Abstract
Context
Evidence regarding the impact of minority, or low frequency, HIV-1 drug-resistant variants on the effectiveness of first-line antiretroviral treatment (ART) is conflicting.
Objective
To evaluate the association of pre-existing HIV-1 minority drug-resistant variants with risk of first-line non-nucleoside reverse transcriptase inhibitor (NNRTI)-based antiretroviral virologic failure.
Data Sources
We searched published and unpublished studies in MEDLINE (1966 through December, 2010), EMBASE (1974 through December, 2010), conference abstracts, and article references. Authors of all studies were contacted for detailed laboratory, ART, and adherence data.
Study Selection and Data Abstraction
Studies involving ART-naive participants initiating NNRTI-based regimens were included. Participants were included if all drugs in their ART regimen were fully active by standard HIV population sequencing. Cox proportional hazard models using pooled patient-level data were used to estimate the risk of virologic failure based on a Prentice weighted case-cohort analysis stratified by study.
Data Synthesis
Individual data from 10 studies and 985 participants were available for the primary analysis. Minority HIV-1 drug resistance mutations were associated with an increased risk of virologic failure (HR 2.3 [95% CI, 1.7–3.3], P<0.001) after controlling for medication adherence, ethnicity, baseline CD4 cell count and plasma HIV-1 RNA levels. The increased risk of virologic failure was most strongly associated with minority variants resistant to NNRTIs (HR 2.6 [95% CI, 1.9–3.5], P<0.001). Among participants from the cohort studies, 35% of those with detectable minority variants experienced virologic failure as compared to 15% of those without minority variants. The presence of minority variants was associated with 2.5–3 times the risk of virologic failure at either ≥95% or <95% overall medication adherence. A dose-dependent increased risk of virologic failure was found in participants with a higher proportion or quantity of drug-resistant variants.
Conclusion
In this pooled analysis, minority HIV-1 resistance mutations, particularly involving NNRTI-resistance, were significantly associated with a dose-dependent increased risk of virologic failure with first-line ART.
Keywords: HIV-1 minority drug resistance mutations, minority drug-resistant variants, virologic failure, NNRTI-based antiretroviral treatment
INTRODUCTION
Genotypic tests for HIV-1 drug resistance employ PCR amplification and population sequencing techniques that detect resistance-associated mutations present at ≥15–25% of the viral population1, 2. Using these traditional assays, the prevalence of transmitted drug resistance mutations is estimated to be between 8% and 16% among HIV-1 infected persons in North America and Europe3, 4. These assays fail to detect the presence of low-frequency resistance mutations present as minority variants within the population of HIV-1 quasispecies in an infected individual. A number of ultra-sensitive assays, including allele-specific PCR and deep sequencing, can detect mutations at a far lower frequency than standard population sequencing5–7. Presence of these minority, or low frequency, variants may adversely affect the response to antiretroviral treatment (ART), but their clinical significance continues to be the subject of considerable debate and uncertainty.
Non-nucleoside reverse transcriptase inhibitor (NNRTI)-based regimens are the most popular first-line HIV treatment regimen both in the United States and world-wide8, 9. Although success rates are high, further improvements would avoid the costs associated with treatment failure and accumulating additional drug resistance mutations. A number of studies have been undertaken to evaluate the impact of baseline NNRTI and nucleoside reverse transcriptase inhibitor (NRTI) resistance mutations on rates of initial ART treatment failure. Results of these studies have been mixed, with some showing that minority drug-resistant variants significantly increase the risk of treatment failure and others showing no significant effect. In contrast, a small number of studies that evaluated the importance of minority resistance mutations on integrase and protease inhibitor-based treatment have generally failed to find a significant association with increased risk of treatment failure10–15.
We performed a systematic review of the literature and performed a pooled analysis to examine the relationship between baseline minority HIV-1 drug resistance mutations and the risk of initial NNRTI-based virologic failure.
METHODS
Data Sources, Study and Participant Selection
A computerized literature search was conducted of Pubmed (1966 through December, 2010) and EMBASE (1974 through December, 2010) using the search terms: “HIV Infections”[mesh] OR “HIV”[mesh] OR “HIV”[tiab] OR “Acquired Immune Deficiency Syndrome Virus”[tiab] OR “Human Immunodeficiency Virus”[tiab] OR “Human Immunodeficiency Viruses”[tiab] OR “AIDS virus”[tiab] AND (“minor”[tiab] OR “minority”[tiab] OR “low abundance”[tiab] OR “low frequency” OR “minorities”[tiab]) AND (“variants”[tiab] OR “variant”[tiab] OR “mutation”[tiab] OR “mutations”[tiab] OR “mutant”[tiab] OR “mutants”[tiab] OR “quasispecies”[tiab]) AND (Drug Resistance, Viral[mesh] OR Treatment Failure[mesh] OR “treatment failure”[tiab] OR “resistance”[tiab] OR “resistant”[tiab]). In addition, experts in the field were contacted, reference lists were reviewed, and abstracts from the International HIV Drug Resistance Workshop and the Conference on Retroviruses and Opportunistic Infections (2007–2010) were searched for additional studies. The inclusion criteria included cohort or case-control studies that evaluated the impact of minority HIV-1 NRTI- and NNRTI-resistance mutations on the rate of virologic failure in treatment-naïve adults receiving an initial NNRTI-based antiretroviral regimen. Studies were excluded if they had no comparison group, did not have treatment outcome data, focused solely on primary infection, or were cross-sectional studies. To assess evidence of publication bias, a funnel plot using study-specific definitions of minority variants and virologic failure was created (RevMan 5.0, Copenhagen). Heterogeneity of the minority variant effect across studies was evaluated with a test of interaction between the presence of minority variants and study.
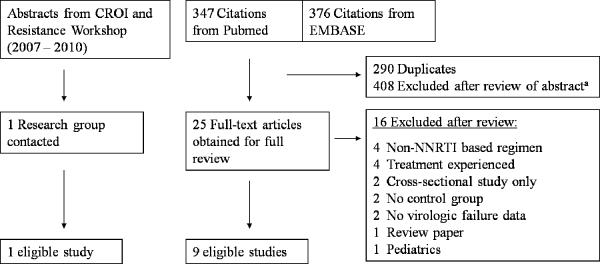
Of the 347 citations obtained from Pubmed and 376 citations from EMBASE, 25 full-text articles were identified as potentially relevant and screened for inclusion (Figure 1). Of these, 16 were excluded because they lacked treatment outcome data (e.g., cross-sectional study only), had no comparison group (i.e., small case series), or on the basis of the study population (e.g., not on NNRTI-based regimen). In addition, one previously unpublished study was identified that matched the inclusion and exclusion criteria. The literature search and review of full-text articles were independently performed by two of the authors (J.Z.L. and R.P.). Investigators from all 10 studies agreed to provide patient-level data (e.g., demographic, laboratory, minority resistance, and adherence data) and to participate in this pooled analysis (Table 1)14–23. Individual patients were excluded with any pre-treatment evidence of reduced NRTI or NNRTI drug susceptibility by standard genotyping based on the Stanford Resistance DB mutation scoring system (score ≥10 for any antiretroviral medication).
Figure 1.
Study Selection Flow Diagram
aCommon reasons for exclusion include: does not involve low-frequency resistance variants, review article, epidemiological study, and treatment experienced patient population only.
Table 1.
Baseline Characteristics of Studies Included in the Pooled-Analysis
| Characteristic | Peuchant15 2008 | Simen14 2009 | Balduin16 2009 | Jakobsen17 2010 | Metzner18 2010 | Goodman19 2011 | Paredes20 2010 | Johnson21 2008 | Geretti22 2009 | Metzner23 2009 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Design | Cohort | Cohort | Cohort | Cohort | Cohort | Cohort | Case cohort | Case control | Case control | Case control | |
| Virologic Failure (N) | 2 | 45 | 7 | 1 | 1 | 44 | 150 | 52 | 14 | 3 | 315 |
| Total Participants (N) | 13 | 70 | 54 | 20 | 56 | 423 | 280 | 240 | 89 | 18 | 1263 |
| Age, mean (yr) | 38±16.8 | 37±8.8 | 41±11.7 | 43±12.3 | 42±11.1 | 38±9.4 | 37±9.6 | 37±9.5 | 38±8.5 | 43±9.5 | 38±9.8 |
| Male Sex % (n/N) | 92 (12/13) | 80 (56/70) | 76 (41/54) | 95 (19/20) | 80 (45/56) | 86 (365/423) | 81 (227/280) | 82 (196/240) | 88 (78/89) | 72 (13/18) | 83 (1052/1263) |
| Ethnicity % (n/N) | |||||||||||
| White | 92 (12/13) | 23 (16/70) | 75 (39/52) | 60 (253/422) | 39 (110/279) | 55 (132/240) | 88 (78/89) | 82 (14/17) | 55 (654/1182) | ||
| Black | 8 (1/13) | 54 (38/70) | 21 (11/52) | 22 (94/422) | 39 (110/279) | 25 (61/240) | 11 (10/89) | 18 (3/17) | 28 (328/1182) | ||
| Hispanic | 0 (0/13) | 20 (14/70) | 0 (0/52) | 14 (61/422) | 19 (54/279) | 18 (42/240) | 0 (0/89) | 0 (0/17) | 14 (171/1182) | ||
| Others | 0 (0/13) | 3 (2/70) | 4 (2/52) | 3 (14/422) | 2 (5/279) | 2 (5/240) | 1 (1/89) | 0 (0/17) | 2 (29/1182) | ||
| CD4+ Count, median (IQR) | 426 (303–522) | 247 (38–344) | 251 (196–326) | 200 (48–278) | 279 (191–368) | 227 (127–319) | 202 (69–331) | 243 (145–327) | 222 (126–299) | 222 (59–249) | 229 (125–324) |
| log10 HIV RNA cp/ml, median (IQR) | 4.4 (4.2–5.3) | 5.3 (4.9–5.8) | 4.7 (4.0–4.9) | 5.1 (4.6–5.8) | 4.9 (4.5–5.3) | 5.0 (4.6–5.4) | 4.8 (4.4–5.4) | 5.1 (4.5–5.5) | 5.2 (4.9–5.5) | 5.4 (4.9–5.9) | 5.0 (4.6–5.4) |
Abbreviations: IQR, Inter-quartile range
Minority Variant and Adherence Information, End Points, and Data Compilation
The most commonly examined mutations across studies included K103N, Y181C, M184V, and K65R (Table 2). For each study, patients with K103N or Y181C minority drug resistance mutations were classified as harboring a minority NNRTI-resistant HIV-1 variant; those with M184V or K65R were classified as having a minority NRTI-resistant variant. In one study, three patients were found to have one of three additional minority mutations associated with NNRTI resistance (G190A, K101E, and P225H) and were included in the analysis as harboring a minority NNRTI-resistant variant14. Minority variant copy numbers were calculated by multiplying the percentage of the minority variant by the plasma HIV-1 RNA level at the time of minority variant measurement. In the analysis of minority variant percentage or copy number, if multiple resistance mutations were present, the minority variant with the highest percentage or copy number was used.
Table 2.
Characteristics of Minority Variants by Study
| Characteristic | Peuchant15 2008 | Simen14 2009 | Balduin16 2009 | Jakobsen17 2010 | Metzner18 2010 | Goodman19 2011 | Paredes20 2010 | Johnson21 2008 | Geretti22 2009 | Metzner23 2009 | Total for Cohort Studiesd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Design | Cohort | Cohort | Cohort | Cohort | Cohort | Cohort | Case cohort | Case control | Case control | Case control | |
| Method of Detectiona | AS-PCR | 454 | AS-PCR | SNaPshot | AS-PCR | AS-PCR | AS-PCR | AS-PCR | AS-PCR | AS-PCR | |
| Limit of Detection (% of viral population) | |||||||||||
| K103N | 0.4 | 1.0 | 0.2 | 2.0 | 0.01 | 0.5 | 0.003 | 0.9 | 0.9 | 0.01 | |
| Y181C | 1.0 | 2.0 | 0.03 | 1.0 | 1.0 | 0.2 | |||||
| M184V | 0.3 | 1.0 | 2.0 | 0.2 | 0.5 | 0.5 | 0.2 | ||||
| K65R | 1.0 | 2.0 | 0.4 | 0.3 | 0.4 | ||||||
| Other NNRTIb | 1.0 | 2.0 | 0.9 | ||||||||
| Participants with MVs and VF / MVsc | |||||||||||
| K103N | 1 / 3 | 1 / 1 | 3 / 13 | 1 / 2 | 0 / 2 | 5 / 14 | 27 / 39 | 1 / 1 | 3 / 3 | 1 / 1 | 17 / 53 |
| Y181C | 0 / 0 | 0 / 0 | 83 / 123 | 1 / 1 | 0 / 0 | 1 / 1 | 25 / 65 | ||||
| M184V | 0 / 3 | 0 / 0 | 1 / 1 | 0 / 3 | 1 / 1 | 0 / 0 | 2 / 2 | 1 / 7 | |||
| K65R | 0 / 0 | 0 / 0 | 0 / 2 | 0 / 0 | 0 / 0 | 0 / 2 | |||||
| Other NNRTIb | 3 / 3 | 0 / 0 | 0 / 0 | 3 / 3 | |||||||
Abbreviations: MVs, Minority Variants; VF, Virologic Failure
AS-PCR, allele-specific PCR; 454, 454 ultradeep pyrosequencing; HIV-SNaPshot is named for a multiplex primer-extension assay for detecting HIV minority variants17.
Other NNRTI minority variants evaluated include G190A (Geretti), G190A/S/E (Jakobsen), and multiple (Simen). Three patients in Simen et al. were found to have other NNRTI minority mutations (G190A 2.1%, K101E 3%, and P225H 3.4%) and were included in the analysis as having an NNRTI minority variant.
Only cases (participants with virologic failure) from the case control studies were used in the primary Cox proportional hazard model and are described here. Data for Paredes et al. includes entire case cohort study (participants in random subcohort and additional virologic failures).
Totals only reflect participants of the cohort studies including the random subcohort from Paredes et al.'s case cohort analysis of A5095.
Data on ART adherence were available from three studies, which in aggregate contributed 78% of the patients used for the primary analysis. ART adherence measurements were based on pill counts19, 4-day self-report20, or 7-day self-report14 and were averaged over the course of the study until the time of virologic failure or censoring. The lower of the NRTI and NNRTI adherence measurements was used as the overall medication adherence rate. Overall adherence was classified as high if the adherence rate was ≥95%.
The definition of virologic failure was standardized for all patients to a plasma HIV-1 RNA level of ≥200 copies/ml at two consecutive time points at least 16 weeks after treatment initiation. Patients were also counted as virologic failures if the last available HIV-1 RNA level was ≥200 copies/ml without a confirmatory measurement.
Statistical Analysis
Cox proportional hazard models stratified by study were used to estimate the risk of virologic failure across multiple factors: with and without minority variants (overall, NNRTI, NRTI), ART regimens (efavirenz versus nevirapine), adherence classifications, and minority variant percentage and copy number categories; tests of interactions were evaluated as appropriate. To avoid bias induced by targeted sampling, non-randomly sampled controls (non-failures) were excluded and non-randomly sampled cases (virologic failures) contributed to the Cox proportional hazard models only at their time of failure. The resulting analysis framework may be considered analogous to a Prentice weighted analysis for a case-cohort study24, 25. For the same reason, Kaplan-Meier failure time distributions were estimated using only patients from randomly sampled cohorts (including analysis of randomized controlled trials and the random subcohort analysis of A5095)14–20. To assess the robustness of the findings, sensitivity analyses were performed using cohort studies14–17, 19, 20, 26, largest cohort studies14, 19, 20, and excluding the study contributing the largest number of participants20. Stratified Wilcoxon rank sum tests were used to compare the distributions of HIV-1 RNA levels and CD4 cell counts between patients with and without minority variants or virologic failure27. The Cochran-Mantel-Haenszel test was used to compare the ethnic distributions between patients with and without minority variants. Only patients from the cohort studies were included in group comparisons by minority variants; the entire dataset was used to compare participants with or without virologic failure with the exception of case-control studies that matched controls based on viral load or CD4 cell count22, 23. The number needed to screen was calculated based on the prevalence of K103N and/or Y181C minority variants detected using the most sensitive resistance test20 and overall virologic failure rates for patients with and without minority NNRTI-resistant variants.
Analysis of minority variant copy numbers excluded three studies using assays that could not provide a percentage17, 21, 22. For the minority variant 1% threshold analysis, one study was excluded due to a limit of detection of 2% for the assay17 and only NNRTI minority variants were evaluated for two studies due to incompatible limits of detection for the NRTI minority variants21, 22. Four studies were excluded from the minority variant analysis using a 0.5% threshold due to higher limits of minority variant detection14, 17, 21, 22. Statistical analysis was performed using SAS 9.2 (SAS Institute, Cary, NC) and PASW Statistics 18 (IBM SPSS, Chicago, IL). Findings with a P-value <0.05 were considered to be statistically significant.
RESULTS
Systematic Review and Baseline Characteristics
In total, 10 studies met the inclusion and exclusion criteria14–17, 19–23, 26. The qualifying studies included six cohort studies14–17, 19, 26, three case-control studies21–23, and one case-cohort study20 (Table 1). Of 1263 patients, 985 were included in the primary Cox proportional hazard analysis. At baseline, the average age of the entire study population was 38 years and 83% were men. The median CD4 cell count was 229 [IQR 125–324] cells/mm3 and median plasma HIV-1 RNA level was 5.0 [IQR 4.6–5.4] log10 copies/mL. All studies evaluated the presence of K103N (Table 2). Other commonly evaluated minority variants included Y181C (N=435) and the NRTI resistance mutations M184V (N=228) and K65R (N=163). Most studies used allele-specific real-time PCR to detect minority variants; one study used the HIV-SNaPshot assay17 and one used deep sequencing (Roche/454 Life Sciences)14. The study that used deep sequencing detected additional minority NNRTI-resistant HIV-1 variants (G190A, K101E, and P225H) in three patients, who were also included in the analysis. The lower limit of detection of minority variants differed widely between assays with an upper range of 2% for the HIV-SNaPshot assay and a lower range of 0.003% for one of the allele-specific PCR assays (Table 2). The assays for three studies were unable to quantify the percentage of minority variants present17, 21, 22. No significant heterogeneity was seen among studies (P=0.77), but there was evidence of limited publication bias (Figure S1).
Minority drug-resistant variants were found in 187 participants including 14% (117/808) of patients in the cohort studies14–17, 19, 20, 26. Patients with minority variants had a baseline median HIV-1 RNA level of 4.79 [IQR 4.4–5.4] log10 copies/mL as compared to 4.95 [IQR 4.6–5.4] log10 copies/mL for those without detectable minority variants (P=0.49). Patients with minority resistance variants had lower CD4 cell counts than those in whom these variants were not detected (median 208 [IQR 50–330] versus 234 [IQR 134–329] cells/mm3, respectively; P=0.03). Patients with or without virologic failure had no significant differences in either baseline plasma HIV-1 RNA levels (median 5.0 [IQR 4.6–5.5] versus 5.0 [IQR 4.6–5.4] log10 copies/mL, respectively; P=0.90) or CD4 cell counts (median 222 [IQR 87–325] versus 235 [135–324] cells/mm3, respectively; P=0.47). Among participants in the cohort studies, the proportion of those harboring HIV drug-resistant minority variants did not differ significantly by ethnicity (P=0.13).
Minority Drug-Resistant HIV-1 Variants and Increased Risk of Virologic Failure
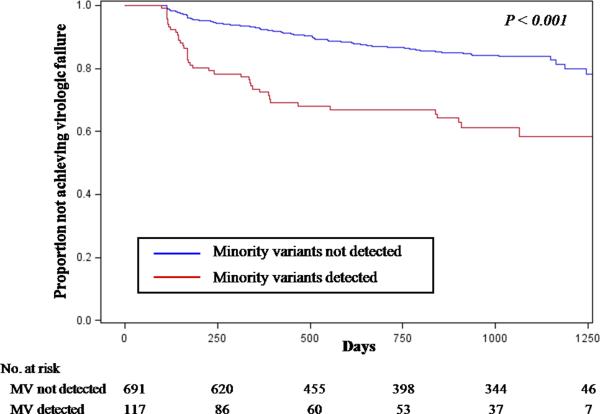
The presence of any minority NNRTI- or NRTI-resistant variant was associated with an increased risk of virologic failure (Hazard Ratio [HR] 2.6 [95% CI, 1.9–3.5], P<0.001). This result was still apparent when the study contributing the largest number of patients with virologic failure20 was excluded (HR 3.6 [95% CI 1.9–6.9], P<0.001) and when the analysis was restricted to include only participants from cohort studies (HR 3.7 [95% CI 2.3–5.9], P<0.001; Figure 2). Specifically, among the 808 participants from cohort studies, 35% of those with detectable minority variants experienced virologic failure as compared to 15% of those without minority variants. A sensitivity analysis that included only the largest cohort studies14, 19, 20 gave similar results, with a virologic failure rate of 40% in those with minority variants versus 17% in those without (HR 3.9 [95% CI 2.3–6.4], P<0.001, N=665).
Figure 2.
Kaplan-Meier Curves for the Proportion of Patients without Virologic Failure by the Presence of Minority HIV-1 Drug-Resistant Variants
Abbreviations: MV, minority variants
Both NNRTI- and NRTI-resistant minority variants are included in this analysis. To avoid bias induced by targeted sampling in case-control studies, Kaplan-Meier failure time distributions were estimated using only date from cohort studies14–20. Kaplan-Meier curves only shown up to 1,250 days due to the small sample sizes thereafter. P-value comparison by Cox proportional hazard analysis. Median follow-up time is 31 months [IQR 12–34 months].
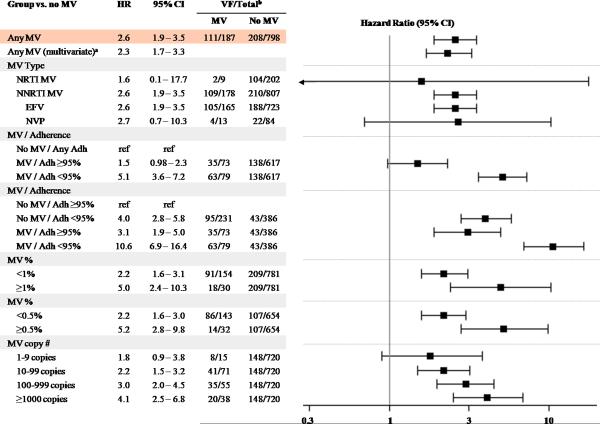
The increased risk of virologic failure was most strongly associated with minority NNRTI-resistant variants (HR 2.6 [95% CI, 1.9–3.5], P<0.001; Figure 3). The presence of only minority NRTI-resistant variants was not associated with an elevated risk of virologic failure (HR 1.6 [95% CI 0.1–17.7]), but only nine participants fell into this category. In participants with minority NNRTI-resistant variants, the overall failure rate among those in the cohort studies was 37% as compared to 15% in those without detectable minority variants (HR 3.8 [95% CI 2.4–6.1], P<0.001). No significant difference was found on the effect of minority NNRTI-resistant variants on the risk of virologic failure with efavirenz- versus nevirapine-based regimens (Interaction P=0.90; Figure 3). There was also no significant difference in the rate of virologic failure between participants with K103N compared to those with Y181C minority variants (HR 0.7 [95% CI 0.4–1.4], P=0.34) among the subset of patients in whom testing for both mutations were performed (N=432).
Figure 3.
Effect of Minority Variants and Antiretroviral Therapy Adherence on Virologic Failure
Abbreviations: MV, Minority Variants; VF, Virologic Failure
aMultivariate Cox regression analysis included adherence, ethnicity, baseline CD4 cell count, and HIV-1 RNA levels.
bNumber of participants with virologic failure / total participants categorized by those with and without detectable minority variants. Participant numbers include additional virologic failure cases from the case-control and case cohort studies20–23.
Cox proportional hazard rations shown are in comparison to those without minority variants unless otherwise noted. Three studies contributed to the adherence analysis14, 19, 20. Analysis of MV copy numbers excluded three studies using assays that could not provide a percentage17, 21, 22. For the MV 1% threshold analysis, one study was excluded due to a limit of detection of 2% for the assay17 and only NNRT1 MVs were evaluated for two studies due to incompatible limits of detection fon the NRTI MVs21, 22. Four studies were excluded from the 0.5% threshold analysis due to higher limits of MV detection14, 17, 21, 22.
Given the virologic failure rates for patients with and without minority NNRTI-resistant variants (37% and 15%, respectively over a median 31 month follow-up period) and using the most sensitive resistance test20, approximately 11 patients would need to be screened prior to initiating an NNRTI-based ART regimen to avoid one case of virologic failure.
Medication Adherence and Minority Variants
Participants with minority drug-resistant variants and ≥95% medication adherence had a significantly lower risk of virologic failure compared to those with minority variants and <95% adherence (HR 0.3 [95% CI, 0.2–0.4], P<0.001). Compared to all participants without minority variants, individuals with minority variants and <95% medication adherence had 5.1 times the risk of virologic failure (95% CI 3.6–7.2, P<0.001). Those with minority variants and ≥95% adherence had 1.5-times the risk of virologic failure (95% CI 0.98–2.3, P=0.06; Figure 3). When compared to participants with ≥95% adherence and no minority variants, both suboptimal adherence and the presence of minority variants were associated with similarly increased risks of virologic failure (HR 4.0 [95% CI 2.8–5.8], P<0.001 and HR 3.1 [95% CI 1.9–5.0], P<0.001, respectively; Figure 3). The combined presence of suboptimal medication adherence and minority drug-resistant variants resulted in a substantially increased risk of virologic failure (HR 10.6 [95% CI 6.9–16.4], P<0.001). Furthermore, within each adherence category, the presence of minority variants was associated with an increased risk of virologic failure (≥95% adherence HR 3.1 [95% CI 1.9–5.0], P<0.001; <95% adherence HR 2.7 [95% CI 1.8–3.8], P<0.001).
Dose-Dependent Association of Minority Drug-Resistant Variants with Increased Risk of Virologic Failure
To evaluate whether a threshold existed for the effect of minority drug-resistant variants, analyses were performed to explore the risk of virologic failure associated with different percentages or absolute numbers of minority drug-resistant variants. Compared to those without minority drug-resistant variants, an increased risk of virologic failure was found when minority drug-resistant variants were present at either <1% or ≥1% of the viral population (HR 2.2 [95% CI 1.6–3.1], P<0.001 and HR 5.0 [2.4–10.3], P<0.001, respectively; Figure 3). However, the presence of minority variants at ≥1% conferred a significantly higher risk of virologic failure as compared to minority variants present at <1% (HR 2.2 [1.0–4.9], P=0.048). Similar results were observed when the proportion of resistant variants in the virus population was stratified as <0.5% versus ≥0.5% (Figure 3, P=0.01 for comparison of <0.5% versus ≥0.5%). A dose-dependent effect on the risk of virologic failure was found when participants were categorized as having 0, 1–9, 10–99, 100–999, and ≥1000 copies of minority drug-resistant variants per mL of plasma (Figure 3). The effect on virologic failure was similar when the analysis was limited to only minority NNRTI-resistant variants (see supplemental data).
Multivariate Analysis
In a multivariate Cox proportional hazard model, the presence of a minority drug-resistant variant (HR 2.3 [95% CI 1.7–3.3], P<0.001; Figure 3), overall medication adherence (HR 0.86 per 5% higher adherence, [95% CI 0.83–0.88], P<0.001), and ethnicity were all significant independent predictors of virologic failure. Compared to whites, participants of black, Hispanic, and other ethnicities all had an increased risk of virologic failure (HR 2.8 [2.0–3.8], P<0.001; HR 2.1 [1.4–3.1], P<0.001; and HR 2.6 [1.0–6.5], P=0.045, respectively). Associations with baseline CD4 cell count and plasma HIV-1 RNA levels were not detected (P=0.59 and P=0.88, respectively).
Time to Virologic Suppression
The effect of minority drug-resistant variants on viral decay dynamics was evaluated using two studies with frequent plasma HIV-1 RNA determinations after ART initiation (N=581)19, 20. The proportion of participants who never reached a plasma HIV-1 RNA level ≤200 copies/mL was significantly higher in the group with minority drug-resistant variants compared to those without detectable minority variants (9% versus 1%, respectively; P<0.001). However, among participants who eventually became suppressed, there was no difference in the median number of days to virologic suppression (57 versus 57 days, respectively; Figure 4).
Figure 4.
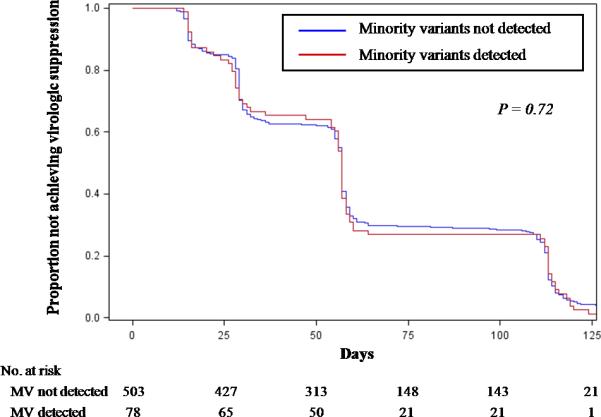
Time to HIV-1 RNA <200 copies/ml in Patients Achieving Virologic Suppression
Abbreviations: MV, minority variants
Two studies with frequent HIV-1 RNA monitoring19,20 were used to determine flie time to HIV-1 RNA <200 copies/ml among individuals who became virologically suppressed. P-value comparison by Cox proportional hazard analysis. Median time to virologic suppression was 57 days [IQR 28–112 days] for those with minority variants and 57 days [IQR 27–111 days] for those without.
DISCUSSION
In this pooled analysis, we found that the presence of minority HIV-1 drug resistant variants was associated with more than twice the risk of virologic failure in patients receiving an initial NNRTI-based ART regimen in an analysis that controlled for medication adherence, ethnicity, baseline CD4 cell count, and HIV-1 viral load. The presence of minority variants was associated with 2.5–3 times the risk of virologic failure at either high or low levels of medication adherence. The association of minority variants with virologic failure was dose-dependent and most prominent in those with NNRTI-resistance mutations.
Multiple factors contribute to the risk of ART failure. Adherence to antiretroviral therapy is a major predictor of viral suppression and disease progression28–30. In this analysis, we found that the risk of virologic failure associated with the presence of minority drug-resistant variants was similar to that conferred by suboptimal medication adherence. Patients with both minority drug-resistant variants and suboptimal medication adherence had a 10-fold risk of virologic failure when compared to those with wild-type virus and excellent adherence. However, optimal medication adherence did not completely compensate for the higher risk of virologic failure in the presence of minority drug-resistant HIV-1 variants.
Interestingly, ethnicity was found to be a significant predictor of virologic failure and in particular, white participants had a lower risk of virologic failure compared to black and Hispanic participants. This risk differential was not due to differing rates of minority variant detection. While some studies have shown no association of race or HIV-1 subtype with initial treatment response31, a secondary analysis of the A5095 trial uncovered an interaction between ethnicity and adherence, and found a greater effect of non-adherence on virologic failure in black participants32. It is interesting to note that the effect of ethnicity on virologic failure seen in our analysis was present even after adjusting for the level of medication adherence. The relationship between ethnicity and virologic failure most likely is mediated by factors such as socioeconomic status, drug and alcohol use, or other factors not accounted for here that may correlate with adherence and could contribute to residual confounding. Another potential explanation for these findings could be related to the recent report that cytochrome P450 polymorphisms affect NNRTI pharmacokinetics and treatment outcome in a race-specific manner33.
Minority drug-resistant variants detected by ultrasensitive assays could arise from a few sources. Those found at higher proportions may represent transmitted drug resistance that have been replaced by wild-type revertants over time34 or resulted from multivariant transmission35, 36, whereas mutations present at extremely low frequencies (much less than 1% of the viral population) could be due to de novo mutations resulting from errors introduced during viral replication37 or laboratory artifacts from reverse transcription and PCR amplification. The presence of spontaneously appearing minority drug-resistance mutations has been described in HIV samples collected in the pre-ART drug era7. It has been proposed that minority drug-resistant variants present at extremely low levels may not have a significant clinical impact. While we found a dose-dependent effect of minority drug-resistant variants on risk of virologic failure, this increased risk was significant even at very low minority variant frequencies (<0.5% and 10–99 copies/mL). A recent study reported a strong correlation between virologic failure and the presence of ≥2,000 copies/ml of K103N-containing HIV-1 whereas patients with <2,000 copies/ml of K103N did not show an increased risk of virologic failure19. One explanation for the difference between these results and those of the current analysis is that the earlier study used an assay with a limit of detection for minority drug-resistant variants of 0.5% of the virus population and therefore identified only a limited number of participants with resistant variants present at low copy numbers. Other possible explanations include the lack of Y181C measurement in that study and differences between studies of the NRTI component of the regimen. Nevertheless, it is clear that not all patients in whom minority drug-resistant variants are identified will experience virologic failure and a frequency-dependent effect of the minority drug-resistant population is clearly evident from the current pooled analysis. Further research is needed to identify additional factors that contribute to the risk of virologic failure.
This analysis has several limitations. In order to combine patient-level data from studies with different study designs, statistical adjustments were required such as limiting the inclusion of patients from case-control studies to only those patients with virologic failure and using a stratified Cox proportional hazard model in which virologic failure patients outside of the cohort studies were only counted at the time of failure. Although this approach has been validated in previous studies24, 25, we confirmed the robustness of our findings in sensitivity analyses limited to data obtained only from the cohort studies. In addition, studies that contributed data to this analysis had differences with regard to assay methodology, sensitivity, and resistance mutations detected. The assay with the highest limit of detection was the HIV-SNaPshot assay (2%)17, whereas allele-specific PCR assays had lower limits of detection (down to 0.003%). The study that contributed the second largest number of participants and the largest proportion of virologic failures utilized the most sensitive assay20. As expected, patients from that study made up the greatest proportion of those with minority drug-resistant variants (72%). Nevertheless, the increased risk of virologic failure associated with presence of minority drug-resistant variants persisted even when this study was removed from the analysis. Visual inspection of the Kaplan-Meier curves (Figure 2) suggests that the increased risk of virologic failure associated with minority variants may be most prominent early in the course of treatment. Such a result would not be unexpected and would mean that the hazard ratios presented (which represent the average hazard ratio over the entire study period) may underestimate the effect of minority drug-resistant variants during the early treatment period. Another limitation involves the types of drug-resistance mutations studied. All studies measured the levels of K103N, but only 6 studies evaluated the presence of Y181C (44% of total patients) and only a minority of the total study population was tested for the presence of M184V (23%) or K65R (17%). Consequently, our ability to detect a significant association of NRTI-resistance mutations and risk of virologic failure or a difference in effect between K103N and Y181C minority variants was limited. Because only a subset of participants were tested for the presence of other NNRTI-resistance mutations, our results most likely underestimate the effect of minority NNRTI-resistant variants on virologic failure as a significant proportion of those categorized as having no detectable minority variants may have had unmeasured Y181C or other NNRTI-resistance mutations.
The findings of this pooled analysis demonstrate that minority HIV-1 drug resistance mutations, and NNRTI resistance mutations in particular, confer a greater than 2-fold risk of virologic failure for individuals on a first-line NNRTI-containing ART regimen. Using the most sensitive test for NNRTI-resistance mutations, approximately 11 patients would need to be screened prior to initiating an NNRTI-based ART regimen to avoid one case of virologic failure. These data provide a rationale for developing standardized clinical assays for the detection of minority NNRTI-resistant variants. As NNRTI-based regimens are the most commonly prescribed first-line antiretroviral therapy, the clinical use of ultra-sensitive HIV drug resistance screening could help identify individuals at greatest risk of virologic failure and allow ART to be tailored appropriately.
Supplementary Material
Acknowledgments
We thank the study participants of all ten studies, including patients from the following trials: FIRST, ACTG A5095, CNA 30021, CNA 30024, GS-01-934, ANRS CO3 Aquitaine Cohort, Swiss HIV Cohort Study, German Truvada Cohort, and the RESINA Cohort, for their contributions to this study. We thank Paul Bain, PhD (Countway Library of Medicine, Harvard Medical School) for his assistance with the systematic review, and Christian Pou (irsiCaixa AIDS Research Institute), Stefano G. Giulieri, MD (Centre Hospitalier Universitaire Vaudois and University of Lausanne), Brian Wine (GlaxoSmithKline), Henry Zhao, PhD (GlaxoSmithKline), and Martin Gartland, PhD (ViiV Healthcare) for their help in the data collection. We wish to thank Christina Lalama, MS (Harvard School of Public Health) and the AIDS Clinical Trials Group for contributing data from ACTG A5095. The findings and conclusions in this manuscript are those of the authors and do not necessarily reflect the official position of the Centers for Disease Control and Prevention.
Dr Li has received research support from Bristol-Myers Squibb and has served as a consultant for Tibotec. Dr Paredes reports having received consulting fees from Pfizer and grant support from Pfizer, Siemens, Merck, and Boehringer Ingelheim. Drs Svarovskaia and Miller are employees and stock-holders of Gilead Sciences, Inc. Dr Metzner has received travel grants and honoraria from Gilead, Roche Diagnostics, GlaxoSmithKline, Bristol-Myers Squibb, Tibotec, and Abbott, and has received a research grant from Gilead. Yale University receives grant support from Merck, Pfizer, Gilead, Abbott, ViiV and Bristol-Myers Squibb for studies that Dr Kozal serves as the principal investigator. Dr Kozal receives royalties from patents owned by Stanford University for some HIV diagnostic tests. Dr Ostergaard has received research grants from Tibotec, Roche, Bristol-Myers Squibb, GlaxoSmithKline, and Merck; educational grants from GlaxoSmithKline and Merck; speaker's fees from Abbott, Tibotec, Bristol-Myers Squibb, Merck, and GlaxoSmithKline. Dr Masquelier received research grants from Pfizer and Janssen-Cilag and speaker's fees from Merck, Pfizer, Gilead, Janssen-Cilag and ViiV Healthcare. Dr Kuritzkes has served as a consultant to and/or has received research grant support from Abbott, Avexa, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead, GlaxoSmithKline, Human Genome Sciences, Merck, Oncolys, Pfizer, Roche, Siemens, Tobira, Vertex, ViroStatistics, and ViiV Healthcare.
Financial Support: Dr Li is the recipient of a Clinical Investigator Training Program Fellowship: Harvard/MIT Health Sciences and Technology - Beth Israel Deaconess Medical Center, in collaboration with Pfizer Inc. and Merck & Co. Dr Paredes was partly supported by the CHAIN, Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project no. 223131, funded by the European Commission Framework 7 Program. Dr Ribaudo is supported in part by grants from the NIH (Statistical and Data Management Center of the AIDS Clinical Trials Group U01 AI068634 and Harvard University CFAR P30 AI060354). Dr. Metzner is supported by the Swiss National Science Foundation (SNF), grants 324700-120793 and CR32I2_127017, Gilead Sciences, and the European Community's Seventh Framework Programme (FP7/2007–2013) under the project “Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN)” - grant agreement n° 223131. Dr Kozal is supported by a VA Merit Award. Dr Kuritzkes is supported in part by grants from the NIH (U01 AI 068636, K24 RR016482) and an ACTG Virology Specialty Laboratory subcontract from the ACTG.
Role of the Sponsors: No sponsors or funding organizations had a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript
Footnotes
Financial disclosures: No other potential conflicts of interest relevant to this article were reported. Dr Li had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Grant RM, Kuritzkes DR, Johnson VA, et al. Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol. 2003 Apr;41(4):1586–1593. doi: 10.1128/JCM.41.4.1586-1593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999 Jul;37(7):2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D, Wheeler W, Ziebell R, et al. Prevalence of transmitted antiretroviral drug resistance among newly-diagnosed HIV-1-infected persons, US, 2007. Paper presented at: 17th Conference on Retroviruses and Opportunistic Infections; San Francisco, CA. February 16–19, 2010. [Google Scholar]
- 4.Vercauteren J, Wensing AM, van de Vijver DA, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis. 2009 Nov 15;200(10):1503–1508. doi: 10.1086/644505. [DOI] [PubMed] [Google Scholar]
- 5.Metzner KJ, Bonhoeffer S, Fischer M, et al. Emergence of minor populations of human immunodeficiency virus type 1 carrying the M184V and L90M mutations in subjects undergoing structured treatment interruptions. J Infect Dis. 2003 Nov 15;188(10):1433–1443. doi: 10.1086/379215. [DOI] [PubMed] [Google Scholar]
- 6.Paredes R, Marconi VC, Campbell TB, Kuritzkes DR. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods. 2007 Dec;146(1–2):136–146. doi: 10.1016/j.jviromet.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JA, Li JF, Wei X, et al. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS One. 2007;2(7):e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinnell JA, Willig JH, Westfall AO, et al. Antiretroviral prescribing patterns in treatment-naive patients in the United States. AIDS Patient Care STDS. 2010 Feb;24(2):79–85. doi: 10.1089/apc.2009.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horberg MA, klein DB. An update on the use of Atripla in the treatment of HIV in the United States. HIV/AIDS - Research and Palliative Care. 2010;2:135–140. doi: 10.2147/hiv.s6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charpentier C, Laureillard D, Piketty C, et al. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS. 2010 Mar 27;24(6):867–873. doi: 10.1097/QAD.0b013e3283367796. [DOI] [PubMed] [Google Scholar]
- 11.Ross LL, Weinberg WG, DeJesus E, et al. Impact of low abundance HIV variants on response to ritonavir-boosted atazanavir or fosamprenavir given once daily with tenofovir/emtricitabine in antiretroviral-naive HIV-infected patients. AIDS Res Hum Retroviruses. 2010 Apr;26(4):407–417. doi: 10.1089/aid.2009.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzner KJ, Rauch P, von Wyl V, et al. Efficient suppression of minority drug-resistant HIV type 1 (HIV-1) variants present at primary HIV-1 infection by ritonavir-boosted protease inhibitor-containing antiretroviral therapy. J Infect Dis. 2010 Apr 1;201(7):1063–1071. doi: 10.1086/651136. [DOI] [PubMed] [Google Scholar]
- 13.Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One. 2010;5(6):e10952. doi: 10.1371/journal.pone.0010952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009 Mar 1;199(5):693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 15.Peuchant O, Thiebaut R, Capdepont S, et al. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS. 2008 Jul 31;22(12):1417–1423. doi: 10.1097/QAD.0b013e3283034953. [DOI] [PubMed] [Google Scholar]
- 16.Balduin M, Oette M, Daumer MP, Hoffmann D, Pfister HJ, Kaiser R. Prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naive patients and their impact on the virological failure. J Clin Virol. 2009 May;45(1):34–38. doi: 10.1016/j.jcv.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Jakobsen MR, Tolstrup M, Sogaard OS, et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis. 2010 Feb 15;50(4):566–573. doi: 10.1086/650001. [DOI] [PubMed] [Google Scholar]
- 18.Metzner KJ, Rauch P, Braun P, et al. Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol. 2010 Nov 3; doi: 10.1016/j.jcv.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Goodman D, Zhou Y, Margot NA, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS. 2011;25:325–333. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- 20.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010 Mar;201(5):662–671. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008 Jul 29;5(7):e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009 Dec;52(5):569–573. doi: 10.1097/QAI.0b013e3181ba11e8. [DOI] [PubMed] [Google Scholar]
- 23.Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009 Jan 15;48(2):239–247. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 24.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. J Clin Epidemiol. 1999 Dec;52(12):1165–1172. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 25.Wacholder S, Boivin JF. External comparisons with the case-cohort design. Am J Epidemiol. 1987 Dec;126(6):1198–1209. doi: 10.1093/oxfordjournals.aje.a114759. [DOI] [PubMed] [Google Scholar]
- 26.Metzner KJ, Rauch P, Braun P, et al. Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol. 2011 Feb;50(2):156–161. doi: 10.1016/j.jcv.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 27.van Elteren P. On the combination of independent two-sample tests of Wilcoxon. Bulletin of the International Statistical Institute. 1960;37:351–361. [Google Scholar]
- 28.Nachega JB, Hislop M, Dowdy DW, Chaisson RE, Regensberg L, Maartens G. Adherence to nonnucleoside reverse transcriptase inhibitor-based HIV therapy and virologic outcomes. Ann Intern Med. 2007 Apr 17;146(8):564–573. doi: 10.7326/0003-4819-146-8-200704170-00007. [DOI] [PubMed] [Google Scholar]
- 29.Lima VD, Harrigan R, Bangsberg DR, et al. The combined effect of modern highly active antiretroviral therapy regimens and adherence on mortality over time. J Acquir Immune Defic Syndr. 2009 Apr 15;50(5):529–536. doi: 10.1097/QAI.0b013e31819675e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnsten JH, Demas PA, Farzadegan H, et al. Antiretroviral therapy adherence and viral suppression in HIV-infected drug users: comparison of self-report and electronic monitoring. Clin Infect Dis. 2001 Oct 15;33(8):1417–1423. doi: 10.1086/323201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geretti AM, Harrison L, Green H, et al. Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis. 2009 May 1;48(9):1296–1305. doi: 10.1086/598502. [DOI] [PubMed] [Google Scholar]
- 32.Schackman BR, Ribaudo HJ, Krambrink A, Hughes V, Kuritzkes DR, Gulick RM. Racial differences in virologic failure associated with adherence and quality of life on efavirenz-containing regimens for initial HIV therapy: results of ACTG A5095. J Acquir Immune Defic Syndr. 2007 Dec 15;46(5):547–554. doi: 10.1097/qai.0b013e31815ac499. [DOI] [PubMed] [Google Scholar]
- 33.Ribaudo HJ, Liu H, Schwab M, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010 Sep 1;202(5):717–722. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little SJ, Frost SD, Wong JK, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008 Jun;82(11):5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010 Jun;84(12):6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Bar KJ, Wang S, et al. High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men. PLoS Pathog. 2010 May;6(5):e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gianella S, Richman DD. Minority variants of drug-resistant HIV. J Infect Dis. 2010 Sep 1;202(5):657–666. doi: 10.1086/655397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.