Abstract
A structural motif, the tryptophan zipper (trpzip), greatly stabilizes the β-hairpin conformation in short peptides. Peptides (12 or 16 aa in length) with four different turn sequences are monomeric and fold cooperatively in water, as has been observed previously for some hairpin peptides. However, the folding free energies of the trpzips exceed substantially those of all previously reported β-hairpins and even those of some larger designed proteins. NMR structures of three of the trpzip peptides reveal exceptionally well-defined β-hairpin conformations stabilized by cross-strand pairs of indole rings. The trpzips are the smallest peptides to adopt an unique tertiary fold without requiring metal binding, unusual amino acids, or disulfide crosslinks.
The design of peptides that have well-defined tertiary structures tests our understanding of the principles governing the folding of larger proteins. Designed sequences (23–28 aa) that adopt the β-β-α “zinc finger” fold have been notable successes (1–3) and are among the smallest protein domains demonstrated to fold cooperatively without assistance from metal ions or disulfide crosslinks (2, 3). Shorter peptides that form three-stranded β-sheets (4) or exhibit partial β-hairpin structure (5–10) undergo extremely broad thermal transitions, suggesting a lower limit of 20–30 aa for a stable tertiary fold. Here we report well-folded peptides of half that size: 12- and 16-residue monomeric β-hairpins, stabilized by tryptophan-tryptophan cross-strand pairs, exhibit reversible and highly cooperative thermal unfolding transitions in water. These tryptophan zippers (trp-zips) are minimal units of β tertiary structure and remarkably, for short β-hairpins, have the thermodynamic properties of typical folded proteins.
Short peptides with significant hairpin structure recently have emerged as β-sheet model systems (5, 6). However, even the best reported examples are only marginally stable (ΔGunf ≈ 0 at 298 K) (5–10). It is not known whether this stability represents an upper limit for these very small peptides and, if it does not, what residue substitutions might further promote hairpin folding. In particular, cross-strand tertiary interactions have not yet been extensively investigated in these systems. To begin to address this, we determined an experimental energy scale for substitutions in a nonhydrogen-bonded (NHB) strand position of a disulfide-cyclized β-hairpin (11). Unexpectedly, tryptophan was much more stabilizing in this site than other amino acids (≥0.5 kcal⋅mol−1) (11). Further studies showed that paired, cross-strand NHB residues in the cyclized hairpin made roughly independent contributions to stability; thus, a tryptophan-tryptophan cross-strand pair was highly stabilizing (and the best NHB residue pair we identified) (12). Accordingly, it seemed likely that introduction of a second tryptophan-tryptophan cross-strand pair would greatly stabilize the hairpin conformation. We thought that this additional stability would be sufficient to drive folding, even in the absence of a covalent disulfide constraint.
We find that this combination of two Trp-Trp nonhydrogen-bonded cross-strand pairs is generally useful in stabilizing β-hairpin structures. We have characterized four variants having different turn sequences and lengths. In each case, the peptides are highly water-soluble, well-structured, and monomeric. High-resolution NMR structures of three of the peptides show the two cross-strand Trp pairs interdigitating in a zipper-like motif on the surface of the folded peptide. This arrangement of the indole side chains confers unusual spectroscopic properties on the folded molecules, and folding therefore can be monitored readily by changes in CD signal. The stabilities of the trpzips are significantly higher than those reported for other small β-structures (ΔGunf = 0.6–1.7 kcal⋅mol−1 at 298 K); on a per-residue basis, the trpzips have stabilities comparable to much larger protein domains (ΔGunf,max = 60–120 cal⋅mol−1⋅residue−1). Because of their small size, unusual stability, and very favorable spectroscopic properties, trpzip peptides should prove useful and quite simple systems in which to study β-structure and folding.
Materials and Methods
Peptide Synthesis.
Peptides were synthesized on a Pioneer synthesizer (PerSeptive Biosystems) as C-terminal amides by using standard fluorenylmethoxycarbonyl chemistry, cleaved from resin with 5% triisopropylsilane in trifluoroacetic acid (TFA), and purified by reversed-phase HPLC (acetonitrile/H2O/0.1% TFA). Peptide identity was confirmed by electrospray MS.
CD Spectroscopy and Analysis of Thermal Denaturation Curves.
Spectra were acquired with an Aviv Instruments (Lakewood, NJ) model 202 spectrophotometer. Peptide concentrations were determined spectrophotometrically as described (13). Melting curves were acquired at 229 nm with 1.5-min equilibration at each temperature and an averaging time of 15 s. Thermal denaturation was reversible, as judged by recovery of CD signal (≥95%) upon cooling. In addition, reverse melting curves were acquired for trpzips 1 and 4 (see Fig. 1B). Reverse and forward curves were identical in shape, with ≤0.5 K shift in Tm. As a model for the unfolded state of the peptides, the melting curve (linear) of an equimolar mixture of the trpzip1 half peptides SWTWEG and NKWTWK was measured. Data for the trpzip peptides then were fit to a two-state unfolding equilibrium as described (14), fixing the unfolded baseline. Folded baselines, Tm, ΔHm (ΔH at Tm), and ΔCp were allowed to vary. For trpzips 5 and 6, the unfolded baseline could be fit directly to the experimental data. ΔSm was calculated from the fit parameters (ΔHm/Tm). Errors in Table 2 were generated by the fitting algorithm (kaleidagraph, Synergy Software, Reading, PA) and are given to indicate the quality of the fits to the particular experimental data set. However, when fitting different data sets, ΔHm and ΔCp values vary by ≈ 10%, as is typical in thermal denaturation experiments (15).
Figure 1.
Folding of trpzips 1–3. (A) CD spectrum of trpzip1. (Inset) The near UV region is shown with a 10-fold expanded y axis. (B) Thermal denaturation of trpzip1 (20 μM) monitored by CD. The forward melting curve is shown as open circles, while the reverse melting curve is shown as the error bars associated with signal averaging during data acquisition. (Inset) The first derivatives of melting curves (20, 50, 100, and 150 μM peptide) are overlaid. (C) Temperature dependence of folding for trpzips 1–3 (calculated from the thermodynamic parameters listed in Table 2).
Table 2.
Thermal unfolding and sedimentation analysis of trpzip peptides
| Parameter | trpzip1 | trpzip2 | trpzip3 | trpzip4 | trpzip5 | trpzip6 |
|---|---|---|---|---|---|---|
| Tm, K | 323.0 ± 0.3 | 345.0 ± 0.1 | 351.8 ± 0.2 | 343.1 ± 0.1 | 315.8 ± 0.2 | 317.7 ± 0.5 |
| ΔHm, cal⋅mol−1 | 10,790 ± 120 | 16,770 ± 60 | 13,020 ± 70 | 21,860 ± 60 | 13,320 ± 140 | 10,290 ± 300 |
| ΔSm, cal⋅mol−1⋅K−1 | 33.4 | 48.6 | 37.0 | 63.7 | 42.2 | 32.4 |
| ΔCp, cal⋅mol−1⋅K−1 | 231 ± 4 | 281 ± 2 | 195 ± 2 | 380 ± 4 | 325 ± 10 | 236 ± 17 |
| σobs/σcalc | 1.02 ± 0.04 | 1.01 ± 0.04 | 1.00 ± 0.04 | n. d. | n. d. | n. d. |
Thermal melts were acquired with 20 μM peptide samples in 20 mM potassium phosphate, pH 7.0. σ ≡ reduced apparent molecular weight, as determined from sedimentation data fit to a nonideal single-species model (see Materials and Methods). n.d. ≡ not determined; the thermal denaturation curve of trpzip4 was identical at 5-fold higher peptide concentration (100 μM vs. 20 μM). Thermal unfolding parameters of ΔH = 11,600 cal⋅mol−1 and ΔS = 39 cal⋅mol−1⋅K−1 have been reported for the gb1 peptide, assuming ΔCp = 0 (7; see also Materials and Methods).
We find that fitting with ΔCp fixed to 0 (as described in studies of the native gb1 peptide) (7, 8) results in significant overestimates of hairpin population at lower temperatures; this portion of the stability curve is especially sensitive to errors in ΔCp (15). Fitting the trpzip denaturation curves in this manner required large shifts in Tm (≈5–10 K higher than the minimum in a derivative plot) and generated fits of lower quality. In addition, van't Hoff plots show clear curvature through the transition region, indicating a nonzero ΔCp. From our data on trpzips 4, 5, and 6, we can estimate ΔCp ≈200 cal⋅mol−1⋅K−1 for the gb1 peptide, which is sufficient to explain the discrepancy between our population estimate for the gb1 hairpin and those previously reported (7, 8). Recently, a nonzero ΔCp (≈100 cal⋅mol−1⋅K−1) was reported for the unfolding of a 12-residue hairpin related to gb1 (10).
Analytical Ultracentrifugation.
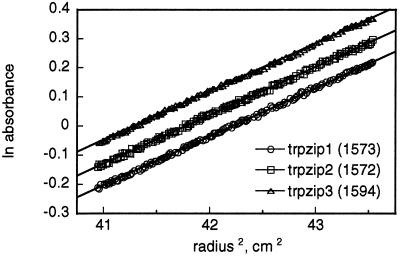
Samples (in 20 mM potassium phosphate, 150 mM KCl, pH 7.1; 277 K) were analyzed in a Beckman XL-A ultracentrifuge at rotor speeds of 40 and 55 krpm. Peptide concentration was monitored by absorbance at or near 290 nm. Data for both speeds and two initial peptide concentrations (60 and 200 μM, 11 data sets total per peptide) were fit simultaneously to a nonideal single species model by using the program nonlin (16). Allowing nonideality improved the fit for the 200 μM samples while only slightly changing the reduced apparent molecular weight σ (≈ +6%). For all three peptides, data from 60-μM samples fit an ideal model (e.g., see Fig. 2) with random residuals. Expected σ values were determined from partial specific volumes based on residue composition, calculated buffer density, and monomer formula weights (17).
Figure 2.
Equilibrium ultracentrifugation of trpzips 1–3. The data shown are for 60 μM peptide samples and a rotor speed of 40 krpm. Apparent molecular weights obtained from the slopes (assuming ideal behavior) are shown; calculated formula weights are 1,608 for trpzips 1 and 2 and 1,648 for trpzip3. Trpzip1 data are offset vertically (ln absorbance −0.085) for clarity.
NMR Spectroscopy and Structure Calculations.
NMR samples contained 1–3 mM peptide in 92% H2O/8% D2O, pH 5.5 (trpzip1 and trpzip2) or pH 6.0 (trpzip4 and gb1,41–56), with 0.1 mM 3-(trimethylsilyl)-1-propane-1,1,2,2,3,3-d6-sulfuonic acid (DSS) as a chemical shift reference. All spectra were acquired on a Bruker DRX-500 or DRX-600 spectrometer at 288 K. Double quantum filtered correlation spectroscopy (2QF COSY), total correlation spectroscopy (τm = 90 ms), rotating-frame Overhauser effect spectroscopy (τm = 150 ms), and nuclear Overhauser effect (NOE) spectroscopy (τm = 100 ms) experiments were acquired as described (18) with gradient coherence selection (19) or excitation sculpting (20) for water suppression. Proton resonances were assigned by standard methods (21). 3JHΝ-Hα were obtained by fitting Lorentzian lines to the antiphase doublets of HN-Hα peaks in 2QF-COSY spectra processed to high-digital resolution in F2. 3JHα−Hβ were extracted from COSY-35 spectra (35° mixing pulse) acquired on D2O solutions of the peptides. Distance and dihedral angle restraints were generated as described (22). Eighty initial structures were calculated by using the hybrid distance geometry/simulated annealing program dgii (23); 50 of those were further refined by restrained molecular dynamics by using the amber all-atom forcefield implemented in discover as described (22). Twenty structures having the lowest restraint violation energy and good geometry were chosen to represent the solution conformation of each peptide. The structure with the lowest rms deviation (rmsd) to the average coordinates of the ensemble was chosen as the representative structure (model 1 in the Protein Data Bank file).
The concentration dependence of the NMR spectra of trpzip2 and trpzip4 was evaluated by one-dimensional 1H NMR (10-fold and 100-fold dilution of samples used to acquire two-dimensional data; final concentrations: 1.2 mM, 120 μM, and 12 μM for trpzip2, and 3.2 mM, 320 μM, and 32 μM for trpzip4). For both peptides, there were small chemical shift changes (in all cases Δδ ≤ 0.08 ppm between concentrated and 10-fold diluted samples, and Δδ ≤ 0.02 ppm between 10-fold and 100-fold diluted samples). For example, the trpzip2 peak with the largest Δδ was that from W4Hɛ3; in the 1.2 mM sample this proton resonates at 5.656 ppm (2.0 ppm upfield from the expected random coil position; ref. 21). This peak shifts 0.043 ppm downfield (120 μM sample), and an additional 0.004 ppm downfield upon further dilution (12 μM sample). In contrast, analytical ultracentrifugation indicates that trpzip2 is monomeric up to at least 200 μM. Taken together, these data imply that limited self-association may be occurring at millimolar concentrations. The fact that the Δδ are extremely small indicates that self-association does not significantly perturb the peptide structure; furthermore, there are no NOEs indicative of a specific interaction between monomers. Thus, the calculated structures accurately represent the monomer conformations.
Results and Discussion
The peptide trpzip1 (Table 1) consists of a representative type II′ turn sequence (EGNK) flanked by the sequence WTW. An additional residue was added to each end of the peptide to permit cross-strand hydrogen bonding between the termini. Residues in hydrogen-bonded positions of the strands were taken from sequences used in our other studies (11, 12) and have not been optimized. Perhaps surprisingly, given that one-third of the residues are tryptophan, the peptide is freely soluble in water at millimolar concentrations. Trpzip1 has an unusual CD spectrum with intense exciton-coupled bands at 215 and 229 nm (Fig. 1A), indicating interaction between the aromatic chromophores (24). Furthermore, the near UV CD spectrum of trpzip1 has well-defined bands at the longer wavelength absorption maxima of tryptophan (Fig. 1A Inset), indicating that the indole side chains are in a defined chiral environment. In proteins, such near UV CD bands are often taken as evidence for fixed tertiary structure.
Table 1.
Sequences of trpzip and gb1 peptides
| trpzip1 | SWTWEGNKWTWK | Type II' turn |
| trpzip2 | SWTWENGKWTWK | Type I' turn |
| trpzip3 | SWTWEpNKWTWK | Type II' turn |
| gb1, 41–56 | GEWTYDDATKTFTVTE | Type I turn |
| trpzip4 | GEWTWDDATKTWTWTE | gb1: Y45W, F52W, V54W |
| trpzip5 | GEWTYDDATKTFTWTE | gb1: V54W |
| trpzip6 | GEWTWDDATKTWTVTE | gb1: Y45W, F52W |
All peptides were synthesized as C-terminal amides; p ≡ d-proline. Residue numbers for the gb1 peptide correspond to those of the parent 56-residue B1 domain.
Trpzip1 has a reversible, cooperative thermal denaturation curve with a midpoint at 323 K (Fig. 1B). The data are of exceptionally high quality for a β-peptide: folding may be monitored sensitively at the 229-nm exciton-coupled band, where sample absorbance causes few problems. Very poor signal-to-noise ratio is frequently a problem in CD-monitored folding studies of other small β-structures; see, for example, ref. 4. Reverse and forward melting curves overlay closely (Fig. 1B), demonstrating that the thermal transition is reversible. The melting temperature does not shift with peptide concentration (20–150 μM; Fig. 1B Inset), suggesting that trpzip1 does not self-associate at these concentrations. The thermal denaturation data fit well to a two-state model and reveal that folding is enthalpically favorable at ambient temperatures, with a significant heat capacity change (Table 2).
We synthesized two variants in which the Gly-Asn turn sequence of trpzip1 was replaced by stronger turn promoting sequences (trpzips 2 and 3; Table 1). Trpzip2 and trpzip3 have CD spectra that overlay closely with that of trpzip1 (not shown) and, likewise, exhibit reversible and cooperative melting behavior. Thermodynamic parameters for trpzips 2 and 3 are similar to those of trpzip1, with stability curves (and Tm) shifted to higher temperatures (Fig. 1C; Table 2). Interestingly, the denaturation curve for trpzip2 (Asn-Gly turn) is distinctly more cooperative than those of trpzips 1 or 3 (d-Pro-Asn turn). Trpzip2 also appears to be more stable than trpzip3 at low temperatures, despite previous conclusions that the d-Pro-Asn turn (and the related II′ turn d-Pro-Gly) are more stabilizing than Asn-Gly (11, 25, 26). Instead, the conformational restriction of the d-proline appears to confer additional stability only at relatively high temperatures. Equilibrium ultracentrifugation confirms that all three trpzip peptides sediment as single species of the expected monomer molecular weights (Fig. 2; Table 2).
The three-dimensional structures of trpzip1 and trpzip2 were determined by NMR. All 1H resonances were assigned by conventional two-dimensional methods at 288 K, pH 5.5. (One-dimensional data are consistent with the peptides being predominantly monomeric at the millimolar concentrations used to acquire the two-dimensional data; see Materials and Methods.) Overall, the NMR data are of unusually high quality for short, linear peptides and provide strong evidence that the molecules are highly structured. The chemical shift dispersion is remarkable, allowing accurate measurement of the majority of HN-Hα and Hα-Hβ coupling constants and unambiguous assignment of nearly all NOE peaks; the number and intensity of observed NOE peaks are comparable to those routinely seen with small, stable proteins. Likewise, in addition to NOE-based distance restraints, numerous backbone dihedral angle restraints (derived from extreme 3JHN−Hα) could be included in the structure calculations. Furthermore, the tryptophan side-chain conformations are all well defined, having χ1 angles of ≈ −60° (indicated by analysis of 3JHα−Hβ and local rotating-frame Overhauser effects). The high population of the folded state under the conditions of the NMR experiments, as well as the quality of the data, validate the high precision of the structures calculated for these peptides.† Trpzip1 adopts a β-hairpin conformation with the expected type II′ β-turn (Fig. 3A). Cross-strand tryptophan rings pack intimately against one another, with less contact between adjacent tryptophan pairs. Analysis of trpzip2 reveals a very similar structure, only deviating from trpzip1 by having a type I′ β-turn at residues 6 and 7 (Fig. 3B; Table 3).
Figure 3.
NMR structures of trpzips 1 and 2. (A) Ensemble of 20 structures of trpzip1 optimally aligned by using backbone atoms of residues 2–11. (B) Representative structures of trpzips 1 and 2 aligned on the backbone atoms of residues 2–5 and 8–11 (rmsd of the mean coordinates of the aligned backbone atoms in the two ensembles is 0.37 Å); the bottom view is rotated 90° relative to the top view. The backbone carbonyl of residue 6 is indicated to emphasize the difference in turn geometry between the two structures (type II′ for trpzip1 vs. type I′ for trpzip2).
Table 3.
NMR structural statistics for trpzip peptides
| Parameter | trpzip1 | trpzip2 | trpzip4 |
|---|---|---|---|
| rmsd from experimental distance restraints (Å) | 0.005 ± 0.001 | 0.004 ± 0.003 | 0.003 ± 0.001 |
| (number of restraints) | (77) | (84) | (117) |
| rmsd from experimental dihedral restraints (°) | 0.14 ± 0.09 | 0.16 ± 0.09 | 0.33 ± 0.08 |
| (number of restraints) | (15) | (15) | (21) |
| Maximum distance violation (Å) | 0.03 ± 0.00 | 0.04 ± 0.03 | 0.03 ± 0.01 |
| Maximum dihedral violation (°) | 0.5 ± 0.3 | 0.6 ± 0.3 | 1.1 ± 0.3 |
| Ramachandran geometry (% in most favored region)* | 71 ± 10 | 85 ± 10 | 82 ± 4 |
| Backbone (N,Cα,C) rmsd from mean coordinates (Å) | 0.40 ± 0.07 | 0.41 ± 0.09 | 0.29 ± 0.06 |
| (residues used for rmsd calculation) | (2–11) | (2–11) | (43–54) |
Resonance assignments and coupling constants for trpzip1, trpzip2, and trpzip4 are available in Tables 4–6.
Ramachandran geometry was evaluated by using the program procheck (31); remainder of the residues for all structures are in the allowed regions of φ, ψ space, with none in the disallowed or generously allowed regions.
The trpzip peptides may be compared with a previously described β-hairpin peptide taken from the B1 IgG-binding domain of protein G. The peptide gb1 (residues 41–56 of the B1 domain) exhibits partial hairpin character, estimated at ≈40% (278 K) by NMR (28). More recently, the estimated hairpin population has doubled, based on fluorescence-monitored folding studies (7) and additional NMR experiments (8). The peptide appears to be stabilized by a cluster of four hydrophobic residues (W43, Y45, F52, and V54) (7, 28, 29). From NOEs observed for the peptide (28), and from the structure of the sequence in the parent protein (30), the hydrophobic strand residues are expected to occupy adjacent nonhydrogen-bonded sites on one face of the hairpin. This is precisely the arrangement of tryptophan residues in the trpzip peptides, allowing the direct comparison of the gb1 and trpzip hydrophobic clusters.
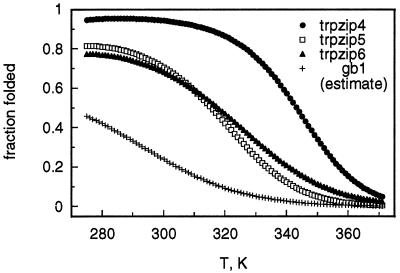
As expected from the stability of trpzips 1–3, replacement of gb1 residues Y45, F52, and V54 with tryptophan yields an exceptionally well-folded β-hairpin (trpzip4; Table 1). The thermal melting curve for trpzip4 is more cooperative than those of trpzips 1–3, yielding thermodynamic parameters that reflect this difference (Table 2). Trpzip4 is also more stable than trpzips 1–3 at low temperatures, resulting in a modest increase in folded population (Figs. 1C and 4). Most importantly, the thermal denaturation curve of trpzip4 is much more cooperative than that of the wild-type gb1 peptide (7, 8), and the melting temperature of trpzip4 is higher by at least 40 K, depending on the method used to estimate the folded population of gb1 (Fig. 4) (7, 8, 28).
Figure 4.
Temperature dependence of folding for trpzips 4–6 (calculated from the thermodynamic parameters listed in Table 2). Our estimated curve for gb1 was calculated by assuming that mutations in trpzip4 (i.e., those present in trpzips 5 and 6) have independent and additive effects on hairpin stability [ΔGunf,gb1 = ΔGunf,trpzip5 − (ΔGunf,trpzip4 − ΔGunf, trpzip6)] (12) and is nearly identical in shape to previously reported gb1 denaturation curves based on fluorescence (7) or NMR (8) measurements. However, our estimated population for the folded conformation of gb1 is much closer to that in the original report (28) than to later estimates (7, 8).
In contrast, when tryptophan residues 4, 9, and 11 of trpzip1 are replaced with the appropriate gb1 residues (Y, F, and V, respectively), we find no evidence by NMR for the hairpin conformation (all 3JHN−Hα < 8 Hz, not shown). Likewise, introduction of the gb1 hydrophobic cluster into a model peptide with a very strong type II′ turn (d-Pro-Gly) yielded only a marginally stable β-hairpin (≈60% folded at 275 K) with an extremely broad thermal denaturation curve (10). These experiments show that the gb1 hydrophobic cluster is not sufficient to maintain a significant hairpin population without additional stabilizing elements. To explore this in more detail, we reintroduced individually into trpzip4 the Phe-Tyr and Trp-Val cross-strand pairs of gb1 (trpzip5 and trpzip6, respectively). Unlike gb1, trpzips 5 and 6 each have one Trp-Trp cross-strand pair, so folding can be monitored by CD (229 nm, as for the other trpzips). We find both trpzip5 and trpzip6 to be much less stably folded than trpzip4 (Table 2; Fig. 4) but more stably folded than wild-type gb1. From our earlier studies in disulfide-cyclized hairpins (12), we expect ≈1 kcal⋅mol−1 loss in stability for each gb1 cross-strand pair (Tyr-Phe or Trp-Val) relative to Trp-Trp. In agreement with this expectation, unfolding free energies (298 K) are 1.69, 0.57, and 0.49 kcal⋅mol−1 for trpzips 4, 5, and 6, respectively. Therefore, assuming additive stabilization from the two pairs (12), we estimate ΔGunf ≈ −0.6 kcal⋅mol−1 for gb1 at 298 K (≈0 kcal⋅mol−1 in ref. 7; see Materials and Methods). Our population estimate for gb1 agrees closely with the lower estimate originally reported (28).
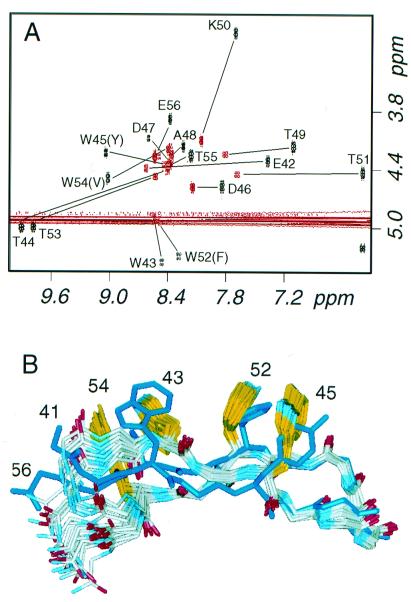
As observed for the other trpzip peptides, the NMR data for trpzip4 are of exceptional quality and support the conclusion that the molecule is well folded (see Tables 4–6, which are published as supplemental material on the PNAS web site, www.pnas.org). The fingerprint region of the trpzip4 COSY spectrum shows dramatic chemical shift dispersion, especially when compared with the spectrum for wild-type gb1 peptide (Fig. 5A). Chemical shifts represent population-weighted averages of all conformations adopted in solution; therefore, the extreme HN and Hα shifts of trpzip4 indicate that the folded conformation is highly populated.‡ From these data, taken together with the thermal denaturation curves (Fig. 4), we conclude that trpzip4 has a much higher folded population than the gb1 peptide and that the cross-strand tryptophan pairs of the trpzip motif are superior to the hydrophobic cluster of gb1.
Figure 5.
NMR analysis of trpzip4. (A) Overlay of the COSY fingerprint regions for the wild-type gb1 peptide (red) and trpzip4 (black); the location of crosspeaks are indicated for both peptides and are labeled for trpzip4. (B) Ensemble of 20 structures of trpzip 4 compared with the minimized mean structure of the B1 domain of protein G (Protein Data Bank code 2GB1) (30); backbone atoms of protein residues 46–52 were superposed on the mean coordinates of the ensemble, yielding a rmsd of 0.67 Å.
The structure of trpzip4 shows Trp-Trp packing and strand orientations similar to those observed in trpzips 1 and 2 (Table 3; Fig. 5B), despite the fact that there are six rather than four intervening turn residues. Trpzip4 extends the strands by another residue and presents a type I β-turn, with K50 adopting a positive φ angle. The turn geometry of trpzip4 is indistinguishable from that of the same turn in the full-length B1 domain (30) within the error of the structure determinations (Fig. 5B). The twist of the two strands, however, is markedly different between the peptide and the protein; the protein is only modestly twisted (2GB1; Θ ≈ 20°), whereas trpzip4 is highly twisted (Θ ≈ 70°) (32). This large twist is within the range observed in natural proteins (32) and still allows good hydrogen-bonding geometry. The high degree of twist would appear to result from the cross-strand Trp-Trp packing, because it is observed in all three trpzip structures. However, the extreme twist may be a general characteristic of two-stranded sheets (32). [Interestingly, in a molecular dynamics study of the wild-type gb1 peptide (33), an initially rather flat starting conformation (derived from the structure of the intact B1 domain) relaxed into more twisted conformations resembling trpzip4.] The backbone coupling constants (see Tables 4–6) for tryptophan residues in the three peptides (7.1–8.2 Hz) are lower than those of the intervening hydrogen-bonded threonine residues (8.9–9.8 Hz), consistent with the alternating less and more negative φ angles that are a hallmark of a twisted sheet (34). The geometry of the trpzip is that expected for an antiparallel β-coiled coil (32, 34, 35).
By generally accepted criteria (36), the trpzip peptides behave as folded proteins; presently, they are the smallest all-natural linear polypeptides to do so. Their per-residue thermodynamic parameters (ΔG, ΔH, and ΔCp) are comparable to those of larger protein domains (15, 37), indicating that, like other proteins, the folding of the trpzip hairpins is driven by burial of hydrophobic surface area (37) (i.e., tryptophan side chains). However, trpzips 1, 2, and 4 are somewhat atypical in the per-residue value of the nonhydrophobic portion of ΔSunf (ΔS110°C = 6.4 ± 0.3 cal⋅mol−1⋅residue−1) when compared with the average for globular proteins (4.2 ± 0.1 cal⋅mol−1⋅residue−1) (37). (With ΔS110°C = 4.5 cal⋅mol−1⋅residue−1, the d-proline-containing trpzip3 is more typical of globular proteins.) The observation of an unusually large configurational component of ΔSunf suggests that the degree of structural definition in the folded state of the trpzips might exceed that of the average protein domain. In the case of trpzip3, this appears to be offset thermodynamically by the d-proline, which might be expected to increase order in the unfolded state.
Despite its utility in stabilizing β-hairpin structures, we have not found examples of the trpzip motif in the protein structure database. It is unlikely that such a stable substructure would be required within a larger globular domain, and if it were present, it might not pack well with other structural elements. For example, in contrast to our results with trpzip4, we expect that mutation of Y45, F52, and V54 to tryptophan in the intact 56-residue B1 domain would disrupt the core of the protein (30). However, when searching sequence databases, we did find several potential trpzips (not shown). One set of examples occurs in some V4 loops from HIV gp120. The V4 loop is highly variable in sequence, and it is not visible in the reported structure of gp120 (38). We synthesized one V4 loop peptide, TWTWNGSAWTWN (GenBank accession no. AAA44126), and found it to exhibit a CD spectrum indistinguishable from those of trpzips 1–3 (not shown). The first tryptophan in the peptide corresponds to a conserved tryptophan at the end of β-strand 18 in the crystal structure (38), suggesting that, in this particular V4 loop, the β-strand is extended slightly, with the trpzip sequence forming a β-hairpin.
Finally, it is interesting to compare the relatively simple design process we have used here with other recently described methods. In the automated procedure described by Mayo and coworkers (2, 3), all residues of the hairpin would be considered to occupy “surface” positions, and, accordingly, would be restricted to small, polar, or charged residues. Our data show clearly that proper arrangement of surface hydrophobic groups can stabilize a specific folded structure without causing aggregation. Such hydrophobic clusters occur on the surface of natural proteins (39) and may well help determine the folded structure. In the case of our hairpin peptides, the additive contributions to stability from cross-strand partners (12) suggests that the observed arrangement of Trp side chains may be especially effective in desolvating the peptide backbone, or the indole rings themselves, rather than contributing a large π-stacking energy.
Another emerging design strategy is to choose sequences or motifs derived from protein structures to fit a target fold (e.g., refs. 4–6, 9, and 28). Although this approach has proven successful, the stabilities of the resulting molecules are not always as high as one might want, and it is difficult to predict whether or how they might be improved. In contrast, the unusual and very specific role of tryptophan in stabilizing β-hairpins could not have been anticipated by analysis of known protein structures. Accordingly, we believe that the experimental determination of folding rules in peptides (11, 12) can add insights to current protein design methods and ultimately to studies of natural proteins.
Supplementary Material
Acknowledgments
We thank P. Harbury and the Stanford Department of Biochemistry for access to the ultracentrifuge.
Abbreviations
- trpzip
tryptophan zipper
- COSY
correlation spectroscopy
- rmsd
rms deviation
- NOE
nuclear Overhauser effect
Footnotes
Data deposition: The peptide structures have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1HRW (trpzip1), 1HRX (trpzip2), and 1HS0 (trpzip4)].
Very recently, the structure of U(1–17)T9D, a variant of the N-terminal β-hairpin of ubiquitin, was reported (Protein Data Bank code 1E0Q) (27). The estimated population of the folded hairpin was only 64%; nevertheless, the NMR resonances were unusually well resolved, and it was possible to attribute particular NOEs to the unfolded state. This deconvolution of the NOE data allowed the calculation of folded-state structures with good geometry and no distance restraint violations > 0.5 Å (27). The precision of the ensemble (backbone rmsd = 0.59 Å) is close to that of the trpzips (see Table 3); however, as those authors note, the structure of the folded state of U(1–17)T9D may be more dynamic than indicated by the final ensemble (27).
One might think that, even in a poorly structured peptide, the observed chemical shift dispersion might result simply from the presence of four Trp residues. We do not believe this is likely. Although it is true that aromatic rings can induce chemical shifts in nearby protons, very large ring-current shifts would require a relatively fixed orientation between them. It should be noted that the partially structured gb1 peptide itself has three aromatic residues (Trp, Tyr, and Phe) at the positions of the four Trp residues of trpzip4, yet the observed dispersion is much less extreme.
References
- 1.Struthers M D, Cheng R P, Imperiali B. Science. 1996;271:342–345. doi: 10.1126/science.271.5247.342. [DOI] [PubMed] [Google Scholar]
- 2.Dahiyat B I, Sarisky C A, Mayo S L. J Mol Biol. 1997;273:789–796. doi: 10.1006/jmbi.1997.1341. [DOI] [PubMed] [Google Scholar]
- 3.Dahiyat B I, Mayo S L. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 4.Kortemme T, Ramirez-Alvarado M, Serrano L. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 5.Gellman S H. Curr Opin Chem Biol. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 6.Ramirez-Alvarado M, Kortemme T, Blanco F J, Serrano L. Bioorg Med Chem. 1999;7:93–103. doi: 10.1016/s0968-0896(98)00215-6. [DOI] [PubMed] [Google Scholar]
- 7.Muñoz V, Thompson P A, Hofrichter J, Eaton W A. Nature (London) 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 8.Honda S, Kobayashi N, Munekata E. J Mol Biol. 2000;295:269–278. doi: 10.1006/jmbi.1999.3346. [DOI] [PubMed] [Google Scholar]
- 9.Maynard A J, Sharman G J, Searle M S. J Am Chem Soc. 1998;120:1996–2007. [Google Scholar]
- 10.Espinosa J F, Gellman S H. Angew Chem Int Ed. 2000;39:2330–2333. doi: 10.1002/1521-3773(20000703)39:13<2330::aid-anie2330>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 11.Cochran A G, Tong R T, Starovasnik M A, Park E J, McDowell R S, Theaker J E, Skelton N J. J Am Chem Soc. 2001;123:625–632. doi: 10.1021/ja003369x. [DOI] [PubMed] [Google Scholar]
- 12.Russell S J, Cochran A G. J Am Chem Soc. 2000;122:12600–12601. [Google Scholar]
- 13.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 14.Minor D L, Jr, Kim P S. Nature (London) 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- 15.Becktel W J, Schellman J A. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 16.Johnson M L, Correia J J, Yphantis D A, Halvorson H R. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 18.Cavanagh J, Fairbrother W J, Palmer A G, Skelton N J. Protein NMR Spectroscopy, Principles, and Practice. San Diego: Academic; 1995. [Google Scholar]
- 19.van Zijl P C M, O'Neil Johnson M, Mori S, Hurd R E. J Magn Reson. 1995;113A:265–270. [Google Scholar]
- 20.Hwang T-L, Shaka A J. J Magn Reson. 1995;112A:275–279. [Google Scholar]
- 21.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 22.Skelton N J, Garcia K C, Goeddel D V, Quan C, Burnier J P. Biochemistry. 1994;33:13581–13592. doi: 10.1021/bi00250a010. [DOI] [PubMed] [Google Scholar]
- 23.Havel T F. Prog Biophys Mol Biol. 1991;56:43–78. doi: 10.1016/0079-6107(91)90007-f. [DOI] [PubMed] [Google Scholar]
- 24.Grishina I B, Woody R W. Faraday Discuss. 1994;99:245–262. doi: 10.1039/fd9949900245. [DOI] [PubMed] [Google Scholar]
- 25.Stanger H E, Gellman S H. J Am Chem Soc. 1998;120:4236–4237. [Google Scholar]
- 26.Syud F A, Espinosa J F, Gellman S H. J Am Chem Soc. 1999;121:11577–11578. [Google Scholar]
- 27.Zerella R, Chen P-Y, Evans P A, Raine A, Williams D H. Protein Sci. 2000;9:2142–2150. doi: 10.1110/ps.9.11.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco F J, Rivas G, Serrano L. Nat Struct Biol. 1994;1:584–590. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi N, Honda S, Yoshii H, Munekata E. Biochemistry. 2000;39:6564–6571. doi: 10.1021/bi000013p. [DOI] [PubMed] [Google Scholar]
- 30.Gronenborn A M, Filpula D R, Essig N Z, Achari A, Whitlow M, Wingfield P T, Clore G M. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 32.Yang A-S, Honig B. J Mol Biol. 1995;252:366–376. doi: 10.1006/jmbi.1995.0503. [DOI] [PubMed] [Google Scholar]
- 33.Roccatano D, Amadei A, Di Nola A, Berendsen H J C. Protein Sci. 1999;8:2130–2143. doi: 10.1110/ps.8.10.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chothia C. J Mol Biol. 1983;163:107–117. doi: 10.1016/0022-2836(83)90031-1. [DOI] [PubMed] [Google Scholar]
- 35.Nishikawa K, Scheraga H A. Macromolecules. 1976;9:395–407. doi: 10.1021/ma60051a004. [DOI] [PubMed] [Google Scholar]
- 36.DeGrado W F, Summa C M, Pavone V, Nastri F, Lombadi A. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 37.Alexander P, Fahnestock S, Lee T, Orban J, Bryan P. Biochemistry. 1992;31:3597–3603. doi: 10.1021/bi00129a007. [DOI] [PubMed] [Google Scholar]
- 38.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tisi L C, Evans P A. J Mol Biol. 1995;249:251–258. doi: 10.1006/jmbi.1995.0294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.