correlation (R2 >0.95) between technical or biological repeats when performed directly on cell lysates.
Time Considerations
The RASL procedure manually performed on one 384 plate takes about 6 hrs. If the assay involves 4 or more 384 plates, we perform the assay on a custom designed robot based the Biomek FX system, which also takes about 6 hrs. The bar-coding process by PCR takes ~1 hr on a Bio-Rad tetrade. After pooling the samples and making the estimate for loading, the sequencing part on HiSeq2000 takes about 3 days from cluster generation to two rounds of sequencing.
Acknowledgments
We thank the members of Dr. Fu’s lab for technical help and stimulating discussion during development of the technology. This work was supported by the Challenge Award from the Prostate Cancer Foundation and by a NIGRI grant (HG004659) to X-D.F.
Literature Cited
- Fan JB, Yeakley JM, Bibikova M, Chudin E, Wickham E, Chen J, Doucet D, Rigault P, Zhang B, Shen R, McBride C, Li HR, Fu XD, Oliphant A, Barker DL, Chee MS. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 2004;14:878–885. doi: 10.1101/gr.2167504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Li HR, Wang-Rodriguez J, Nair TM, Yeakley JM, Kwon YS, Bibikova M, Zheng C, Zhou L, Zhang K, Downs T, Fu XD, Fan JB. Two-dimensional transcriptome profiling: identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66:4079–4088. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, Lee DU, Fu XD, Dong X. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD. Profiling alternative splicing on fiber-optic arrays. Nat Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]