Abstract
AIM: To investigate the relationship between polymorphisms present in the vitamin D receptor (VDR) gene and colorectal cancer risk, a systematic meta-analysis of population-based studies was performed.
METHODS: A total of 38 relevant reports published between January 1990 and August 2010 were identified, of which only 23 qualified for this meta-analysis based on our selection criteria. Five polymorphic variants of the VDR gene, including Cdx-2 (intron 1e) and FokI (exon 2) present in the 5’ region of the gene, and BsmI (intron 8), ApaI (intron 8), and TaqI (exon 9) sites present in the 3’ untranslated region (UTR), were evaluated for possible associations with colorectal cancer risk. Review manager 4.2 was used to perform statistical analyses.
RESULTS: In the meta-analysis performed, only the BsmI polymorphism was found to be associated with colorectal cancer risk. In particular, the BsmI B genotype was found to be related to an overall decrease in the risk for colorectal cancer [BB vs bb: odds ratio (OR) = 0.87, 95% CI: 0.80-0.94, P = 3 × 10-4; BB vs Bb + bb: OR = 0.90, 95% CI: 0.84-0.97, P = 5 × 10-4]. Moreover, in subgroup analyses, the BsmI B genotype was significantly associated with colon cancer, and not rectal cancer. An absence of between-study heterogeneity was also observed.
CONCLUSION: A meta-analysis of 23 published studies identified the BsmI polymorphism of the VDR gene to be associated with an increased risk of colon cancer.
Keywords: Vitamin D receptor, Polymorphism, Meta-analysis, Colorectal cancer
INTRODUCTION
Colorectal cancer represents the third most common cancer worldwide, second only to lung cancer and gastric cancer[1]. Furthermore, it is estimated that there are more than 370 000 cases of colon and rectal cancer diagnosed in Europe every year, with 200 000 cases resulting in death[2]. However, the underlying etiology of colorectal cancer, including cancerous growths of the colon, rectum, and appendix, remains poorly understood. It has been proposed that some categories of external agents, including physical, chemical, and biological carcinogens, may contribute to the development of this disease, and the role of these factors in carcinogenesis would depend largely on genetic factors. Correspondingly, a recent study showed that insufficient levels of vitamin D may result in colorectal cancer[3]. Furthermore, genetic variations in genes controlling vitamin D activity would be hypothesized to play an important role in determining susceptibility to colorectal cancer.
In vivo, vitamin D helps bones and muscles grow, and may also help prevent many diseases, such as prostate cancer and breast cancer. The biological activity of vitamin D is mediated by the vitamin D receptor (VDR)[4], which interacts with other cell signaling pathways to influence cell behavior. Expression of VDR has been detected in various organs and tissues of the human body, including the kidney and bone cells. VDR is also expressed in normal colon mucosa[5]. In the intestine, VDR plays an important role in regulating cell proliferation, differentiation, and the induction of apoptosis[6]. Furthermore, VDR may be associated with the effects of calcium on colorectal epithelial proliferation[7].
Molecular epidemiological studies have shown that polymorphisms in the VDR gene may be linked to biological functions of vitamin D. At the 5’ end of the VDR gene, a FokI polymorphism (rs2228570/rs10735810, exon 2) has been associated with a frameshift in the VDR protein[8]. Moreover, polymorphisms in the 3’ untranslated region (UTR), including BsmI (rs1544410, intron 8), ApaI (rs7975232, intron 9), and TaqI (rs731236, exon 9) sites, have been shown to influence gene transcription and mRNA stability[9]. Additionally, these polymorphisms have exhibited the potential for strong linkage disequilibrium (LD)[10,11], and functional differences have been associated with the associated haplotypes[11,12]. Given that polymorphisms in the VDR gene could potentially influence the binding of 1, 25(OH)2D3 and the anti-proliferative effects of vitamin D, VDR polymorphisms have been hypothesized to be associated with colorectal cancer risk.
In 2001, the first report of an association between colorectal cancer and the VDR gene was published by Kim and colleagues[13]. They identified a random subset of 393 cases of colorectal adenomas and 406 colonoscopy-negative controls from a clinic-based, case-control study conducted in the United States between 1991 and 1994. Based on their analysis, the BsmI BB genotype was found to be associated with a reduced risk of colorectal adenoma when intake of calcium and vitamin D was reduced. In addition to the BsmI site[12,14-23]. Other polymorphic sites present in the VDR gene, including Cdx-2[12,19,21,24,25], FokI[12,15,16,18,19,21-24,26-33], ApaI[12,16,19,21,34], and TaqI[12,16,19,23,24,29,31,33-35], have been evaluated in genetic association studies. However, the results are inconsistent. Since it can be difficult for individual studies to achieve sufficient statistical power to detect associations between VDR polymorphisms and colorectal cancer risk, a meta-analysis that combines data from all published studies may detect genetic associations more accurately. In addition, a reduced probability of false-negatives might also be achieved[36]. Therefore, a systematic meta-analysis of population-based studies was performed to investigate the association between VDR polymorphisms and the risk of colorectal cancer. Based on the search strategy and criteria used, 23 studies were analyzed which identified several important polymorphic variants.
MATERIALS AND METHODS
Search strategy and data extraction
To examine the association between VDR polymorphisms and colorectal cancer risk, a search of the MEDLINE database (from January 1990 to August 2010) and the US National Library of Medicine’s PubMed database (http://www.ncbi.nlm.nih.gov/pubmed) was performed. In addition, various scientific research tools available on the web were used to search relevant references such as Google (http://scholar.google.com/) and Scirus (http://www.scirus.com/). In particular, data relevant to five well-characterized polymorphic variants was identified, including: Cdx-2, FokI, BsmI, ApaI, and TaqI sites within VDR. Keywords used in searches included “vitamin D receptor” in combination with “polymorphism”, “vitamin D”, “genotype”, “allele”, “colorectal cancer”, or “risk”.
Papers selected for this meta-analysis included a case-control study and complete data, including the authors’ names; the subjects’ region/country; year of publication; numbers of cases and controls; mean age (or range) of the case/control group; diagnostic criteria used; and number of subjects with the VDR genotype in both case and control groups. All relevant references that met these inclusive criteria and that were published as articles or abstract containing original data, were included in this study. In contrast, case-only studies, studies with incomplete data, or studies with inadequate control groups were excluded. In addition, the data extracted needed to conform to the guidelines of MOOSE, a proposal for reporting meta-analyses of observational studies[37]. If the same or overlapping data were reported in multiple publications, the most recent publication was selected[38].
Statistical analysis
For each data set included in this study, the odds ratios (ORs) and corresponding 95% CI for the incidence of cancer in subjects with or without particular restriction sites (lowercase vs uppercase lettering), was compared. Furthermore, deviations from the Hardy-Weinberg equilibrium for each control group were assessed using the goodness-of-fit test. To estimate associations with colorectal cancer risk, various genotypic models were selected, including codominant, additive, recessive, and dominant. Both the Peto Mantel-Haenszel fixed-effects model and the DerSimonian Laird random-effects model (with weights based on the inverse variance) were used to calculate summary ORs, and both within- and between-study variations were considered[39]. A P-value less than 0.10 was considered statistically significant when comparing trials showing heterogeneity, and random-effects analysis was selected. In contrast, fixed-effects analysis was used for comparing trials exhibiting homogeneity. Inverted funnel plots were also used to examine asymmetry, in which the ORs were plotted on a logarithmic scale against the inverse of their corresponding standard errors[40]. In the presence of publication bias, the funnel plot was asymmetric and the data showed remarkable skewness. There may be many reasons for this, most notably that some studies with negative findings are not published. In contrast, the plots were symmetric when bias was absent.
All statistical analyses were performed using the Statistical Package for Social Sciences (SPSS, version 13.0) and Review Manager (version 4.2, The Cochrane Collaboration), and all P-values were two-sided.
RESULTS
Characteristics of case-control studies included in the meta-analysis
According to the criteria defined above, 38 published studies relevant to the VDR gene and colorectal cancer risk were reviewed. Fifteen of these papers were excluded due to insufficient clarity in data presentation, repeated literature, or significant differences were present in their study design compared with the other papers identified[41]. The remaining 23 eligible case-control studies are listed in Table 1, and were included in a meta-analysis to investigate possible associations between Cdx-2, FokI, BsmI, ApaI, and TaqI polymorphisms present in the VDR gene and the risk of colorectal cancer.
Table 1.
Characteristics of case-control studies included in the meta-analysis
| First author | Year | Country | Racial descent | Mean age in cases | Mean age in controls | Cases/controls | Genotyping method | Quality control | Adjusted | Studied polymorphisms |
| Ingles et al[11] | 1998 | United States | American | 62.3 | 62.2 | 373/394 | PCR-RFLP | Yes | Yes | FokI |
| Kim et al[13] | 2001 | United States | American | 58.0 ± 9.7 | 53.0 ± 11.0 | 393/406 | TaqMan | Yes | Yes | BsmI |
| Peters et al[27] | 2001 | United States | American | 18-74 | 18-74 | 208/184 | PCR-RFLP | Yes | NR | FokI |
| Slatter et al[23] | 2001 | United States | American | NR | NR | 424/366 | PCR-RFLP | NR | Yes | FokI, BsmI, TaqI |
| Speer et al[14] | 2001 | Hungary | European | 64 | 63 | 56/112 | PCR-RFLP | NR | NR | BsmI |
| Grau et al[29] | 2003 | United States | American | 60.8 ± 9.0 | 60.9 ± 9.0 | 372/379 | PCR-RFLP | Yes | Yes | FokI, TaqI |
| Wong et al[28] | 2003 | Singapore | Asian | 66 | 56.5 | 217/890 | PCR-RFLP | Yes | Yes | FokI |
| Peters et al[35] | 2004 | United States | American | 62.9 | 62.3 | 763/774 | PCR-RFLP | Yes | NR | TaqI |
| Slattery et al[15] | 2004 | United States | American | 30-79 | 30-79 | 1936/2130 | PCR-RFLP | NR | Yes | FokI, BsmI |
| Murtaugh et al[30] | 2006 | United States | American | 30-79 | 30-79 | 2450/2821 | PCR-RFLP | NR | Yes | FokI |
| Park et al[16] | 2006 | South Korea | Asian | 55 | 55 | 190/354 | PCR-RFLP | NR | Yes | FokI, BsmI, ApaI, TaqI |
| Flügge et al[12] | 2007 | Germany | European | 61.9 ± 10.0 | 62.2 ± 11.2 | 256/256 | PCR-RFLP | Yes | Yes | Cdx-2, FokI, BsmI, ApaI, TaqI |
| Kadiyska et al[19] | 2007 | Bulgaria | European | 59 | 59 ± 5 | 133/94 | PCR-RFLP | NR | Yes | BsmI |
| Slattery et al[18] | 2007 | United States | American | 30-79 | 30-79 | 2380/2990 | TaqMan | Yes | Yes | FokI, BsmI |
| Yaylim-Eraltan et al[31] | 2007 | Turkey | European | 59.1 ± 4.0 | 52.0 ± 0.8 | 26/52 | PCR-RFLP | NR | Yes | FokI, TaqI |
| Grünhage et al[32] | 2008 | Germany | European | 65 ± 9 | 63 ± 8 | 192/220 | PCR-RFLP | NR | Yes | FokI |
| Hubner et al[17] | 2008 | United Kingdom | European | NR | NR | 137/409 | TaqMan | Yes | Yes | Cdx-2, FokI, BsmI, ApaI, TaqI |
| Ochs-Balcom et al[24] | 2008 | United States | American | 62.8 ± 10.2 | 58.5 ± 12.1 | 250/246 | TaqMan | Yes | Yes | Cdx-2, FokI, TaqI |
| Parisi et al[20] | 2008 | Spain | European | NR | NR | 170/120 | PCR-RFLP | NR | Yes | BsmI |
| Theodoratou et al[21] | 2008 | United Kingdom | European | 62.0 ± 10.8 | 62.4 ± 10.5 | 3005/3072 | Microarray | Yes | Yes | Cdx-2, FokI, BsmI, ApaI |
| Wang et al[33] | 2008 | China | Asian | 38-78 | 19.6 ± 1.3 | 69/218 | PCR-RFLP | NR | Yes | FokI |
| Jenab et al[22] | 2009 | Europe | European | NR | NR | 1248/1248 | TaqMan | Yes | Yes | FokI, BsmI |
| Mahmoudi et al[34] | 2010 | Iran | Asian | 52.7 ± 14.0 | 44.4 ± 17.7 | 160/180 | PCR-RFLP | Yes | Yes | ApaI, TaqI |
NR: Not reported.
In 21/23 studies, data regarding the 5’ end of the VDR gene were provided. In four of these studies, 2639 cases and 2948 controls were analyzed for the Cdx-2 polymorphism, while 17 studies included 13 301 cases and 15 942 controls analyzed for the FokI polymorphism. In addition, the 3’ UTR region of the VDR gene has been analyzed. For example, 12 studies containing 10 083 cases and 11 242 controls analyzed the BsmI polymorphism, 5 studies including 2739 cases and 3200 controls analyzed the ApaI polymorphism, and 9 studies including 2580 cases and 3016 controls analyzed the TaqI polymorphism.
Among controls, the frequency of the c allele at the Cdx-2 site ranged from 65.6% in Berlin-Bush populations of Germany, to 80.0% in a United Kingdom population[12,19,21,24,25]. In contrast, the frequency of the f allele of FokI among controls ranged from 31.7% in Turkey, to 47.2% in a Singapore population[12,15,16,18,19,21-24,26-33]. The frequency of the b allele at BsmI among controls ranged from 56.1% in a Bulgarian population, to 94.7% in a Korean population[12,14-23], while the frequency of the a allele at ApaI among controls ranged from 23.0% in a Korean population to 49.3% in a population of the United Kingdom[12,16,19,21,34]. Lastly, the frequency of the t allele at TaqI among controls ranged from 8.8% in a Korean population to 43.6% in a population of the United States[12,16,19,23,24,29,31,33-35].
Qualitative assessment of included studies
Genotyping of the Cdx-2, FokI, BsmI, ApaI and TaqI polymorphisms was performed using the polymerase chain reaction-restriction fragment length polymorphism technique in 75% of the studies included in this meta-analysis. Due to the low sensitivity of this classic technology, quality control of this genotyping was required, and included blindness to the case-control status, random repeats of samples, or validation using a different genotyping method. However, only 38.9% (7/18) of the eligible studies provided sufficient quality control. Regarding sample size, only 5/24 (20.8%) studies employed more than 1000 cases or controls. Moreover, most of these studies were associated with poor statistical power due to sample sizes that were less than 500 and in some cases contained less than 100 cases or controls.
Assessment of Hardy-Weinberg proportion is regarded as an important criterion for evaluating genetic association studies[38]. Most of the studies included in this meta-analysis reported genotype frequencies in their control groups that were consistent with Hardy-Weinberg proportions (P > 0.05). For example, deviations from Hardy-Weinberg proportions in controls were observed in three studies for FokI[29,31,32], two studies of BsmI[19,21], one study of ApaI[19], and two studies of TaqI[23,24].
Funnel plotting was performed to evaluate whether publication bias was present in the meta-analysis performed. As shown in Figure 1, the shapes of the funnel plots obtained appear to be symmetrical in codominant, dominant, and recessive models, suggesting that publication bias is absent in the meta-analysis performed.
Figure 1.
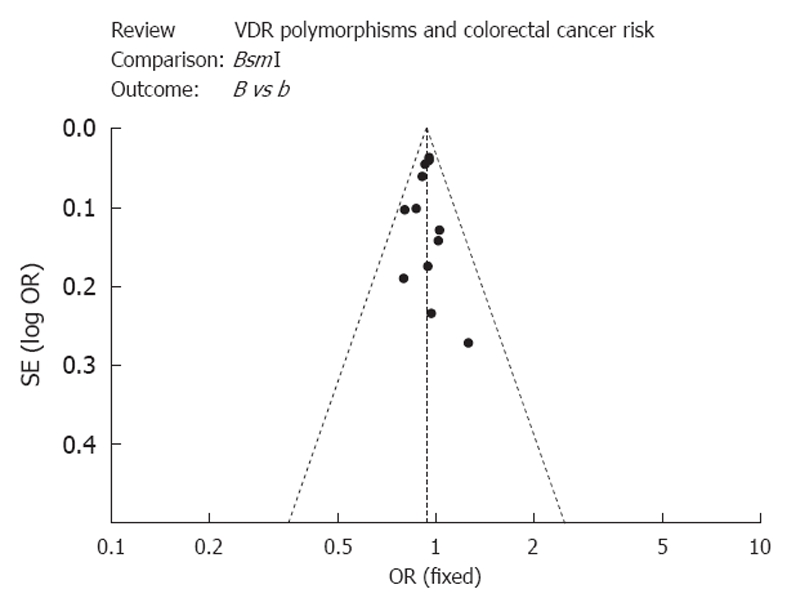
A funnel plot was used to estimate the publication bias of the studies included in the meta-analysis performed.
Cdx-2, FokI, BsmI, ApaI, TaqI polymorphisms and colorectal cancer risk
A heterogeneity test of potential associations between the Cdx-2, FokI, BsmI, ApaI, and TaqI polymorphisms and risk of colorectal cancer are presented in Tables 2 and 3.
Table 2.
Summary odds ratios and 95% CI in the vitamin D receptor gene
| SNP | Model | Total No. cases | Total No. controls | OR (95% CI)1 | P value2 | P value3 |
| Cdx-2 | Codominant (CC vs cc) | 152/1561 | 146/1820 | 1.25 (0.98-1.59) | 0.07 | 0.26 |
| Codominant (Cc vs cc) | 926/2487 | 982/2802 | 1.09 (0.97-1.22) | 0.15 | 0.64 | |
| Codominant (C vs C) | 1230/4048 | 1279/4622 | 1.10 (1.01-1.21) | 0.03 | 0.47 | |
| Dominant (CC + Cc vs cc) | 1078/1561 | 1208/1820 | 0.98 (0.88-1.09) | 0.72 | < 0.001 | |
| Recessive (CC vs Cc + cc) | 152/2487 | 146/2802 | 1.22 (0.96-1.54) | 0.10 | 0.23 | |
| FokI | Codominant (ff vs FF) | 1844/5068 | 2377/5982 | 0.94 (0.87-1.01) | 0.09 | 0.001 |
| Codominant (Ff vs FF) | 6189/11 257 | 7583/13 565 | 0.98 (0.93-1.03) | 0.34 | 0.001 | |
| Codominant (f vs F) | 9867/16 320 | 12 190/19 329 | 0.97 (0.94-1.00) | 0.07 | < 0.001 | |
| Dominant (ff + Ff vs FF) | 8033/5068 | 9960/5982 | 0.96 (0.92-1.01) | 0.15 | < 0.001 | |
| Recessive (ff vs FF + Ff) | 1844/11 257 | 2377/13 565 | 0.95 (0.89-1.02) | 0.13 | 0.01 | |
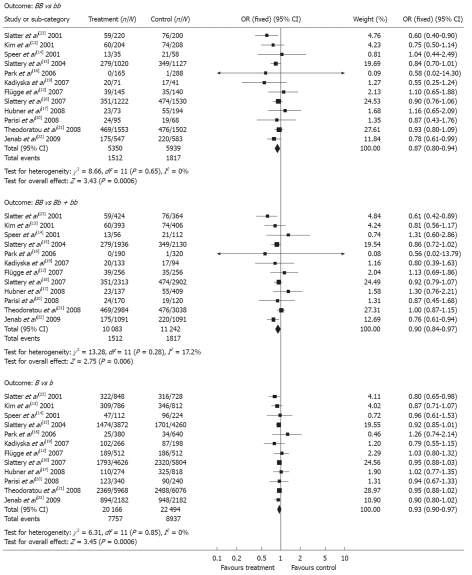
| BsmI | Codominant (BB vs bb) | 1512/3838 | 1817/4122 | 0.87 (0.80-0.94) | < 0.001 | 0.65 |
| Codominant (Bb vs bb) | 4733/8571 | 5303/9425 | 0.94 (0.88-0.99) | 0.03 | 0.64 | |
| Codominant (B vs b) | 7757/12 409 | 8937/13 577 | 0.93 (0.90-0.97) | < 0.001 | 0.85 | |
| Dominant (BB + Bb vs bb) | 6245/3838 | 7120/4122 | 0.92 (0.87-0.97) | 0.003 | 0.84 | |
| Recessive (BB vs Bb + bb) | 1512/8571 | 1817/9425 | 0.90 (0.84-0.97) | 0.006 | 0.28 | |
| ApaI | Codominant (AA vs aa) | 748/578 | 1004/603 | 0.85 (0.73-0.99) | 0.03 | 0.06 |
| Codominant (Aa vs aa) | 1378/1956 | 1593/2196 | 0.91 (0.79-1.04) | 0.18 | 0.39 | |
| Codominant (A vs a) | 2944/2534 | 3601/2799 | 0.92 (0.85-0.99) | 0.02 | 0.04 | |
| Dominant (AA + Aa vs Aa) | 2161/578 | 2597/603 | 0.89 (0.78-1.01) | 0.07 | 0.12 | |
| Recessive (AA vs Aa + aa) | 783/1956 | 1004/2196 | 0.89 (0.80-1.00) | 0.05 | 0.21 | |
| TaqI | Codominant (tt vs TT) | 382/1112 | 398/1320 | 1.05 (0.89-1.24) | 0.58 | 0.07 |
| Codominant (Tt vs TT) | 1086/2198 | 1298/2618 | 0.93 (0.83-1.05) | 0.23 | 0.30 | |
| Codominant (t vs T) | 1850/3310 | 2098/3938 | 0.99 (0.92-1.08) | 0.86 | 0.10 | |
| Dominant (tt + Tt vs TT) | 1468/1112 | 1696/1320 | 0.95 (0.85-1.07) | 0.42 | 0.37 | |
| Recessive (tt vs Tt + TT) | 382/2198 | 398/2618 | 1.07 (0.92-1.25) | 0.38 | 0.01 |
Based on fixed effects model;
Test for overall effect;
Test for heterogeneity. OR: Odds ratios.
Table 3.
The BsmI effect odds ratios stratified by anatomical site
| Model | Colon cancer | Rectal cancer | ||||
| Total No. cases/controls | OR (95% CI) | P value1 | Total No. cases/controls | OR (95% CI) | P value1 | |
| Codominant (BB vs bb) | 1365/1581 | 0.80 (0.68-0.93) | 0.004/0.21 | 659/790 | 0.92 (0.73-1.15) | 0.46/0.90 |
| Codominant (Bb vs bb) | 2257/2438 | 0.99 (0.88-1.12) | 0.92/0.95 | 1029/1240 | 0.94 (0.80-1.11) | 0.48/0.21 |
| Codominant (B vs b) | 5328/5972 | 0.91 (0.84-0.98) | 0.01/0.25 | 2440/2966 | 0.95 (0.85-1.06) | 0.38/0.99 |
| Dominant (BB + Bb vs bb) | 2664/2986 | 0.94 (0.84-1.05) | 0.25/0.65 | 1220/1483 | 0.94 (0.77-1.16) | 0.59/0.40 |
| Recessive (BB vs Bb + bb) | 2664/2986 | 0.80 (0.69-0.92) | 0.002/0.19 | 1220/1483 | 0.93 (0.80-1.09) | 0.40/0.51 |
Test for overall effect and heterogeneity, respectively. OR: Odds ratios.
Cdx-2 polymorphism: Currently, only four studies have investigated the relationship between the VDR Cdx-2 polymorphism and colorectal cancer risk, and all of these studies were in Hardy-Weinberg equilibrium[12,19,21,24,25]. Furthermore, in the overall and subgroup analyses performed, the Cdx-2 polymorphism did not appear to be linked to colorectal cancer risk.
FokI polymorphism: Seventeen studies included in the meta-analysis performed found the FokI polymorphism to be statistically heterogeneous in all genetic models (P ≤ 0.01)[12,15,16,18,19,21-24,26-33]. Moreover, no significant association was found between FokI and colorectal cancer risk in overall and subgroup analyses.
BsmI polymorphism: A total of 12 studies examined the association between colorectal cancer and the BsmI polymorphism, and there was little statistical evidence of heterogeneity among the studies (P ≥ 0.28)[12,14-23]. Individuals with the BB genotype (OR = 0.87; 95% CI = 0.80-0.94, P = 3 × 10-4; P = 0.65 for heterogeneity), or the Bb genotype (OR = 0.94; 95% CI: 0.88-0.99, P = 0.03; P = 0.48 for heterogeneity), were associated with a significant decrease in colorectal cancer risk compared with patients carrying the bb genotype. The Dominant model (BB + Bb vs bb) and the recessive model (BB vs Bb + bb) also showed a significant association with colorectal cancer risk, with the associated ORs being 0.92 (95% CI: 0.87-0.97, P = 0.003; P = 0.84 for heterogeneity) and 0.90 (95% CI: 0.84-0.97, P = 0.006; P = 0.28 for heterogeneity), respectively (Figure 2). Although two of these studies were not consistent with Hardy-Weinberg proportions[17,21], the effect was negligible. In addition, the BB genotype showed a decreased risk for colon cancer compared with the bb (OR = 0.80, 95% CI: 0.68-0.93, P = 0.004; P = 0.21 for heterogeneity), or Bb + bb genotypes (OR = 0.80, 95% CI: 0.69-0.92, P = 0.002; P = 0.19 for heterogeneity). However, no significant differences were observed between these polymorphisms and rectal cancer risk (Table 3).
Figure 2.
Forest plot of the meta-analysis performed to investigate the association between the BsmI polymorphism of the vitamin D receptor gene and colorectal cancer risk (fixed-effects model).
ApaI polymorphism: The association between the ApaI polymorphism and colorectal cancer was investigated in five studies, of which only one study was not consistent with Hardy-Weinberg proportions[12,16,19,21,34]. Moreover, although the codominant model (AA vs aa, OR = 0.83; 95% CI: 0.71-0.97, P = 0.02) showed a significant association with colorectal cancer risk, the P value of 0.05 suggested that this genetic model was statistically heterogeneous.
TaqI polymorphism. Except for the recessive model (tt vs Tt + TT, P = 0.06), there was little evidence of statistical heterogeneity among the nine studies that investigated an association between the TaqI polymorphism and colorectal cancer risk (P ≥ 0.35)[12,16,19,23,24,29,31,33-35]. When the two studies in which controls were not in Hardy-Weinberg equilibrium were excluded, the pooled ORs for all genetic models for TaqI were shifted, yet the results remained null[23,24].
DISCUSSION
This study was undertaken to assess whether VDR polymorphisms in both the 5’ (Cdx-2 and FokI) and 3’ (BsmI, ApaI and TaqI) regions of the VDR gene are associated with colorectal cancer risk. A total of 38 reports had previously evaluated a possible genetic association, and only 23 of these were eligible for this study based on the selection criteria employed. The pooled ORs (95% CI) for these studies were identical according to both fixed- and random-effects models. Moreover, only the polymorphic variant, BsmI, was found to be associated with increased risk for colorectal cancer. The meta-analysis performed also showed that the BsmI B genotype was related to a significant decrease in overall risk for colorectal cancer, with the co-dominant BB and the BB + Bb vs bb dominant model exhibiting 0.87- and 0.92-fold increases in the risk for disease, respectively. The BB vs Bb + bb recessive model also had a 90% decreased risk for colorectal cancer.
In addition, subgroup analyses by anatomical site identified the BsmI polymorphism to be significantly associated with colon cancer, and not rectal cancer. Furthermore, compared with the bb or Bb + bb genotypes, the BB genotype was associated with a 90% decrease in the risk for colon cancer. However, it remains unclear why the BsmI site is related to the risk of colon cancer, and not rectal cancer. It is possible that the difference in epithelial cells between the two cancers plays a role, with ciliated columnar epithelial cells being present in the lining of the colon, while squamous epithelial cells are present in the rectum. The role of the micro-environment may also be a contributing factor, since physical damage as a result of oxygen or poisonous food residues is more likely to influence the rectum than the colon. In addition, genetic factors may have a more important role in colon cancer than rectal cancer.
Based on the distribution differences observed between cases of colorectal cancer and controls, we hypothesized that the BsmI B allele might have a protective effect against tumorigenesis. Correspondingly, of the 12 relevant reports reviewed, 9 studies supported this hypothesis. In these studies, the populations analyzed were from Asia (n = 1), Europe (n = 4), and the United States (n = 4). In addition, Jenab et al[22] evaluated different Caucasian populations from 23 centers in Denmark, France, Greece, Germany, Italy, the Netherlands, Norway, Spain, Sweden, and the United Kingdom. In this study, the BB genotype, rather than the wild-type bb genotype, was associated with a reduced risk of colorectal cancer. The BsmI BB genotype was also found to be associated with a reduced risk of colorectal cancer among non-aspirin/NSAID users[42]. Thus, multiple lines of evidence support the hypothesis that the BsmI B allele mediates a protective effect against the development of cancers in the digestive tract, especially colon cancer.
The BsmI polymorphism is located in the 3’ UTR of the VDR gene, and does not alter the amino acid sequence of the VDR protein. Thus, for a single BsmI polymorphism, there is a low probability that it directly influences VDR function[9]. The BsmI polymorphism also does not affect mRNA or protein levels of VDR[20], or levels of 25(OH)2D3 or 125(OH)2D3[43]. However, the BsmI site does exhibit strong LD with other VDR polymorphisms, including eTru9I, ApaI, TaqI, and Poly(A) microsatellites. Based on these results, it appears that the BsmI polymorphism affects some type of biological function, and these could potentially include regulation of VDR transcription, translation, or RNA processing[9]. Other unidentified SNPs in the VDR gene, as well as SNPs in other genes such as CYP3A5[44], may also affect the function of the BsmI. Furthermore, patients carrying the BsmI allele have also been shown to have significantly higher levels of erbB-2 expression, suggesting other tumor-related molecules may also be involved in the function of the BsmI polymorphism[14].
Although the BsmI B allele has been associated with a protective effect, the frequency of this effect has been found to be lower in Asian populations than in Caucasian populations. However, this is inconsistent with the incidence of colorectal cancer identified in recent epidemiological data[2]. Moreover, in the meta-analysis performed in the present study, no significant association was found between VDR genotypes and the risk of colorectal cancer in group analyses (not shown). In combination, these results suggest that other factors may be involved. For example, environment, food, and lifestyle may play a more significant role, in combination with genetic factors, in the occurrence and development of colorectal cancer than previously thought, which would potentially account for the inconsistent results obtained from previous studies.
COMMENTS
Background
Colorectal cancer is one of the most common cancers worldwide, and its incidence is increasing with each year. However, the underlying etiology of colorectal cancer remains unclear. Several epidemiologic studies have reported that 1, 25(OH)2D3 can reduce the risk of colorectal cancer, and thus, vitamin D receptor (VDR), a crucial mediator of the cellular effects of 1, 25(OH)2D3, may play an important role in the occurrence and development of colorectal cancer.
Research frontiers
Recently, several polymorphic variants of the VDR gene have been reported to be associated with the risk of colorectal cancer. However, the published findings remain inconsistent. In this study, the authors conducted a systematic meta-analysis to evaluate the evidence regarding this association.
Innovations and breakthroughs
In the present study, all relevant reports published between January 1990 and August 2010 were reviewed, with a focus on five well-characterized polymorphic variants of VDR: Cdx-2, FokI, BsmI, ApaI, and TaqI. In the meta-analysis performed, BsmI was found to be associated with colorectal cancer, while the Cdx-2, FokI, ApaI, and TaqI sites did not exhibit any significant association. Moreover, the BsmI ‘B’ genotype was associated with a significant decrease in the risk of colorectal cancer, especially colon cancer. Based on these results, it is hypothesized that BsmI may mediate a protective effect on tumorigenesis.
Applications
The results of this meta-analysis have the potential to partly explain the genetics that influence the pathogenesis of colorectal cancer. This study also helps provide a basis for clinical diagnosis and methods for early intervention.
Peer review
The authors performed a systematic meta-analysis of population-based studies to investigate the association between VDR polymorphisms and colorectal cancer risk. The authors found a BsmI site in the 3’ UTR of the VDR gene to be associated with colon cancer risk, and then analyzed the underlying mechanism. The results are interesting and may help explain the genetic mechanism of colorectal carcinogenesis.
Footnotes
Supported by Zhejiang provincial top key discipline in surgery
Peer reviewer: Mark S Pearce, Institute of Health and Society, Newcastle University, Sir James Spence Institute, Royal Victoria Infirmary, Newcastle upon Tyne NE1 4LP, United Kingdom
S- Editor Gou SX L- Editor A E- Editor Zheng XM
References
- 1.World Health Organization. Cancer. Fact sheet N°297. February. 2012. Available from: http://www.who.int/mediacentre/factsheets/fs297/en. [Google Scholar]
- 2.American Cancer Society. Cancer Facts and Figures. 2009. Available from: http://www.cancer.org/Research/CancerFactsFigures/index. [Google Scholar]
- 3.Garland CF, Garland FC. Do sunlight and vitamin D reduce the likelihood of colon cancer? Int J Epidemiol. 1980;9:227–231. doi: 10.1093/ije/9.3.227. [DOI] [PubMed] [Google Scholar]
- 4.Garland CF, Garland FC, Gorham ED. Calcium and vitamin D. Their potential roles in colon and breast cancer prevention. Ann N Y Acad Sci. 1999;889:107–119. doi: 10.1111/j.1749-6632.1999.tb08728.x. [DOI] [PubMed] [Google Scholar]
- 5.Vandewalle B, Adenis A, Hornez L, Revillion F, Lefebvre J. 1,25-dihydroxyvitamin D3 receptors in normal and malignant human colorectal tissues. Cancer Lett. 1994;86:67–73. doi: 10.1016/0304-3835(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 6.Thomas MG, Tebbutt S, Williamson RC. Vitamin D and its metabolites inhibit cell proliferation in human rectal mucosa and a colon cancer cell line. Gut. 1992;33:1660–1663. doi: 10.1136/gut.33.12.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt PR. Dairy foods and prevention of colon cancer: human studies. J Am Coll Nutr. 1999;18:379S–391S. doi: 10.1080/07315724.1999.10718902. [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto K, Kesterson RA, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S, Inoue Y, Morita K, Takeda E, Pike JW. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol. 1997;11:1165–1179. doi: 10.1210/mend.11.8.9951. [DOI] [PubMed] [Google Scholar]
- 9.Whitfield GK, Remus LS, Jurutka PW, Zitzer H, Oza AK, Dang HT, Haussler CA, Galligan MA, Thatcher ML, Encinas Dominguez C, et al. Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol Cell Endocrinol. 2001;177:145–159. doi: 10.1016/s0303-7207(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 10.Bai Y, Yu Y, Yu B, Ge J, Ji J, Lu H, Wei J, Weng Z, Tao Z, Lu J. Association of vitamin D receptor polymorphisms with the risk of prostate cancer in the Han population of Southern China. BMC Med Genet. 2009;10:125. doi: 10.1186/1471-2350-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingles SA, Coetzee GA, Ross RK, Henderson BE, Kolonel LN, Crocitto L, Wang W, Haile RW. Association of prostate cancer with vitamin D receptor haplotypes in African-Americans. Cancer Res. 1998;58:1620–1623. [PubMed] [Google Scholar]
- 12.Flügge J, Krusekopf S, Goldammer M, Osswald E, Terhalle W, Malzahn U, Roots I. Vitamin D receptor haplotypes protect against development of colorectal cancer. Eur J Clin Pharmacol. 2007;63:997–1005. doi: 10.1007/s00228-007-0367-4. [DOI] [PubMed] [Google Scholar]
- 13.Kim HS, Newcomb PA, Ulrich CM, Keener CL, Bigler J, Farin FM, Bostick RM, Potter JD. Vitamin D receptor polymorphism and the risk of colorectal adenomas: evidence of interaction with dietary vitamin D and calcium. Cancer Epidemiol Biomarkers Prev. 2001;10:869–874. [PubMed] [Google Scholar]
- 14.Speer G, Cseh K, Winkler G, Takács I, Barna I, Nagy Z, Lakatos P. Oestrogen and vitamin D receptor (VDR) genotypes and the expression of ErbB-2 and EGF receptor in human rectal cancers. Eur J Cancer. 2001;37:1463–1468. doi: 10.1016/s0959-8049(01)00139-3. [DOI] [PubMed] [Google Scholar]
- 15.Slattery ML, Murtaugh M, Caan B, Ma KN, Wolff R, Samowitz W. Associations between BMI, energy intake, energy expenditure, VDR genotype and colon and rectal cancers (United States) Cancer Causes Control. 2004;15:863–872. doi: 10.1007/s10552-004-1048-6. [DOI] [PubMed] [Google Scholar]
- 16.Park K, Woo M, Nam J, Kim JC. Start codon polymorphisms in the vitamin D receptor and colorectal cancer risk. Cancer Lett. 2006;237:199–206. doi: 10.1016/j.canlet.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 17.Hubner RA, Muir KR, Liu JF, Logan RF, Grainge MJ, Houlston RS. Dairy products, polymorphisms in the vitamin D receptor gene and colorectal adenoma recurrence. Int J Cancer. 2008;123:586–593. doi: 10.1002/ijc.23536. [DOI] [PubMed] [Google Scholar]
- 18.Slattery ML, Wolff RK, Herrick JS, Caan BJ, Potter JD. IL6 genotypes and colon and rectal cancer. Cancer Causes Control. 2007;18:1095–1105. doi: 10.1007/s10552-007-9049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadiyska T, Yakulov T, Kaneva R, Nedin D, Alexandrova A, Gegova A, Savov A, Mitev V, Kremensky I. Vitamin D and estrogen receptor gene polymorphisms and the risk of colorectal cancer in Bulgaria. Int J Colorectal Dis. 2007;22:395–400. doi: 10.1007/s00384-006-0163-0. [DOI] [PubMed] [Google Scholar]
- 20.Parisi E, Reñé JM, Cardús A, Valcheva P, Piñol-Felis C, Valdivielso JM, Fernández E. Vitamin D receptor levels in colorectal cancer. Possible role of BsmI polymorphism. J Steroid Biochem Mol Biol. 2008;111:87–90. doi: 10.1016/j.jsbmb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Theodoratou E, Farrington SM, Tenesa A, McNeill G, Cetnarskyj R, Barnetson RA, Porteous ME, Dunlop MG, Campbell H. Modification of the inverse association between dietary vitamin D intake and colorectal cancer risk by a FokI variant supports a chemoprotective action of Vitamin D intake mediated through VDR binding. Int J Cancer. 2008;123:2170–2179. doi: 10.1002/ijc.23769. [DOI] [PubMed] [Google Scholar]
- 22.Jenab M, McKay J, Bueno-de-Mesquita HB, van Duijnhoven FJ, Ferrari P, Slimani N, Jansen EH, Pischon T, Rinaldi S, Tjønneland A, et al. Vitamin D receptor and calcium sensing receptor polymorphisms and the risk of colorectal cancer in European populations. Cancer Epidemiol Biomarkers Prev. 2009;18:2485–2491. doi: 10.1158/1055-9965.EPI-09-0319. [DOI] [PubMed] [Google Scholar]
- 23.Slatter ML, Yakumo K, Hoffman M, Neuhausen S. Variants of the VDR gene and risk of colon cancer (United States) Cancer Causes Control. 2001;12:359–364. doi: 10.1023/a:1011280518278. [DOI] [PubMed] [Google Scholar]
- 24.Ochs-Balcom HM, Cicek MS, Thompson CL, Tucker TC, Elston RC, J Plummer S, Casey G, Li L. Association of vitamin D receptor gene variants, adiposity and colon cancer. Carcinogenesis. 2008;29:1788–1793. doi: 10.1093/carcin/bgn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slattery ML, Herrick J, Wolff RK, Caan BJ, Potter JD, Sweeney C. CDX2 VDR polymorphism and colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2752–2755. doi: 10.1158/1055-9965.EPI-07-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingles SA, Wang J, Coetzee GA, Lee ER, Frankl HD, Haile RW. Vitamin D receptor polymorphisms and risk of colorectal adenomas (United States) Cancer Causes Control. 2001;12:607–614. doi: 10.1023/a:1011292002475. [DOI] [PubMed] [Google Scholar]
- 27.Peters U, McGlynn KA, Chatterjee N, Gunter E, Garcia-Closas M, Rothman N, Sinha R. Vitamin D, calcium, and vitamin D receptor polymorphism in colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2001;10:1267–1274. [PubMed] [Google Scholar]
- 28.Wong HL, Seow A, Arakawa K, Lee HP, Yu MC, Ingles SA. Vitamin D receptor start codon polymorphism and colorectal cancer risk: effect modification by dietary calcium and fat in Singapore Chinese. Carcinogenesis. 2003;24:1091–1095. doi: 10.1093/carcin/bgg059. [DOI] [PubMed] [Google Scholar]
- 29.Grau MV, Baron JA, Sandler RS, Haile RW, Beach ML, Church TR, Heber D. Vitamin D, calcium supplementation, and colorectal adenomas: results of a randomized trial. J Natl Cancer Inst. 2003;95:1765–1771. doi: 10.1093/jnci/djg110. [DOI] [PubMed] [Google Scholar]
- 30.Murtaugh MA, Sweeney C, Ma KN, Potter JD, Caan BJ, Wolff RK, Slattery ML. Vitamin D receptor gene polymorphisms, dietary promotion of insulin resistance, and colon and rectal cancer. Nutr Cancer. 2006;55:35–43. doi: 10.1207/s15327914nc5501_5. [DOI] [PubMed] [Google Scholar]
- 31.Yaylim-Eraltan I, Arzu Ergen H, Arikan S, Okay E, Oztürk O, Bayrak S, Isbir T. Investigation of the VDR gene polymorphisms association with susceptibility to colorectal cancer. Cell Biochem Funct. 2007;25:731–737. doi: 10.1002/cbf.1386. [DOI] [PubMed] [Google Scholar]
- 32.Grünhage F, Jungck M, Lamberti C, Berg C, Becker U, Schulte-Witte H, Plassmann D, Rahner N, Aretz S, Friedrichs N, et al. Association of familial colorectal cancer with variants in the E-cadherin (CDH1) and cyclin D1 (CCND1) genes. Int J Colorectal Dis. 2008;23:147–154. doi: 10.1007/s00384-007-0388-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Li BQ, Zhou HH. [Polymorphism of vitamin D receptor Fok I and colorectal cancer risk in Chinese] Zhongnan Daxue Xuebao Yixueban. 2008;33:399–403. [PubMed] [Google Scholar]
- 34.Mahmoudi T, Mohebbi SR, Pourhoseingholi MA, Fatemi SR, Zali MR. Vitamin D receptor gene ApaI polymorphism is associated with susceptibility to colorectal cancer. Dig Dis Sci. 2010;55:2008–2013. doi: 10.1007/s10620-009-0989-8. [DOI] [PubMed] [Google Scholar]
- 35.Peters U, Hayes RB, Chatterjee N, Shao W, Schoen RE, Pinsky P, Hollis BW, McGlynn KA. Circulating vitamin D metabolites, polymorphism in vitamin D receptor, and colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev. 2004;13:546–552. [PubMed] [Google Scholar]
- 36.Egger M, Juni P, Bartlett C, Holenstein F, Sterne J. How important are comprehensive literature searches and the assessment of trial quality in systematic reviews? Empirical study. Health Technol Assess. 2003;7:1–76. [PubMed] [Google Scholar]
- 37.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 38.Little J, Bradley L, Bray MS, Clyne M, Dorman J, Ellsworth DL, Hanson J, Khoury M, Lau J, O’Brien TR, et al. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156:300–310. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 39.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Oxman AD, Guyatt GH. The science of reviewing research. Ann N Y Acad Sci. 1993;703:125–133; discussion 133-134. doi: 10.1111/j.1749-6632.1993.tb26342.x. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Li Y, Gao LB, Wang YY, Zhou B, Lv ML, Lu HM, Zhang L. Vitamin D receptor gene polymorphisms and the risk of colorectal cancer in a Chinese population. Dig Dis Sci. 2009;54:634–639. doi: 10.1007/s10620-008-0375-y. [DOI] [PubMed] [Google Scholar]
- 42.Slattery ML, Samowitz W, Hoffman M, Ma KN, Levin TR, Neuhausen S. Aspirin, NSAIDs, and colorectal cancer: possible involvement in an insulin-related pathway. Cancer Epidemiol Biomarkers Prev. 2004;13:538–545. [PubMed] [Google Scholar]
- 43.Holt PR, Arber N, Halmos B, Forde K, Kissileff H, McGlynn KA, Moss SF, Kurihara N, Fan K, Yang K, et al. Colonic epithelial cell proliferation decreases with increasing levels of serum 25-hydroxy vitamin D. Cancer Epidemiol Biomarkers Prev. 2002;11:113–119. [PubMed] [Google Scholar]
- 44.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]