Abstract
BACKGROUND
Genome-wide association studies (GWAS) of prostate cancer have identified single nucleotide polymorphism (SNP) markers in a region of chromosome 11q13.3 in men of European decent. A fine-mapping analysis with tag SNPs in the Cancer Genetic Markers of Susceptibility (CGEMS) study identified three independent loci, marked by rs10896438, rs12793759, and rs10896449. This study further annotates common and uncommon variation across this region.
METHODS
A next generation resequence analysis of a 122.9kb region of 11q13.3 (68,642,755-68,765,690) was conducted in 78 unrelated individuals of European background, 1 CEPH trio, and 1 YRI trio.
RESULTS
In total, 644 polymorphic loci were identified by our sequence analysis. Of these, 166 variants – 118 SNPs and 48 insertion-deletion polymorphisms (indels) – were novel, namely not present in the 1000 Genomes or International HapMap Projects. We identified 22, 25, 6, and 4 variants strongly correlated (r2 ≥ 0.8) with rs10896438, rs10896449, rs12793759, and rs11228565, respectively. HapMap SNPs were in linkage disequilibrium (r2 ≥ 0.8) with 48%, 69%, 14%, and 60% of SNPs marking bins by rs10896438, rs10896449, rs12793759, and rs11228565, respectively.
CONCLUSIONS
Our next generation resequence analysis compliments publicly available datasets of European descent (HapMap, build 28 and 1000 Genome, Pilot 1, Oct 2010), underscoring the value of targeted resequence analysis prior to initiating functional studies based on public databases alone. Increasing the number of common variants enables investigators to better prioritize variants for functional studies designed to uncover the biological basis of the direct association(s) in the region.
Keywords: Resequence, 11q13, prostate cancer, SNP
Introduction
Prostate cancer is the most common non-cutaneous cancer in men (1). Prior to the age of genome-wide association studies (GWAS), established risk factors included age, family history, and ethnic background. Previous studies have estimated that genetic risk factors overall could account for up to 42% of risk, a figure which may be higher in men of African-American background (2). GWAS of prostate cancer have identified at least 35 common genetic variants associated with prostate cancer to date (3-9). Two prostate cancer GWAS identified a pair of highly correlated common single nucleotide polymorphisms (SNP), rs10896449 and rs7931342 (r2 = 0.966, D’=1.000, HapMap 3 release 28 CEU), in the human chromosome 11q13.3 region (4,6). A subsequent fine-mapping study identified a second locus, rs12418451, 60kb centromeric to the previously identified loci, which is independently associated with prostate cancer (10). This independent association is corroborated by an existence of a recombination hotspot separating rs12418451 from the others. A second study reported rs11228565 as a refinement SNP which remained significant after adjustment for rs10896450 or rs7931342 (5). A recent fine-mapping study by the Cancer Genetic Markers of Susceptibility (CGEMS) study confirmed the second locus with rs10896438 (r2 = 0.958, D’=1.000 with rs12418451, HapMap 3 release 28 CEU) and also identified a third independent locus, rs12793759, using ~10,000 case/control pairs (11).
The markers for the prostate cancer susceptibility loci reported within the 11q13.3 region map to an intergenic region flanked by TPCN2 and MYEOV. None of the common SNPs are in high linkage disequilibrium (LD) with common genetic variations in known or putative functional places in either gene. Interestingly, two coding SNPs within TPCN2 (two-pore segment channel 2) were reported to be associated with blond versus brown hair color (12). The nearest gene flanking the three prostate susceptibility loci on the telomeric side, MYEOV, is frequently over-expressed in different cancers, such as multiple myeloma, squamous cell carcinoma, breast cancer, and oral cancer (13-14). Often co-amplified and overexpressed with MYEOV is CCND1 (cyclin D1), a cell cycle regulator gene, which is ~340kb telomeric to prostate cancer susceptibility loci. Recently, two independent risk loci for kidney and breast cancer have also been identified by GWAS between MYEOV and CCND1 but these are not in appreciable LD with prostate susceptibility loci (15-16).
While GWAS have successfully identified multiple prostate cancer susceptibility loci, these only capture a fraction of the total heritability (17-18), and additional loci are estimated to be identified by larger GWAS (19). It was recently proposed that multiple rare variants may create ‘synthetic association’ in GWAS in association with one of the alleles of a common surrogate marker (20). A comprehensive assessment of common and rare genetic variations and prioritization of variant set represent important next steps following GWAS discovery, especially if the region has a complex genomic architecture. In this study, we used next-generation sequencing technology to re-sequence a region of 11q13.3 (68,642,755-68,765,690; UCSC genome build hg18) defined by the linkage disequilibrium pattern, in 80 unrelated individuals of European background drawn from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) (21) cohort used in the initial GWAS for CGEMS.
Materials and methods
Samples
Eighty individuals, all of European ancestry, were selected from the NCI Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) (21). Six additional individuals were included, a Yoruba trio Y005 (NA18503, NA18504, and NA18505) and trio from a CEPH pedigree 1350 (NA10855, NA10856, and NA11824). All but NA11824 were directly genotyped in The International HapMap Project (http://hapmap.org).
Region selection
A 122.9 kb region of chromosome 11q13, spanning 68,642,755-68,765,690 (UCSC genome build hg18), was selected for next generation sequence analysis based on the observed LD pattern flanking rs10896449, the most notable marker in the CGEMS prostate cancer GWAS using HapMap CEU data (release 22, phase II) (6). The boundaries of this region within 11q13 include the six previously reported markers, rs10896438, rs12418451, rs12793759, rs11228565, rs7931342, and rs10896449. The most telomeric side of the region is approximately 51 kb from the MYEOV gene; the centromeric side extends ~28 kb beyond the TPCN2 gene.
Primer design, PCR and sequencing
A Nimblegen capture probe pool was designed to cover the 122.9 kb targeted region. The capture probes were approximately 60 bp in size, and the probe pool was designed for amplicon overlap (100 bp in average). Primers were designed using Nimblegen Proprietary Capture probe design software followed by in silico quality assessment for uniqueness, possible sequence paralogy, and DNA repeat sequences using the BLAT of the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgBlat). For primer secondary structure and PCR efficiency check, NetPrimer (http://www.premierbiosoft.com/netprimer/index.html) was used. Primers were ordered from Integrated DNA Technologies (Coralville, IA, USA; http://www.idtdan.com). The BED file is available upon request for this region. After performing the Nimblegen solution-based sequence capture method, sequence analysis was performed on the 454 Genome Sequencer FLX system (http://www.454.com/products-solutions/product-list.asp).
Detection of Genetic Variation
An in-house automated computational pipeline was developed to process sequence reads generated by 454 FLX Genome Sequencers. Whenever applicable, sequence reads from the same sample were pooled based on barcodes provided by Roche/454. Quality check (QC) was performed using vendor-supplied software; sequence reads that passed QC were aligned to the target genomic region (11q13: 68,642,755-68,765,690, UCSC genome build hg18) by MOSAIK aligner (http://bioinformatics.bc.edu/marthlab/Mosaik). The resulting assembly was analyzed column-by-column and both putative polymorphic sites and most likely genotypes were called based on a set of heuristic rules. All possible indels were checked individually. The minimal sequence coverage depth was set to 20 reads for each nucleotide position and the ratio (r) of forward and reverse reads was determined. To avoid directional bias, an optimal range of r was set between 10 and 90%. Homozygous genotype calls were made when the most frequent allele was present in at least 85% of the reads. Heterozygous genotype calls were made when the two most frequent alleles were represented in 30-70% of reads. No genotype calls were made if the above criteria were not met. For quality assurance (QA), NextGENe software (http://www.softgenetics.com) and Consed (http://bozeman.mbt.washington.edu/consed/consed.html) were used to resolve ambiguous calls.
Descriptive statistics and data quality control
Genotype completion, concordance, minor allele frequency (MAF) estimations, deviations from fitness for Hardy-Weinberg proportion (HWP), pair-wise LD, and tag SNP analysis were performed using the GLU software package (Genotype Library and Utilities; http://code.google.com/p/glu-genetics/). To check concordance, variant calls of the five individuals genotyped in HapMap were compared with HapMap data (release 28). Matched variants were identified in the 1000 Genome Project data (pilot 1 low-coverage CEU data, October 2010 release) and allele frequencies were compared using a two-group χ2 test of equal proportions (22), followed by a correction for multiple testing using an R-based software package, QVALUE (23).
In silico genomic analysis
Putative functional elements within the resequenced region were assessed using the UCSC genome browser (http://genome.ucsc.edu/), a publically available bioinformatics website. Specifically, human mRNA and spliced EST tracks for any known transcripts, ENCODE Integrated Regulation tracks for putative enhancer/promoter regions, and conservation tracks were assessed by scanning 500 bp-window over the entire 122.9 kb resequenced region.
Results
Coverage and depth
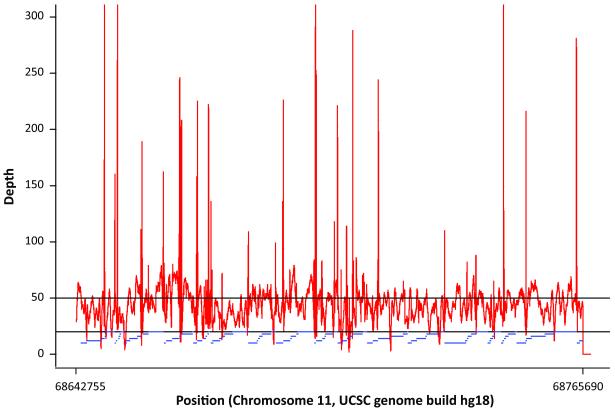
Sequence coverage and depth averaged over all samples in the targeted genomic region (chr11: 68,642,755-68,765,690, hg18) are shown in Figure 1. No gaps in coverage were observed. The average read depth was approximately 50-fold (range from 2- to 470-fold, median 44). A cumulative length of approximately 4.2 kb was observed with an average coverage depth of less than 20-fold (Supplementary Table 1).
Figure 1. Coverage and depth averaged over all samples in the targeted region of 11q13.3 (68,642,755-68,765,690, 122.9kbps).
The horizontal line at 50-fold represents the average depth. The blue horizontal bars represent amplicons from long range PCR
Polymorphism detection and quality control
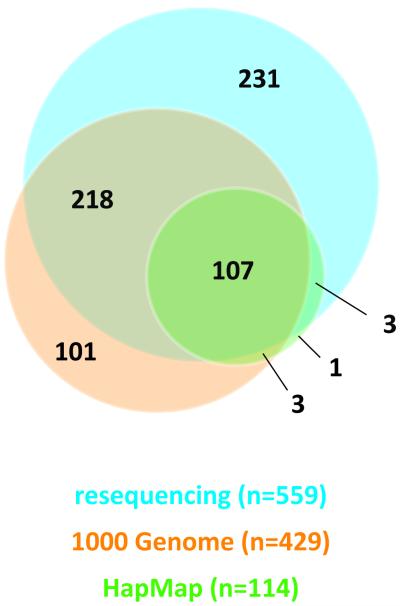
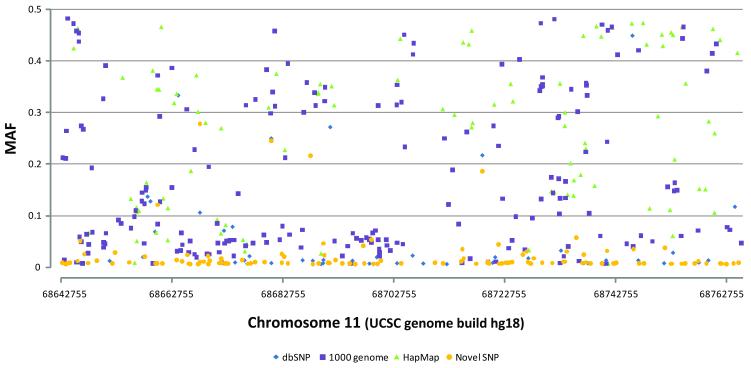
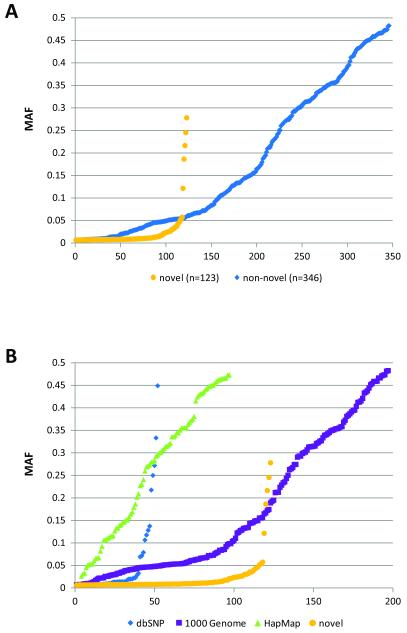
Genotypes were called for 888 possible segregating sites in 80 samples from the National Cancer Institute’s PLCO Cancer Screening Trial (21), one trio from the CEPH pedigree 1350, and one trio from a Yoruba pedigree Y005. The concordance between the sequence data and HapMap data for the five samples was 99.66%. During the data QC assessment, 244 loci were excluded due to monomorphism (n=214), no reads (n=19), or substantial violation of fitness for HWP for called genotypes (P < 0.001, n=11). Two unexpected duplicate samples from PLCO were excluded in the formal analysis. No loci were dropped due to low per locus completion rates. The final genotype dataset contained 84 individuals (78 from PLCO, 1 CEPH trio, and 1 Yoruba trio) and 644 polymorphic loci detected by 454 analysis (Supplementary Table 2). These include 118 novel SNPs, 48 novel indels and 478 variant loci previously described in NCBI’s dbSNP (build 132) database (http://www.ncbi.nlm.nih.gov/projects/SNP/) and/or reported in HapMap data (release 28) and 1000 Genome data (pilot 1 low-coverage CEU of 10-2010 release). From the analysis including only 80 unrelated individuals of European origin (78 from PLCO + 2 founders of a CEPH trio), 559 loci were polymorphic, and when compared with 1000 Genome CEU data and HapMap CEU data (Figure 2), 231 polymorphic loci (41.3%) were uniquely identified by our study. For subsequent analyses, loci with completion rates ≥ 40%, which included 469 polymorphic loci (103 novel SNPs, 20 novel indels, 346 loci previously described in NCBI’s dbSNP build 132, 1000 Genome data, or HapMap data),were considered (Table 1, Figure 3 and 4). The average genotype call rate was 74.7% (range 40%-100%, median 76.3%) (Supplementary Table 2); the average of computed MAF estimates was 13.7% (range 0.6-48.2%, median 5.4%) (Table 1). Allele frequencies were compared in 308 SNPs detected both in this study and in the 1000 Genome CEU data. No significant difference in allele frequency was observed except for one locus (rs1542335, q-value = 0.0001), which was excluded in subsequent analyses. Since our insertion/deletion calling algorithm is under refinement, indels detected in this study should be considered preliminary.
Figure 2. Comparison of bi-allelic polymorphisms (SNPs and indels) among 3 datasets.
The 1000 Genome CEU data (pilot 1 low-coverage, 10-2010 release), HapMap CEU data (release 28), and our resequencing data within the 122.9kb resequenced region (chr11:68,642,755-68,765,690, UCSC genome build hg18) were compared. Combined number of reported bi-allelic polymorphisms is 664 (575 SNPs and 89 indels)
Table 1.
MAF distribution of resequencing discovered SNPs and indels with regard to dbSNP, 1000 Genome, and HapMap inclusion
| SNP | Indel | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Content | count | MAF |
count | MAF |
count | MAF |
|||
| average | (min-max), median | average | (min-max), median | average | (min-max), median | ||||
| dbSNPa | 47 | 0.040 | (0.006-0.449), 0.013 | 5 | 0.167 | (0.029-0.272), 0.217 | 52 | 0.052 | (0.006-0.449), 0.014 |
| 1000 Genomeb | 179 | 0.167 | (0.007-0.481), 0.103 | 18 | 0.222 | (0.019-0.482), 0.130 | 197 | 0.172 | (0.007-0.482), 0.103 |
| HapMapc | 97 | 0.258 | (0.007-0.473), 0.280 | 0 | - | - | 97 | 0.258 | (0.007-0.473), 0.280 |
| Noveld | 103 | 0.011 | (0.006-0.051), 0.008 | 20 | 0.076 | (0.017-0.278), 0.031 | 123 | 0.021 | (0.006-0.278), 0.008 |
|
| |||||||||
| Totale | 426 | 43 | 469 | 0.137 | (0.006-0.482), 0.054 | ||||
dbSNP (b132) reported, exclusive of 1000 Genome or HapMap reported ones
1000 Genome CEU (pilot 1 low-coverage data, 10-2010 release) reported, exclusive of HapMap CEU (release 28) reported ones
HapMap CEU reported SNPs
Novel polymorphisms previously not reported by dbSNP or 1000 Genome CEU data
Polymorphism counts and MAF estimation was performed after exclusion of 90 variants with call rate ≤ 40% in 80 (78 PLCO + 2 CEU founders) unrelated Europeans
Figure 3. MAF distributions of 469 polymorphic variants (minimum call rate ≥ 0.4) by position.
The data characteristics are the same as presented in Table 1.
Figure 4. MAF distributions of 469 polymorphic variants (minimum call rate ≥ 0.4) by frequency rank.
A: newly discovered polymorphisms vs. previously reported polymorphisms by dbSNP (build 132), The 1000 Genome (pilot 1 low-coverage, 10-2010 release), and/or HapMap (release 28)
B: by the same data characteristics as Table 1
Linkage disequilibrium (LD) and tag SNP analysis
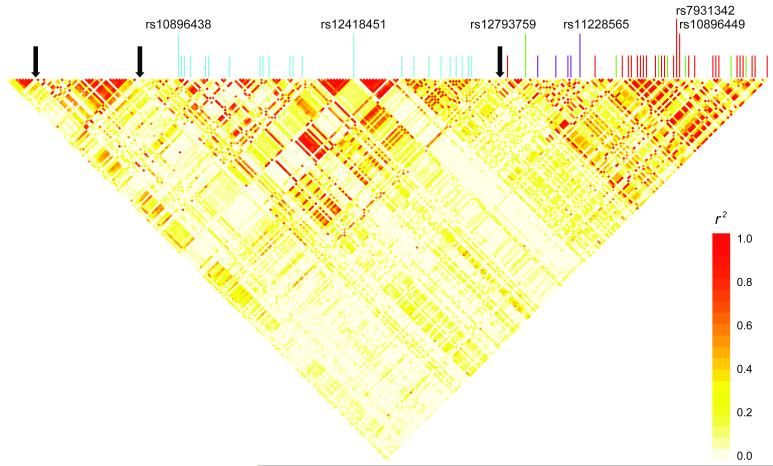
Based on our data (call rate ≥ 0.4, MAF ≥ 0.5, n=253), the linkage disequilibrium pattern across the sequenced region indicates 4 complex block structure defined by 3 inferred recombination hotspots (Figure 5). The two telomeric blocks, defined by two recombination hotspots (chr11:68,659,036-68,662,036 and chr11:68,727,036-68,729,036), include the previously reported prostate cancer susceptibility loci. Notably, rs10896438 and rs12418451 (r2 = 0.897, D’=0.999) and their surrogates are separated by a recombination hotspot from other susceptibility loci, corroborating their independent contribution to prostate cancer susceptibility (10-11).
Figure 5. Linkage disequilibrium, recombination hotspots, and prostate cancer GWAS hits with surrogates (r2 ≥ 0.8) within a 122.9kb resequenced region.
Pairwise linkage disequilibrium was calculated and the heatmap was drawn using resequencing identified polymorphisms with MAF ≥ 0.05, genotype completion rate ≥ 0.4 (n=253) in 80 unrelated individuals of European ancestry. Vertical color lines indicate location of previously reported prostate cancer GWAS hits in the region to date and their surrogates identified by resequencing. Light-blue lines represent rs10896438/rs12418451 (bin2, n=23), green lines represent surrogates of rs12793759 (bin3, n=7), purple lines represent surrogates of rs11228565 (bin4, n=5), and red lines represent surrogates of rs10896449/rs7931342 (bin1, n=26). Black solid arrows indicate location of recombination hotspots
A tagging analysis was performed with 80 unrelated samples of European origin for the 122.9kb sequenced region (r2 ≥ 0.8-1.0, MAF ≥ 0.01 and 0.05, minimum call rate > 0.40) using the TagZilla program implemented in GLU (Genotype Library and Utilities). Based on the loci with MAF ≥ 0.05 (n=253), at an r2 ≥ 0.8, 65 tags are required to tag 100%, at an r2 ≥ 0.9, 84 tags, and at an r2 = 1.0, 175 tags are required (Table 2, Supplementary Table 3). Within the region, 90 SNPs with MAF ≥ 0.05 are common in our resequencing data, HapMap III CEU (release 28), and 1000 Genome CEU (pilot 1 low-coverage data, 10-2010 release). In analysis restricted to the SNPs at an r2 ≥ 0.8, 29 bins monitoring 206 loci were covered (81.4%), while using variants reported in the 1000 Genome CEU data (n=233), 60 bins monitoring 248 loci (98.0%) were covered, whereas 5 singleton bins were exclusively covered by resequencing data (Table 2). For loci with MAF ≥ 0.01 (n=358), at an r2 ≥ 0.8, 117 tags are required to tag 100% (r2 ≥ 0.9, 136 tags, and r2 = 1.0, 259 tags are required), of which 41 tags representing 52 loci (14.5%) were not covered by HapMap or 1000 Genome data (Table 2).
Table 2.
Number of tags and coverage using variants with call rate ≥ 0.4 by various r2 thresholds by category
| MAF ≥ 0.05 | r2 ≥ 0.8 | r2 ≥ 0.9 | r2 ≥ 1.0 | |||
|---|---|---|---|---|---|---|
| category* | number of tags | coverage (%) | number of tags | coverage (%) | number of tags | coverage (%) |
| HapMap (n=90) | 29 | 81.4 | 36 | 76.3 | 72 | 56.1 |
| 1000 Genome (n=233) | 60 | 98.0 | 78 | 97.6 | 165 | 96.0 |
| resequence (n=253) | 65 | 100 | 84 | 100 | 175 | 100 |
| MAF ≥ 0.01 | r2 ≥ 0.8 | r2 ≥ 0.9 | r2 ≥ 1.0 | |||
|---|---|---|---|---|---|---|
| category* | number of tags | coverage (%) | number of tags | coverage (%) | number of tags | coverage (%) |
| HapMap (n=94) | 31 | 65.1 | 37 | 61.2 | 75 | 41.3 |
| 1000 Genome (n=282) | 76 | 85.5 | 93 | 84.9 | 196 | 82.1 |
| resequence (n=358) | 117 | 100 | 136 | 100 | 259 | 100 |
HapMap CEU (release 28) and 1000 Genome CEU (pilot 1 low-coverage data, 10-2010 release) were compared with variants discovered by resequencing Number of matched variants available in each dataset is in parenthesis. 1000 Genome is inclusive of all HapMap reported SNPs
Assessment of variants for follow-up studies
To date, six prostate cancer susceptibility loci (rs10896449, rs7931342, rs12418451, rs10896438, rs12793759, and rs11228565) have been previously reported in independent GWAS and fine-mapping studies (4-6,10-11). Based on the high degree of LD across this region, it is a priority to catalogue highly correlated common variants in the region prior to conducting the bioinformatic analysis in search of putative functional elements across this non-coding region. We performed tag analysis in the 122.9kb resequenced region with all 6 prostate cancer susceptibility loci within 11q13.3 as obligate includes using our data, HapMap CEU (release 28), and the 1000 Genome CEU (pilot 1 low-coverage data, 10-2010 release), then catalogued all possible surrogates (r2 ≥ 0.8) (Table 3, Supplementary Table 3, 4). Of the six loci, high correlation between pairs existed: rs10896449 and rs7931342 (r2=0.967, D’=1.000), rs10896438 and rs12418451 (r2=0.895, D’=1.000), and rs12793759 and rs11228565 (r2=0.728, D’=1.000). Our resequencing data catalogued 24, 21, 6, and 4 highly correlated surrogates (r2 ≥ 0.8) with rs7931342/rs10896449 (bin1), rs10896438/rs12418451 (bin2), rs12793759 (bin3), and rs11228565 (bin4), respectively, which included 6 indel polymorphisms (Table 3, Supplementary Table 4). The 1000 Genome CEU data (pilot 1 low-coverage data, October 2010 release) catalogued 100% of all possible surrogates for rs7931342/rs10896449, rs12793759, and rs11228565, while cataloging 57.7% of all possible surrogates of rs10896438 (Table 3). HapMap CEU data (release 28) catalogued only 59.3% of rs7931342/rs10896449 surrogates, 34.6% of rs12418451/rs10896438 surrogates, 50% of rs11228565 surrogates, and none of rs12793759 surrogates, with no indel polymorphisms included (Table 3).
Table 3.
Number of correlated variants (r2 ≥ 0.8) per bin containing prostate cancer susceptibility loci in 11q13.3
| Correlation Bin | Susceptibility loci | Resequence | 1000 genome | HapMap | Total |
|---|---|---|---|---|---|
| Bin 1 | rs7931342/rs10896449 | 26 (3) | 29 (3) | 18 | 29 (3) |
| Bin 2 | rs12418451/rs10896438 | 23 (2) | 17 (2) | 11 | 28 (4) |
| Bin 3 | rs12793759 | 7 (1) | 9 (2) | 1 | 9 (2) |
| Bin 4 | rs11228565 | 5 | 5 | 3 | 5 |
|
| |||||
| Total | 61 (6) | 60 (6) | 33 | 71 (8) | |
HapMap CEU (release 28) and 1000 Genome CEU (pilot 1 low-coverage data, 10-2010 release) were compared with variants discovered by resequencing
In silico genomic analysis
The previously reported 6 prostate cancer risk loci and all possible surrogates at r2 ≥ 0.8 were primarily assessed for existence of potential regulatory elements using the UCSC genome browser ENCODE Integrated Regulation track. On the centromeric side of the recombination hotspot (chr11:68,727,036-68,729,036), rs10896438 localizes to an alternative TPCN2 transcript (RefSeq accession: NM_139075) as well as a spliced EST AL137479, whereas rs12418451 maps to two known spliced ESTs – BC843531 and BI826779 (Supplementary Figure 1). On the telomeric side of the recombination hotspot, rs7931342 and rs10896449 are located within 5kb centromeric to the spliced EST DB036467. None of the 6 risk loci directly overlap with transcription factor binding sites reported by ENCODE Transcription Factor ChIP-seq data, but 12 surrogate markers (8 surrogate markers of rs10896438, 2 surrogates of rs7931342/rs10896449, and 2 surrogates of rs12793759) overlap transcription factor binding sites of interest meriting further follow-up (Supplementary Table 5).
Discussion
In this study, we have characterized common genetic variants, namely, SNPs and indels, across a 122.9kb region (11q13: 68,642,755-68,765,690, UCSC genome build hg18) by next-generation resequencing technology and catalogued a comprehensive set of surrogates of previously reported prostate cancer susceptibility loci. Comparison of our resequence results with the current public datasets (1000 Genome CEU and HapMap CEU) revealed a substantial number of common and uncommon variants (with MAF between 1% and 10%). In total we called 664 polymorphic sites where 107 SNPs were identified by all three datasets with a median MAF of 0.295 (range 0.007-0.5), whereas resequence analysis determined 218 variants previously not included in HapMap but with a lower median MAF of 0.118. When we examined the 332 variants exclusively reported by sequence analysis, 231 variants (MAF median=0.013, average=0.066) were unique to our resequencing analysis, as compared to 101 variants (MAF median=0.046, average=0.093) observed uniquely in the 1000 Genome CEU data. This difference can be attributed to the number of chromosomes analyzed and the depth of coverage per base.
Indel polymorphisms represent an important type of genetic variant that are, thus far, not well annotated in large data sets, mainly because consensus calling methods for indels are not as robust as for single base pair substitutions. Moreover, they appear to contribute to the genetic architecture of human diseases by altering functional elements (24-25). Overall, we observed that 13.4% (n=89) of the 664 reported variants are indels, 58.5% (n=52) of which were uniquely identified by our resequencing study. Twenty indel polymorphisms (MAF median=0.134, average=0.225) were identified by our study and the 1000 Genome CEU, while 17 indel polymorphisms (MAF median=0.125, average=0.169) were unique to the 1000 Genome CEU data. In an in silico assessment, one indel polymorphism, rs11357679 (GT/T), a surrogate of rs7931342/10896449 at an r2 ≥ 0.8, maps to a transcription factor glucocorticoid receptor (GR) binding site according to the ENCODE Transcription Factor ChIP-seq data from the UCSC genome browser (26). Although this study extended the list of indel polymorphisms by reporting 72 indels, which involve 1 to 9 bases insertions or deletions, further validation is needed to confirm the current analytical algorithm for detection.
Using the three available data sets, we conducted an analysis of tagging SNPs to determine the extent of coverage for each data set. Restricting the analysis to all SNPs with MAF ≥ 5% and a threshold for binning of r2 ≥ 0.8 for variants, we note that 65 tags are required; an increased number of tags is needed for higher r2 thresholds (r2 ≥ 0.9, 84 tags, and r2 ≥ 1.0, 175 tags). When we only looked at the content of HapMap reported SNPs, 18.6% of the variants with MAF ≥ 5% within the region cannot be monitored at an r2 ≥ 0.8, whereas the 1000 Genome coverage approximates our re-sequence analysis (98%). As we lower the filter for tagging to SNPs with MAF between 1% and 5%, the resequence analysis provides approximately one third more coverage than HapMap and 14.5% more than the 1000 Genome data. We also note that as the 1000 Genome Project expands and more subjects are analyzed with deeper coverage these estimates will shift slightly.
Our study provides important insights into the next steps required to map GWAS regions, especially since the majority of reported SNP markers have MAFs well above 10%, while a small proportion have MAFs between 5 and 10% due to inadequate power to detect small effects and the limited number of low MAF SNPs with current data sets (19,27). In the case of 11q13, so far, all of the known SNP markers have MAFs that exceed 15% (4-6,10-11). Pursuing the recent hypothesis of ‘synthetic association’ will be particularly difficult in this region because the notable variants appear to map to a non-genic region (20). On the other hand, others have argued that this is probably less common than suggested (28). Nonetheless, mapping and functional studies should provide insights into the specific underpinnings of GWAS signals.
A bioinformatic analysis of the variants suggests interesting sites to pursue for functional analysis, such as the set of variants that cluster near an alternative transcript of TPCN2 (RefSeq accession: NM_139075, chr11:68,596,959–68,686,483, 89.525kb, 15 exons, UCSC genome browser) that extends 72kb telomeric of the protein-coding TPCN2 transcript (RefSeq accession: NM_139075.3). Two spliced ESTs (BC043531, chr11:68,671,272–68,695,606; BI826779, chr11:68,671,430–68,695,608), both detected in brain tissue, localize to the telomeric side of NM_139075, but in the opposite direction (negative strand). More than a half of rs10896438/rs12418451 surrogates (19 out of 28) reside in the vicinity of these transcripts; 6 of the 19 reside in transcription factor binding sites, but further work is needed to demonstrate that these are functionally active. rs3019748 maps to multiple transcription factor binding sites, including p300, notable for its binding to putative enhancers (29). The local region is also enriched for H3K4Me1 sites in the HMEC (human mammary epithelial cell) cell line. rs12275055 and rs11228580, two of the eight rs12793759 surrogates at an r2 ≥ 0.8, are located on transcription factor NFkB binding sites; rs11228580 is also located within DB036467, a spliced EST.
The LD across this region is quite interesting, particularly as it relates to the signals detected for breast and renal cancers: in recent GWAS, rs7105934 (chr11:68,948,922, ~198kb telomeric to rs10896449) was recently identified in renal cancer (p=7.8×10−14) (16), while rs614367 (chr11:69,037,945, ~287kb telomeric to rs10896449) was associated with breast cancer risk (p=3.2×10−15) (15). Though it was suggested that one of the previously reported prostate cancer risk loci, rs7931342, might be associated with breast cancer (OR, 0.95 with 95% CI 0.91-0.99, p=0.028) in a candidate gene analysis prior to the GWAS, (30), this signal was not confirmed conclusively in the GWAS. The complex LD across the region could account for the above suggestion, as there is minimal correlation between rs7931342 and rs614367 (r2=0.001 in HapMap CEU).
Conclusions
In this study, we have conducted a resequence analysis of a 122.9kb region that harbors three distinct loci for prostate cancer risk and identified a large annotated set of variants that should be considered for follow-up studies. We have shown that additional resequence analysis supplements the public databases and affords investigators the opportunity to discover and characterize variants that directly account for the association signals observed in large-scale GWAS studies of cancer.
Supplementary Material
Supplementary Figure 1 A snapshot of 11q13.3 resequenced region on the UCSC genome browser and location of 6 prostate cancer susceptibility loci
Acknowledgments
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health (NIH). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organization indicate endorsement by the U.S. Government. The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, NCI, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr. Thomas Riley and staff at Information Management Services, Inc., and Ms. Barbara O’Brien and staff at Westat, Inc. for their contributions to the PLCO Cancer Screening Trial. We thank Marie-Josephe Horner for editorial support. Finally, we acknowledge the study participants for donating their time and making this study possible.
Footnotes
Disclosure Statement None of the authors listed have any significant or perceived conflicts of interest relating to the publishing of this manuscript.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 3.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, Hopper JL, Southey MC, Muir KR, English DR, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Sawyer E, Lophatananon A, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper C, Donovan JL, Hamdy FC, Neal DE, Eeles RA, Easton DF. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 4.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G, Easton DF. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41(10):1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 7.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni JF, Jr., Hunter DJ, Thomas G, Chanock SJ. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41(10):1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 9.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, Nakagawa H. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42(9):751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 10.Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, Smith S, Pruett K, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Johansson JE, Liu W, Kim JW, Chang BL, Duggan D, Carpten J, Rodriguez C, Isaacs W, Gronberg H, Xu J. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung CC, Ciampa J, Yeager M, Jacobs KB, Berndt SI, Hayes RB, Gonzalez-Bosquet J, Kraft P, Wacholder S, Orr N, Yu K, Hutchinson A, Boland J, Chen Q, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson BE, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs WB, Isaacs SD, Grunberg H, Wiklund F, Xu J, Vatten LJ, Hveem K, Njolstad I, Gerhard DS, Tucker M, Hoover RN, Fraumeni JF, Jr., Hunter DJ, Thomas G, Chatterjee N, Chanock SJ. Fine Mapping of a Region of Chromosome 11q13 Reveals Multiple Independent Loci Associated with Risk of Prostate Cancer. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, Palsson S, Sigurgeirsson B, Thorisdottir K, Ragnarsson R, Benediktsdottir KR, Aben KK, Vermeulen SH, Goldstein AM, Tucker MA, Kiemeney LA, Olafsson JH, Gulcher J, Kong A, Thorsteinsdottir U, Stefansson K. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40(7):835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 13.Janssen JW, Cuny M, Orsetti B, Rodriguez C, Valles H, Bartram CR, Schuuring E, Theillet C. MYEOV: a candidate gene for DNA amplification events occurring centromeric to CCND1 in breast cancer. Int J Cancer. 2002;102(6):608–614. doi: 10.1002/ijc.10765. [DOI] [PubMed] [Google Scholar]
- 14.Janssen JW, Imoto I, Inoue J, Shimada Y, Ueda M, Imamura M, Bartram CR, Inazawa J. MYEOV, a gene at 11q13, is coamplified with CCND1, but epigenetically inactivated in a subset of esophageal squamous cell carcinomas. J Hum Genet. 2002;47(9):460–464. doi: 10.1007/s100380200065. [DOI] [PubMed] [Google Scholar]
- 15.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den Ouweland A, Houlston R, Ross G, Langford C, Pharoah PD, Stratton MR, Dunning AM, Rahman N, Easton DF. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, Prokhortchouk E, Wu X, Kiemeney LA, Gaborieau V, Jacobs KB, Chow WH, Zaridze D, Matveev V, Lubinski J, Trubicka J, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Bucur A, Bencko V, Foretova L, Janout V, Boffetta P, Colt JS, Davis FG, Schwartz KL, Banks RE, Selby PJ, Harnden P, Berg CD, Hsing AW, Grubb RL, 3rd, Boeing H, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Duell EJ, Quiros JR, Sanchez MJ, Navarro C, Ardanaz E, Dorronsoro M, Khaw KT, Allen NE, Bueno-de-Mesquita HB, Peeters PH, Trichopoulos D, Linseisen J, Ljungberg B, Overvad K, Tjonneland A, Romieu I, Riboli E, Mukeria A, Shangina O, Stevens VL, Thun MJ, Diver WR, Gapstur SM, Pharoah PD, Easton DF, Albanes D, Weinstein SJ, Virtamo J, Vatten L, Hveem K, Njolstad I, Tell GS, Stoltenberg C, Kumar R, Koppova K, Cussenot O, Benhamou S, Oosterwijk E, Vermeulen SH, Aben KK, van der Marel SL, Ye Y, Wood CG, Pu X, Mazur AM, Boulygina ES, Chekanov NN, Foglio M, Lechner D, Gut I, Heath S, Blanche H, Hutchinson A, Thomas G, Wang Z, Yeager M, Fraumeni JF, Jr., Skryabin KG, McKay JD, Rothman N, Chanock SJ, Lathrop M, Brennan P. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43(1):60–65. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 18.Kim ST, Cheng Y, Hsu FC, Jin T, Kader AK, Zheng SL, Isaacs WB, Xu J, Sun J. Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic Variation. Prostate. 2010;70(16):1729–1738. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes RB, Sigurdson A, Moore L, Peters U, Huang WY, Pinsky P, Reding D, Gelmann EP, Rothman N, Pfeiffer RM, Hoover RN, Berg CD. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res. 2005;592(1-2):147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med. 1998;17(8):873–890. doi: 10.1002/(sici)1097-0258(19980430)17:8<873::aid-sim779>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins FS, Drumm ML, Cole JL, Lockwood WK, Vande Woude GF, Iannuzzi MC. Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science. 1987;235(4792):1046–1049. doi: 10.1126/science.2950591. [DOI] [PubMed] [Google Scholar]
- 25.Warren ST, Zhang F, Licameli GR, Peters JF. The fragile X site in somatic cell hybrids: an approach for molecular cloning of fragile sites. Science. 1987;237(4813):420–423. doi: 10.1126/science.3603029. [DOI] [PubMed] [Google Scholar]
- 26.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung CC, Magalhaes WC, Gonzalez-Bosquet J, Chanock SJ. Genome-wide association studies in cancer--current and future directions. Carcinogenesis. 2010;31(1):111–120. doi: 10.1093/carcin/bgp273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson CA, Soranzo N, Zeggini E, Barrett JC. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9(1):e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song H, Koessler T, Ahmed S, Ramus SJ, Kjaer SK, Dicioccio RA, Wozniak E, Hogdall E, Whittemore AS, McGuire V, Ponder BA, Turnbull C, Hines S, Rahman N, Eeles RA, Easton DF, Gayther SA, Dunning AM, Pharoah PD. Association study of prostate cancer susceptibility variants with risks of invasive ovarian, breast, and colorectal cancer. Cancer Res. 2008;68(21):8837–8842. doi: 10.1158/0008-5472.CAN-08-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 A snapshot of 11q13.3 resequenced region on the UCSC genome browser and location of 6 prostate cancer susceptibility loci