Abstract
AIM: To characterize the colon microbiota in two women histologically diagnosed with collagenous colitis using a culture-independent method.
METHODS: Biopsies were taken from the ascending colon and the total DNA was extracted. Universal bacterial primers were used to amplify the bacterial 16S rRNA genes. The amplicons were then cloned into competent Escherichia coli cells. The clones were sequenced and identified by comparison to known sequences.
RESULTS: The clones could be divided into 44 different phylotypes. The microbiota was dominated by Firmicutes and Bacteroidetes. Seven phylotypes were found in both patients and constituted 47.5% of the total number of clones. Of these, the most dominating were clones similar to Bacteroides cellulosilyticus, Bacteroides caccae, Bacteroides thetaiotaomicron, Bacteroides uniformis and Bacteroides dorei within Bacteroidetes. Sequences similar to Faecalibacterium prausnitzii and Clostridium citroniae were also found in both patients.
CONCLUSION: A predominance of potentially pathogenic Bacteroides spp., and the presence of clones showing similarity to Clostridium clostridioforme were found but the overall colon microbiota showed similarities to a healthy one. Etiologies for collagenous colitis other than an adverse bacterial flora must also be considered.
Keywords: Microscopic colitis, Collagenous colitis, Lymphocytic colitis, Colonic microbiota, 16S rRNA sequencing
INTRODUCTION
Collagenous colitis (CC), an idiopathic inflammatory bowel disease, is a subtype of microscopic colitis (MC) together with lymphocytic colitis (LC)[1]. It is considered as a common cause of chronic diarrhea. In Sweden the incidence is approx four to five cases per 100 000[2]. The incidence for both CC and LC in Europe and North America is almost as high as for Crohn’s disease and ulcerative colitis[2].
CC is clinically characterized by chronic non-bloody diarrhea, often combined with abdominal pain and weight loss[2]. The colonic mucosa appears macroscopically normal or near-normal and the diagnosis is made by microscopic examination of mucosal biopsies that reveals diagnostic histopathological changes. CC was first described in 1976 by Lindström[3] in a woman with chronic watery diarrhea in whom histological examination revealed a thick subepithelial collagenous deposition in the rectum. In 1989, Lazenby et al[4] proposed the term lymphocytic colitis in a group of patients with chronic diarrhea and normal colonoscopy with only minor histological changes, where the microscopic evaluation of colonic biopsy specimens revealed modestly increased inflammation in the lamina propria without subepithelial collagen deposition or other mucosal changes.
The peak incidence of MC is in individuals between 55 years and 70 years of age. The female:male ratio is about 7:1 for CC. For LC the female predominance is less pronounced, with a female:male ratio of 2-3:1[5]. However, the disease can occur at all ages, and a few children with CC have been reported[6,7]. Bile acid malabsorption is found in about 27%-44% of patients with CC and 9%-60% in patients with LC[5,8-9]. Treatment with bile acid binding medications is effective in patients with bile-acid malabsorption but can also be effective in patients without bile-acid malabsorption[10].
Both etiology and pathogenesis of MC are uncertain. The most widely held hypothesis is that a noxious agent in the lumen, probably originating from the bacterial microflora, may have a major pathogenic role in the chronic intestinal inflammation. This is supported by regression of symptoms and histopathological changes after diversion of the fecal stream, and recurrence after restoration of intestinal continuity[11,12]. Other observations supporting this hypothesis are the sudden onset of diarrhea and that treatment with antibiotics may have positive effects[2,13]. The increased infiltration of lymphocytes in the mucosa also indicates a proinflammatory component in the lumen. There are case reports of linking pathogenic bacteria such as Clostridium difficile, Yersinia enterocolitica, Campylobacter jejuni and Aeromonas hydrophila to MC[2,7,14-16].
The human microbiota in healthy persons as well as in patients with inflammatory bowel disease has been analyzed in several studies using culture-independent methods[17-19]. However, to our knowledge no such studies have been performed on patients diagnosed with CC. The aim of the present study was to characterize the mucosa-associated microbiota in the ascending colon in two women histologically diagnosed with CC, by cloning and sequencing of the bacterial 16S rRNA genes.
MATERIALS AND METHODS
Subjects and samples
Two female patients, 51 years and 60 years old (A and B) with a known diagnosis of MC, took part in the study. Patient A, otherwise healthy, started to experience watery, non-bloody diarrhea after an antibiotic treatment for gastroenteritis 10 years earlier. Colonoscopy was performed and she was diagnosed with LC. She was treated with Loperamid® (Merck NM AB, Stockholm, Sweden). Two years later she had a relapse of watery, non-bloody diarrhea and a second colonoscopy was performed, still indicating LC. This time she improved spontaneously. At the time of the present study, after a period of stress and a viral gastroenteritis, she started to lose weight and had frequent, watery, non-bloody diarrhea. The present colonoscopy showed a slightly swollen mucosa and increased vascular pattern. The histological examination revealed a thickened subepithelial collagen layer as well as inflammation in the lamina propria and a damaged surface epithelial layer. Patient B had a history of chronic thyreoiditis but was otherwise healthy. She was diagnosed with CC as well as with bile acid malabsorption 4 years before the study. At that time she improved spontaneously but had a recurrence after a period of major stress. Previously, she was treated with non-steroidal anti-inflammatory drugs due to muscular stiffness and actually experienced an improvement of her bowel function by this treatment. At the time of the present colonoscopy her symptoms had improved due to dietary fat reduction. Colonoscopy showed an increased vascular pattern in the right colon but was otherwise normal. Histological examination could verify a collagenous colitis.
Neither patient had any medication at the time of the colonoscopy. Celiac disease had been excluded in both women. They were both non-smokers.
The patients were asked to avoid fiber-rich foods such as fruits, vegetables, grains and seeds some days before the colonoscopy. The day before the examination they ate a plain breakfast, and no solid food was allowed after noon. Intestinal cleansing was carried out with Phosphoral® (Clean Chemical Sweden AB), a salt preparation with osmotic effects. Colonoscopy was performed and serial biopsies throughout the colon as well as two extra biopsies from the right colon were collected. The histological examination followed routine procedures. The latter were placed in tubes with TE-buffer [10 mmol Tris-HCl, 1 mmol ethylenediaminetetraacetic acid (EDTA), pH 8.0], frozen immediately in liquid nitrogen and stored at -80 °C. The study was approved by the Ethics Committee at Lund University. The women gave written, informed consent before entering the study.
DNA extraction and amplification
Frozen tissue samples were thawed on ice and a single biopsy was transferred to a 1.5 mL tube with 190 μL Buffer G2 (DNA Tissue Kit; Qiagen, Gmbh, Hilden, Germany) and 10 μL of Proteinase K (Qiagen). Eight to ten sterile glass (2 mm) beads were added and the cells were lysed at 56 °C for 3-4 h in a shaking water bath. Tubes were cooled on ice and shaken for 30 min on an Eppendorf Mixer 5432 (Eppendorf, Hamburg, Germany) at 4 °C to disintegrate all bacteria. After centrifugation at 300 × g for one minute, the solution was transferred to a Qiagen sample tube, and total DNA was extracted by using Biorobot EZ1 (Qiagen) according to the manufacturer’s instructions. DNA was eluted in 200 μL.
Polymerase chain reaction amplification and cloning
The bacterial 16S rRNA genes were amplified by the universal primers ENV1 and ENV2 annealing to positions 8-27 and 1492-1511, respectively, according to Escherichia coli (E. coli) numbering[20]. The reaction mixture contained 5 μL of 10× polymerase chain reaction (PCR) buffer (100 mmol Tris-HCl, 15 mmol MgCl2, 500 mmol KCl, pH 8.3), each deoxynucleotide phosphate at a concentration of 200 μmol, 2.5 U of Tag DNA Polymerase (Roche Diagnostics, GmbH, Mannheim, Germany) and 10 pmol of each primer. To each tube, 5 μL of extracted sample DNA was added and sterile water was added to 50 μL. As negative controls, water was added to the reaction mixture instead of DNA. Amplification was performed on an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany). Initially, the reaction was heated to 94 °C for 3 min, followed by 25 cycles of denaturing at 94 °C for 1 min, annealing at 50 °C for 45 s and elongation at 72 °C for 2 min. Finally, the reaction was held at 72 °C for 7 min before cooling down to 4 °C. Six PCR tubes were prepared from each sample and then pooled. Forty-two μL of the pooled reaction mixture from one sample was separated on a 1.5% (w/v) agarose gel (Agarose Type III; Sigma Aldrich, St Louis, Mo., United States) in TBE-buffer (89 mmol Tris, 89 mmol boric acid and 2.5 mmol EDTA, pH 8.3). The agarose gel was stained with ethidium bromide (0.5 mg/L) and the band was cut out from the gel. DNA was purified by using Wizard® SV Gel and PCR Clean-Up System (Promega Corp., Madison, WI, United States). For cloning Promega pGEM®-T Vector System and E. coli JM 109 (Promega Corp.) competent cells were used as described previously[20]. Colonies were selected randomly and recultivated on LB-agar containing ampicillin, and then harvested and stored in freezing buffer at -80 °C.
Sequencing
Selected clones were single-strand sequenced by MWG Biotech (Ebersberg, Germany). ENV1 primer was used as sequencing primer. Sequences were edited using Bioedit Sequence Alignment editor 7.0.5.3[21]. Sequences were identified by comparing them to sequences using the option “seqmatch” available at the Ribosomal Database Project[22]. Sequences were checked for chimeric artifacts by using the Bellerophon server[23] and by creating phylogenetic trees of both 5’- and 3’- ends of the sequences. DNAdist calculations were performed using the Phylip DNAdist program using the “similarity table” option (available at: http://mobyle.pasteur.fr/ cgi-bin/portal.py?form=dnadist)[24]. Sequences representing the different phylotypes have been submitted to Genbank and the accession numbers are HQ992999- HQ993042.
Diversity calculations
Shannon and Simpson’s indices were used for diversity calculations. The Shannon index is based on the proportional abundance of species and accounts for both evenness and species richness. Simpson’s index is the dominance measure where the abundance of commonest species is considered more than species richness[25]. The Simpson’s index was expressed as 1/D.
RESULTS
Two clone libraries were constructed, one for patient A with 87 clones and one for patient B with 90 clones. Five clones were suspected chimeras and were removed from the dataset before analysis. The lengths of the sequenced fragments were approximately 750 bp. Sequences showing > 98% similarity to each other were assigned to a single phylotype and a total of 44 phylotypes were identified (Table 1).
Table 1.
Sequences grouped into phylotypes at 98% similarity
| Phylotype No. | Closest type strain | Acc. No1 | Similarity (%)2 | No. of clones3 | Distribution of clones4 | Assignment of clones |
| 1 | Faecalibacterium prausnitzii | AJ413954 | 98.4-98.5 | 9 | 8 (A); 1 (B) | Ruminococcaceae |
| 2 | Faecalibacterium prausnitzii | AJ413954 | 98.4-99.1 | 4 | 4 (B) | Ruminococcaceae |
| 3 | Subdoligranulum variabile | AJ518869 | 97.0 | 1 | 1 (B) | Ruminococcaceae |
| 4 | Anaerofilum agile | X98011 | 90.6 | 1 | 1 (B) | Ruminococcaceae |
| 5 | Ruminococcus lactaris | L76602 | 94.0 | 1 | 1 (B) | Ruminicoccaceae |
| 6 | Oscillibacter valericigenes | AB238598 | 92.5 | 1 | 1 (B) | Ruminicoccaceae |
| 7 | Ruminococcus lactaris | L76602 | 95.9 | 2 | 2 (A) | Ruminococcaceae |
| 8 | Ruminococcus lactaris | L76602 | 95.1 | 2 | 2 (B) | Ruminococcaceae |
| Clostridium jejuense | AY494606 | 94.7 | Lachnospiraceae | |||
| 9 | Marvinbryantia formatexigens | AJ505973 | 95.7 | 1 | 1 (B) | Lachnospiraceae |
| 10 | Roseburia intestinalis | AJ312385 | 94.1 | 1 | 1 (B) | Lachnospiraceae |
| 11 | Anaerostipes caccae | AJ270487 | 99.2 | 2 | 2 (A) | Lachnospiraceae |
| 12 | Anaerostipes caccae | AJ270487 | 95.8 | 1 | 1 (B) | Lachnospiraceae |
| 13 | Roseburia intestinalis | AJ312385 | 100.0 | 2 | 2 (A) | Lachnospiraceae |
| 14 | Roseburia faecis | AY305310 | 96.9 | 2 | 2 (B) | Lachnospiraceae |
| Roseburia intestinalis | AJ312385 | 97.1 | Lachnospiraceae | |||
| 15 | Pseudobutyrivibrio ruminis | X95893 | 94.2-94.3 | 2 | 2 (A) | Lachnospiraceae |
| 16 | Dorea longicatena | AJ132842 | 94.9-95.2 | 3 | 3 (A) | Lachnospiraceae |
| 17 | Dorea longicatena | AJ132842 | 96.4-97.0 | 5 | 5 (A) | Lachnospiraceae |
| 18 | Dorea longicatena | AJ132842 | 100.0 | 3 | 3 (B) | Lachnospiraceae |
| 19 | Dialister pneumosintes | X82500 | 99.6 | 1 | 1 (A) | Veillonellaceae |
| 20 | Eubacterium plautii | AY724678 | 91.5 | 1 | 1 (B) | Eubacteriaceae |
| 21 | Streptococcus thermophilus | AY188354 | 99.9 | 1 | 1 (B) | Streptococcaceae |
| 22 | Blautia wexlerae | EF036467 | 99.1-99.9 | 23 | 23 (B) | Insertae cedis XIV |
| 23 | Clostridium citroniae | DQ279737 | 95.1-96.2 | 9 | 7 (A); 2 (B) | Unclass Clostridiales |
| Clostridium asparagiforme | AJ582080 | 95.1-95.5 | Unclass Clostridiales | |||
| 24 | Clostridium clostridioforme | M59089 | 95.0-95.2 | 5 | 5 (A) | Unclass Clostridiales |
| Clostridium citroniae | DQ279737 | 95.0 | Unclass Clostridiales | |||
| 25 | Clostridium clostridioforme | M59089 | 99.4-99.7 | 3 | 3 (A) | Unclass Clostridiales |
| 26 | Clostridium aldenense | DQ279736 | 99.1 | 1 | 1 (A) | Unclass Clostridiales |
| 27 | Clostridium asparagiforme | AJ582080 | 95.7 | 1 | 1 (A) | Unclass Clostridiales |
| 28 | Clostridium asparagiforme | AJ582080 | 96.5 | 1 | 1 (B) | Unclass Clostridiales |
| 29 | Clostridium clostridioforme | M59089 | 95.1-95.9 | 5 | 5 (B) | Unclass Clostridiales |
| 30 | Clostridium ramosum | X73440 | 100.0 | 2 | 2 (A) | Unclass firmicutes |
| 31 | Escherichia fergusonii | AF530475 | 99.7-99.9 | 2 | 2 (A) | Gammaproteobacteria |
| 32 | Barnesiella intestinihominis | AB267809 | 99.1-99.3 | 2 | 2 (B) | Porphyrmonadaceae |
| 33 | Barnesiella viscericola | AB267809 | 92.1 | 1 | 1 (B) | Porphyrmonadaceae |
| 34 | Barnesiella viscericola | AB267809 | 90.0 | 1 | 1 (A) | Porphyrmonadaceae |
| 35 | Parabacteroides distasonis | AB238922 | 99.4-100.0 | 4 | 4 (B) | Porphyrmonadaceae |
| 36 | Bacteroides cellulosilyticus | AJ583243 | 97.6-98.9 | 9 | 4 (A); 5 (B) | Bacteroidaceae |
| 37 | Bacteroides caccae | X83951 | 99.4-99.9 | 16 | 2 (A); 14 (B) | Bacteroidaceae |
| 38 | Bacteroides xylanisolvens | AM230650 | 97.7 | 1 | 1 (A) | Bacteroidaceae |
| 39 | Bacteroides thetaiotaomicron | AE015928 | 99.9 | 6 | 4 (A); 2 (B) | Bacteroidaceae |
| 40 | Bacteroides thetaiotaomicron | AE015930 | 99.3 | 1 | 1 (B) | Bacteroidaceae |
| 41 | Bacteroides uniformis | AB050110 | 99.7-100.0 | 6 | 3 (A); 3 (B) | Bacteroidaceae |
| 42 | Bacteroides dorei | AB242142 | 97.3-98.7 | 29 | 26 (A); 3 (B) | Bacteroidaceae |
| 43 | Alistipes putredinis | L16497 | 92.4-92.7 | 2 | 2 (B) | Rikenellaceae |
| 44 | Alistipes onderdonkii | AY974071 | 99.7 | 1 | 1 (B) | Rikenellaceae |
The type strain showing the highest similarity to the sequence is shown. Assignment of the clones to bacterial family level was done using the “sequence match” option in the Ribosomal data base[22].
Accession number for the type strain;
Similarity to the closest type strain;
The total number of clones assigned to the phylotype;
Number of clones found in patient A and B, respectively.
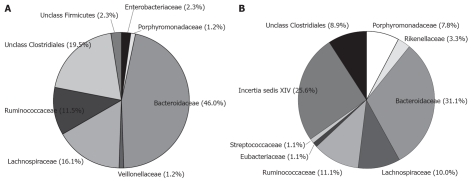
Sequences could be grouped into 22 phylotypes in patient A and 29 phylotypes in patient B. Shannon’s and Simpsons diversity indices were calculated and both the patients showed similar values. The Shannon index was 2.61 for patient A and 2.78 for patient B, and the Simpson index was 8.13 for A and 9.29 for patient B. Firmicutes and Bacteroidetes were the dominating phyla with 50.6% and 47.2% in patient A and 57.8% and 42.2% in patient B, respectively (Figure 1).
Figure 1.
Distribution of clones at family level. Assignment of the clones were done using the Ribosomal Data Base Project Release 10 and the option “seqmatch”[22]. A: Patient A; B: Patient B.
In patient A Porphyromonadaceae constituted 1.2% of the clones and in patient B, Porphyromonadaceae and Rikenellaceae constituted 11.1% of the clones. Only two clones (2.3%) similar to Enterobacteriaceae were found in patient A.
The most common phylotypes were sequences similar to Blautia wexlerae (23 clones), Faecalibacterium prausnitzii (13 clones) and Clostridium citroniae (9 clones) within Firmicutes, and Bacteroides dorei (29 clones), Bacteroides caccae (16 clones) and Bacteroides cellulosilyticus (9 clones) within Bacteroidetes (Table 1). These phylotypes showed > 97% similarity to the closest type strain except for C. citroniae. Out of the 44 phylotypes identified, the two patients had 7 in common and 5 of these were assigned to Bacteroidetes and two to the Firmicutes. The phylotypes in common constituted 84 clones (47.5%) of the total number of clones. Sequences similar to F. prausnitzii and C. citroniae were found in both patients (Table 1). Within Bacteroidetes the shared phylotypes were most similar to, Bacteroides thetaiotaomicron, Bacteroides uniformis, B. cellulosilyticus, B. caccae and B. dorei.
DISCUSSION
In the present study the microbiota of the ascending colon in the two female patients with CC showed similarities to a normal colon microbiota with Firmicutes and Bacteroidetes as dominating phyla, making up 97.7% and 100.0% of the clones in patient A and B, respectively. Only two clones close to Enterobacteriaceae were found in patient A. In several studies, the microbiota of healthy persons have been analyzed by sequencing of the 16S rRNA genes using either fecal samples or tissue samples from the intestinal mucosa[17,18,26-28]. All these studies showed a predominance of Firmicutes and Bacteroidetes while Verrucomicrobia, Actinobacteria and gamma proteobacteria were detected at lower frequency.
The proportion of clones belonging to Bacteroides was 47.0% in patient A and 31.1% in patient B. These were higher figures than Wang et al[18], using a similar methodology, found in biopsies taken from the ascending colon from a healthy, 54-year old woman where Bacteroides constituted 24.4% of the clones. Hayashi et al[17] analyzed fecal samples of 3 healthy men aged 27, 34 and 54 years, and the proportion of Bacteroides was 4.2%, 3.4% and 14.9%, respectively. In another study of fecal samples from a healthy 40-year old man, Bacteroides constituted 14.4% of the total number of clones[26]. Delgado et al[27] analyzed clones from the descending colon from a healthy 45-year old man and found one clone out of 20 (5%) belonging to Bacteroides. Of the 44 phylotypes found here, the two patients had only 7 in common. However, these shared phylotypes constituted 47.5% of the total number of clones. Within Bacteroides five phylotypes were common to both patients. Of these the most dominating were clones similar to B. caccae and B. dorei making up 25.4% of the total number of clones (Table 1). Both species belong to the Bacteroides fragilis group that are opportunistic pathogens isolated from a variety of anaerobic infections and cause about 50% of all anaerobic bacteremias[29,30].
A subgroup of B. fragilis, enterotoxigenic B. fragilis (ETBF), that can secrete a proinflammatory enterotoxin, has been found to be implicated in traveller´s diarrhea[31]. In a study by Zhang et al[32], significantly more ETBF were found in patients with watery diarrhea (26.8%) than in the control group (12.4%). ETBF was also found at a higher frequency in patients over 30 years of age compared to the control group. Additonally, it was shown that 27.0% of patients over the age of 60 carried ETBF compared to 3.7% for the control group. It has been suggested that Bacteroides fragilis toxin can bind to receptors on the epithelial cells, leading to a signal cascade and clevage of cadherine promoting an increased intestinal permeability[33]. An increased intestinal permeability was shown in one patient with CC, using an Ussing chamber[12]. Permeability was measured on biopsies taken from the sigmoid colon and it was shown that the intestinal integrity was improved after a fecal diversion by an ileostomy, but after restoration of the bowel continuity the permeabilty increased again[12]. Some improvement has been reported when CC patients were treated with metronidazole, penicillin or erythromycin[34]. Bacteroides are sensitive to metronidazole and that might point to Bacteroides as a possible disease-provoking agent[30]. On the other hand, the positive effect shown with penicillin and erythromycin speaks against Bacteroides[34].
It has been shown that Akkermansia mucinciphila and strains of Clostridium, Prevotella and Bacteroides are able to degrade mucin[35,36]. The type strain B. thetaiotaomicron NCTC 10582 was shown to express glycosidases and glycosulphatase and could degrade pig gastric mucin[37]. In the present study 4 clones from patient A and 3 clones from patient B showed high similarity (99.3%-99.9%) to the type strain B. thetaiotaomicron NCTC 10582 (Table 1). Clones belonging to Akkermansia muciniphila were not found. However, it has been shown that this species represents only about one percent of the microbiota in healthy children and adults[38]. One might speculate that specific components present within the microbiota of the CC patients, i.e., Bacteroides spp., that has an impact both on the colonic mucin layer and the intestinal permeability, leading to an immune response.
The clones resembling Clostridium clostridioforme, Clostridium citroniae, Clostridium aspargiforme and Clostridium aldenense were distributed into 7 phylotypes showing 95%-99.7% similarity to the different type strains. Four clones from patient A showed high similarity to C. clostridioforme and C. aldenense. Also in patient B, 5 clones resembling C. clostridioforme were found, but they showed lower similarity to the type strain. They are all related and belong to cluster XIVa as defined by Collins et al[39], Warren et al[40] and Mohan et al[41]. Strains of C. clostridioforme and closely related species have been involved in a variety of infections[42]. In a study of autistic children, all of whom had gastrointestinal symptoms, high counts of fecal isolates showing 95% similarity to C. clostridioforme were found in the diseased children but not in the controls. It cannot be excluded that the presence of sequences resembling C. clostridioforme might play a role in the disease in the patients analyzed here.
Clones identified as F. prausnitzii of the Ruminococcaceae family were found in both patients and constituted about 7% of the total number of clones. These bacteria together with Eubacterium rectale and Roseburia spp. are known as butyrate producers and usually make up about 5%-10% of the human microbiota and can be regarded as commensals[43]. No clones resembling Lactobacillus nor Actinobacteria or Verrucomicrobia were found. This can probably be explained by the fact that too few clones were sequenced and that they usually constitute a minor part of the microbiota. Previously published case reports have suggested Clostridium difficile, Yersinia enterocolitica, Campylobacter jejuni and Aeromonas hydrophila to CC as possible pathogens[2,7,14-16]. This could not be confirmed in the present study. As different pathogens are described, and the fact that the colonic microbiota was similar to a healthy one, the etiology to CC may not primarily depend on abnormal microbiota, and antibiotics may not be the treatment of choice in this entity, as it is sometimes considered[34].
This study has some limitations. Only two patients were examined and the method applied here only detects the dominant bacteria. Future research needs to examine the presence of common pathogens in the bowel, but also etiologies of CC other than bacteria must be considered.
To the best of our knowledge, this is the first study of the intestinal microbiota in patients with a histologically diagnosed CC, by a culture-independent method. The overall composition of the colonic microbiota was similar to a healthy one with dominance of Firmicutes and Bacteroidetes. Due to the fact that only two patients were analyzed it is difficult to draw any conclusions, but in both patients a high proportion of potentially pathogenic species of Bacteroides and clones related to C. clostridioforme were found.
ACKNOWLEDGMENTS
We thank Martin Olesen, MD, PhD for characterizing the patients histologically and Ingrid Palmquist, RN for excellent technical assistance.
COMMENTS
Background
Collagenous colitis (CC) is an idiopathic inflammatory bowel disease characterized by chronic non-bloody diarrhea. CC is regarded as a subtype of microscopic colitis. The etiology is unknown but a noxious agent, probably originating from the microbiota, in the intestinal lumen has been proposed to have a pathogenic role. However, no attempt to analyze the microbiota in diseased patients has been done.
Research frontiers
The intestinal mucosa is colonized by a huge number of bacteria that are important for health and disease. In several studies the gut microbiota has been analyzed by culture-independent methods in patients with intestinal inflammatory diseases such as ulcerative colitis and Crohn’s disease.
Innovations and breakthroughs
Having the opportunity to obtain histologically well-defined collagenous colitis samples, the authors have characterized the dominant microbiota in two diseased patients.
Applications
Culture-independent methods can be used for analyzing the dominant mucosa-associated microbiota in collagenous colitis.
Terminology
The meaning of the word microbiota here is synonymous to the bacterial flora in the intestine.
Peer review
It is well organized. Several papers have been presented in support of microbiota from controls, but no speculation have been made about findings in this paper and clinical applications, limitations of the study and future of research in this field.
Footnotes
Supported by Grants from Development Foundations of Region Skåne and from Skåne University Hospital, Malmö
Peer reviewer: Antonio Gasbarrini, Professor, Internal Medi-cine Institute, Catholic University, Largo Agostino Gemelli 8, 00168 Roma, Italy
S- Editor Gou SX L- Editor A E- Editor Li JY
References
- 1.Olesen M, Eriksson S, Bohr J, Järnerot G, Tysk C. Microscopic colitis: a common diarrhoeal disease. An epidemiological study in Orebro, Sweden, 1993-1998. Gut. 2004;53:346–350. doi: 10.1136/gut.2003.014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nyhlin N, Bohr J, Eriksson S, Tysk C. Systematic review: microscopic colitis. Aliment Pharmacol Ther. 2006;23:1525–1534. doi: 10.1111/j.1365-2036.2006.02913.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindström CG. Collagenous colitis. Leber Magen Darm. 1991;21:103–104, 106. [PubMed] [Google Scholar]
- 4.Lazenby AJ, Yardley JH, Giardiello FM, Jessurun J, Bayless TM. Lymphocytic (“microscopic”) colitis: a comparative histopathologic study with particular reference to collagenous colitis. Hum Pathol. 1989;20:18–28. doi: 10.1016/0046-8177(89)90198-6. [DOI] [PubMed] [Google Scholar]
- 5.Temmerman F, Baert F. Collagenous and lymphocytic colitis: systematic review and update of the literature. Dig Dis. 2009;27 Suppl 1:137–145. doi: 10.1159/000268134. [DOI] [PubMed] [Google Scholar]
- 6.Mahajan L, Wyllie R, Goldblum J. Lymphocytic colitis in a pediatric patient: a possible adverse reaction to carbamazepine. Am J Gastroenterol. 1997;92:2126–2127. [PubMed] [Google Scholar]
- 7.Camarero C, Leon F, Colino E, Redondo C, Alonso M, Gonzalez C, Roy G. Collagenous colitis in children: clinicopathologic, microbiologic, and immunologic features. J Pediatr Gastroenterol Nutr. 2003;37:508–513. doi: 10.1097/00005176-200310000-00020. [DOI] [PubMed] [Google Scholar]
- 8.Ung KA, Gillberg R, Kilander A, Abrahamsson H. Role of bile acids and bile acid binding agents in patients with collagenous colitis. Gut. 2000;46:170–175. doi: 10.1136/gut.46.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Bañares F, Esteve M, Salas A, Forné TM, Espinos JC, Martín-Comin J, Viver JM. Bile acid malabsorption in microscopic colitis and in previously unexplained functional chronic diarrhea. Dig Dis Sci. 2001;46:2231–2238. doi: 10.1023/a:1011927302076. [DOI] [PubMed] [Google Scholar]
- 10.Tysk C, Bohr J, Nyhlin N, Wickbom A, Eriksson S. Diagnosis and management of microscopic colitis. World J Gastroenterol. 2008;14:7280–7288. doi: 10.3748/wjg.14.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Järnerot G, Tysk C, Bohr J, Eriksson S. Collagenous colitis and fecal stream diversion. Gastroenterology. 1995;109:449–455. doi: 10.1016/0016-5085(95)90332-1. [DOI] [PubMed] [Google Scholar]
- 12.Münch A, Söderholm JD, Wallon C, Ost A, Olaison G, Ström M. Dynamics of mucosal permeability and inflammation in collagenous colitis before, during, and after loop ileostomy. Gut. 2005;54:1126–1128. doi: 10.1136/gut.2004.058750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Järnerot G, Bohr J, Tysk C, Eriksson S. Faecal stream diversion in patients with collagenous colitis. Gut. 1996;38:154–155. doi: 10.1136/gut.38.1.154-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erim T, Alazmi WM, O’Loughlin CJ, Barkin JS. Collagenous colitis associated with Clostridium difficile: a cause effect? Dig Dis Sci. 2003;48:1374–1375. doi: 10.1023/a:1024127713979. [DOI] [PubMed] [Google Scholar]
- 15.Perk G, Ackerman Z, Cohen P, Eliakim R. Lymphocytic colitis: a clue to an infectious trigger. Scand J Gastroenterol. 1999;34:110–112. doi: 10.1080/00365529950172925. [DOI] [PubMed] [Google Scholar]
- 16.Mäkinen M, Niemelä S, Lehtola J, Karttunen TJ. Collagenous colitis and Yersinia enterocolitica infection. Dig Dis Sci. 1998;43:1341–1346. doi: 10.1023/a:1018836614448. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Wang M, Molin G, Ahrné S, Adawi D, Jeppsson B. High proportions of proinflammatory bacteria on the colonic mucosa in a young patient with ulcerative colitis as revealed by cloning and sequencing of 16S rRNA genes. Dig Dis Sci. 2007;52:620–627. doi: 10.1007/s10620-006-9461-1. [DOI] [PubMed] [Google Scholar]
- 20.Olsson C, Ahrné S, Pettersson B, Molin G. The bacterial flora of fresh and chill-stored pork: analysis by cloning and sequencing of 16S rRNA genes. Int J Food Microbiol. 2003;83:245–252. doi: 10.1016/s0168-1605(02)00372-0. [DOI] [PubMed] [Google Scholar]
- 21.Hall TA. Bioedit a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 22.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. 1993. [Google Scholar]
- 25.Magurran AE. Ecological diversity and it’s measurement. New Jersey: Princeton University Press; 1988. pp. 34–41. [Google Scholar]
- 26.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado S, Suárez A, Mayo B. Identification of dominant bacteria in feces and colonic mucosa from healthy Spanish adults by culturing and by 16S rDNA sequence analysis. Dig Dis Sci. 2006;51:744–751. doi: 10.1007/s10620-006-3201-4. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Heazlewood SP, Krause DO, Florin TH. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol. 2003;95:508–520. doi: 10.1046/j.1365-2672.2003.02005.x. [DOI] [PubMed] [Google Scholar]
- 29.Brook I. The role of anaerobic bacteria in bacteremia. Anaerobe. 2010;16:183–189. doi: 10.1016/j.anaerobe.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Nagy E, Urbán E, Nord CE. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe: 20 years of experience. Clin Microbiol Infect. 2011;17:371–379. doi: 10.1111/j.1469-0691.2010.03256.x. [DOI] [PubMed] [Google Scholar]
- 31.Jiang ZD, Dupont HL, Brown EL, Nandy RK, Ramamurthy T, Sinha A, Ghosh S, Guin S, Gurleen K, Rodrigues S, et al. Microbial etiology of travelers’ diarrhea in Mexico, Guatemala, and India: importance of enterotoxigenic Bacteroides fragilis and Arcobacter species. J Clin Microbiol. 2010;48:1417–1419. doi: 10.1128/JCM.01709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G, Svenungsson B, Kärnell A, Weintraub A. Prevalence of enterotoxigenic Bacteroides fragilis in adult patients with diarrhea and healthy controls. Clin Infect Dis. 1999;29:590–594. doi: 10.1086/598639. [DOI] [PubMed] [Google Scholar]
- 33.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349–369, Table of Contents. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohr J, Tysk C, Eriksson S, Abrahamsson H, Järnerot G. Collagenous colitis: a retrospective study of clinical presentation and treatment in 163 patients. Gut. 1996;39:846–851. doi: 10.1136/gut.39.6.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 36.Stanley RA, Ram SP, Wilkinson RK, Roberton AM. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl Environ Microbiol. 1986;51:1104–1109. doi: 10.1128/aem.51.5.1104-1109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai HH, Hart CA, Rhodes JM. Production of mucin degrading sulphatase and glycosidases by Bacteroides thetaiotaomicron. Lett Appl Microbiol. 1991:13: 97–101. [Google Scholar]
- 38.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 40.Warren YA, Tyrrell KL, Citron DM, Goldstein EJ. Clostridium aldenense sp. nov. and Clostridium citroniae sp. nov. isolated from human clinical infections. J Clin Microbiol. 2006;44:2416–2422. doi: 10.1128/JCM.00116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohan R, Namsolleck P, Lawson PA, Osterhoff M, Collins MD, Alpert CA, Blaut M. Clostridium asparagiforme sp. nov., isolated from a human faecal sample. Syst Appl Microbiol. 2006;29:292–299. doi: 10.1016/j.syapm.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Finegold SM, Song Y, Liu C, Hecht DW, Summanen P, Könönen E, Allen SD. Clostridium clostridioforme: a mixture of three clinically important species. Eur J Clin Microbiol Infect Dis. 2005;24:319–324. doi: 10.1007/s10096-005-1334-6. [DOI] [PubMed] [Google Scholar]
- 43.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]