Abstract
When examining complex problems, such as the folding of proteins, coarse grained descriptions of the system drive our investigation and help us to rationalize the results. Oftentimes collective variables (CVs), derived through some chemical intuition about the process of interest, serve this purpose. Because finding these CVs is the most difficult part of any investigation, we recently developed a dimensionality reduction algorithm, sketch-map, that can be used to build a low-dimensional map of a phase space of high-dimensionality. In this paper we discuss how these machine-generated CVs can be used to accelerate the exploration of phase space and to reconstruct free-energy landscapes. To do so, we develop a formalism in which high-dimensional configurations are no longer represented by low-dimensional position vectors. Instead, for each configuration we calculate a probability distribution, which has a domain that encompasses the entirety of the low-dimensional space. To construct a biasing potential, we exploit an analogy with metadynamics and use the trajectory to adaptively construct a repulsive, history-dependent bias from the distributions that correspond to the previously visited configurations. This potential forces the system to explore more of phase space by making it desirable to adopt configurations whose distributions do not overlap with the bias. We apply this algorithm to a small model protein and succeed in reproducing the free-energy surface that we obtain from a parallel tempering calculation.
Statistical mechanics connects the micro and macro scales by showing how thermodynamic state functions, such as free energy, can be calculated from the classical Hamiltonians that govern the motions of atoms and molecules. These equations allow us to calculate ensemble averages, the relative stabilities of structures, and in some cases reaction mechanisms. At first glance the 3N-dimensional integrals over configuration space make the equations of statistical mechanics appear unsolvable. However, all of them involve integrals over distributions in which the probability of a microstate is related to its energy. Therefore, because the vast majority of phase space is energetically inaccessible, only a relatively small number of configurations make nonnegligible contributions (1–4). Hence, the problem is not so much the integrals, but rather it is determining which are the low energy states that significantly contribute to them.
Molecular dynamics (MD)—using Newton’s equations to calculate a trajectory for the system—is a technique that we can use to find the energetically accessible portions of phase space. The configurations visited during an MD simulation are distributed according to the canonical ensemble so ensemble averages can be calculated by just averaging over the trajectory. However, to do so, one has to assume ergodicity, i.e., that all relevant configurations have been visited during the simulation. This assumption is problematic whenever the energy landscape contains long-lived stable/metastable minima separated by high barriers (5). These features dramatically decrease the rate at which phase space is sampled and so introduce a characteristic timescale for phenomena, which, for protein folding and phase transitions, is typically on the order of milliseconds or more. Studying these processes using unbiased MD is difficult because when using this technique it is only possible to simulate the system for very short (≈1 μs) periods of time. Admittedly, this limit can be extended (to ≈1 ms) by using specialized hardware, but doing so forces one to limit the form of the Hamiltonian (6).
It is possible to increase the frequency with which the barriers separating metastable basins are crossed by introducing a bias potential that makes the energies in the basins comparable with the energies at the transition states (7). Furthermore, because we know the form of the bias function, we can reweight the biased trajectory and obtain the unbiased free-energy surface (8–11). These so-called enhanced sampling methods are now commonplace and applying them to simple chemical problems is relatively straightforward (12–14). The problem comes when the chemistry is more complex, in large part because it is then not obvious how to construct the biasing potential using chemical/physical intuition.
Bias potentials are typically constructed as a function of a small number of collective variables (CVs). Selecting these CVs is the most difficult part of any investigation, so we have recently begun to develop an automated strategy based on machine learning. The first step in this strategy is to obtain a very thorough sampling of the accessible portion of phase space using an algorithm, which adaptively constructs a bias as a function of a large number (D) of collective variables (15). By applying dimensionality reduction—in particular our recently developed sketch-map algorithm (16)—to the trajectory obtained from this calculation, one can obtain a lower, d-dimensional, representation of the accessible portion of phase space. Herein we present the final step of the process in which we adaptively construct a bias potential as a function of the sketch-map coordinates and thereby obtain a thorough sampling of phase space from which we can extract free energies through reweighting. In what follows, we present the mathematical concepts and demonstrate the application of the algorithm on a simple model potential. We then apply it to the alanine 12 system that we examined in our two previous articles (15, 16) and show that we can use our metadynamics algorithm to reproduce the free-energy surface (FES) obtained via parallel tempering.
Background
In all chemical systems the shape of the potential energy surface makes large portions of phase space inaccessible by placing energetic constraints on the geometry of the system (5). In many of the commonly used biasing methods we assume that this accessible portion of phase space lies on a low-dimensionality manifold that is embedded in the full dimensionality space. For many methods, vectors (CVs) that describe this manifold are selected through chemical/physical intuition. However, this process of finding appropriate CVs is often far from straightforward (17) and so there is a strong temptation to look to see whether an automated process can be devised.
An ideal CV for biased dynamics should produce a map of phase space in which all the significant basins in the free-energy surface are well-separated. In addition the CVs should be constructed so that, during the biased dynamics, the system will be pushed along the lowest-lying transition pathways. Dimensionality reduction and manifold learning algorithms are tools that, at least in theory, allow us to develop such CVs. These algorithms construct a d-dimensional representation of a set of data points distributed in a D-dimensional space, by projecting points in the low-dimensionality space in a way that reproduces the pairwise distances between the points in the high-dimensionality space. In the high-dimensionality space, these pairwise distances can be the Pythagorean distance (multidimensional scaling) (18), the geodesic distance (isomap) (19, 20), a nonlinear transformation of the Pythagorean distance (kernel principal component analysis) (21, 22) or the diffusion distance (diffusion maps) (23–27). In contrast, in the low-dimensional space, the Pythagorean metric is used and points are distributed so that the distances between them are approximately equal to their corresponding high-dimensional value. Additionally, in the vast majority of applications, this process of distance matching is not done by iteratively minimizing the discrepancies between the distances in the high- and low-dimensional spaces. Instead some algebra is performed on the matrix of D-dimensional distances which makes the optimization process deterministic (18).
The problem with these methods is that it is difficult to come up with general D-dimensional metrics that will by necessity produce a set of distances that can be reproduced in a low-dimensional, linear space (28, 29). As an example, consider mapping the surface of a sphere in two dimensions, as one has to do to draw a map of the world. The resulting representation will inevitably provide a distorted view of the original. Furthermore, discontinuities can only be avoided if one incorporates a nonlinear feature—the periodicity—in the low-dimensional representation. Worse still, and more relevant to the problem at hand, is the fact that in our previous paper we provided evidence that certain features in typical trajectory data are characteristic of a distribution of points in the full-dimensionality space (16). These realizations led us to develop an algorithm, sketch-map, for performing dimensionality reduction on trajectory data. In developing this algorithm we imagine that the free-energy surface is composed of a network of energetic basins, connected by a spider’s web of narrow transition pathways. Points distributed on this surface display high-dimensionality features because the fluctuations within each basin take place in the full-dimensional space, and because the basins are scattered across the D dimensions. Thus, in sketch-map we try to qualitatively reproduce the spider’s web of connections by transforming the distances in both the D-dimensional and d-dimensional spaces. This transformation ensures that the algorithm focuses on reproducing the distances that lie within a particular range—the length scale that corresponds to the transition pathways between basins. For the remainder of the distances we only insist that if the points are close together in the high-dimensionality space they should be projected close together, and if they are far apart they should be projected far apart.
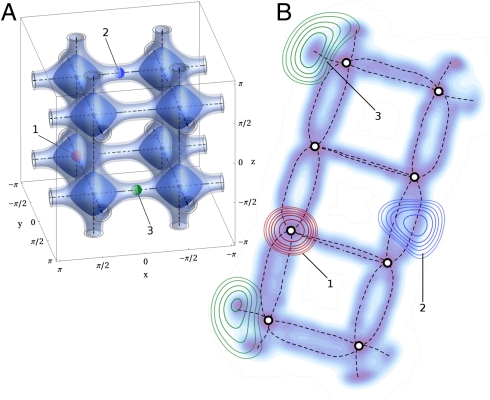
Sketch-map produces a low-dimensional map of phase space in which the various basins in the high-dimensionality free-energy surface are well-separated (16). As such sketch-map coordinates satisfy one of the conditions we require for a good collective variable, and free-energy surfaces projected as a function of them are highly revealing. Where they fall short somewhat is in their description of the transition pathways between basins. These failures are to a certain extent unavoidable—representing complex features in a lower dimensionality space introduces distortions, which inevitably concentrate in poorly sampled regions such as the transition states. To clarify this issue a potential pitfall is illustrated in Fig. 1. This figure shows a three-dimensional potential energy surface with periodic boundary conditions that contains eight energetic basins and twenty-four transition pathways. In projecting this landscape we must map a three-dimensional, toroidal space into a two-dimensional plane. It is impossible to do this mapping without introducing distortions much as it is impossible to map the surface of the Earth on a flat surface rather than on the surface of a globe. Fig. 1 also shows the two-dimensional representation of the surface generated by sketch-map. This projection is nevertheless revealing as it nicely separates the basins while mapping out most of the transition pathways in this free-energy surface. However, the mapping is imperfect, four of the transition pathways are distorted to the extent that points which are adjacent in the three-dimensional representation are projected at opposite ends of the two-dimensional representation. Consequentially, certain portions of the high-dimensionality space are not mapped out properly and will present a problem when this projection is used inside an enhanced sampling algorithm. As we will discuss in the next section we have remedied this problem by developing a more versatile framework for enhanced sampling, which exploits more of the information we obtain when we perform projections from the D-dimensional to the d-dimensional space.
Fig. 1.
A complex free-energy surface that is periodic in three directions (A) and its sketch-map projection (B). B shows how one can use functions of the sketch-map coordinates to describe the position in the three-dimensional space. The fields generated for the three marked points are shown. Where sketch-map reproduces the topology (point 1) the field is sharply peaked and is roughly Gaussian shaped. Where sketch-map provides a less good description the field has multiple peaks because there are multiple points where it is reasonable to project (point 3).
Enhanced Sampling Algorithm
To enhance the sampling along the sketch-map coordinates using metadynamics we must be able to calculate the projection (x) of any arbitrary point (X) in the D-dimensional space. Using a set of N landmark points Xi and their projections xi one could compute a weight for each landmark based on the distance |X - Xi| and then compute x as a weighted average. This idea is the basis of path collective variables (30) and a recently proposed method based on Isomap (31). It assumes that the Xis represent a dense sampling of the high-dimensionality manifold and that the manifold is quasilinear in the neighborhood of each landmark point. These assumptions are not valid for sketch-map coordinates, which endeavor to describe poorly sampled, highly noneuclidean space. Hence, as discussed in our previous paper (16), a better approach for finding out-of-sample projections is to minimize the stress function
 |
[1] |
where wi is the weight of the ith landmark point, and F(x) and f(x) are the sigmoid functions that were used to construct the sketch-map projection. Minimizing this function is problematic because in the vicinity of transition states where the sketch-map projection is poor there may be multiple nearly degenerate minima in the above. Consequentially, the d-dimensional projection of a trajectory in the D-dimensional space contains discontinuities and poor descriptions of some of the conformational transitions. We thus require an alternative approach that incorporates a better description of these problematic regions and in which any discontinuities are smoothed out. In our solution to this problem each high-dimensional configuration X is associated with a d-dimensional field, ϕX(x), which is given by
 |
[2] |
This field replaces the usual representation based on d-dimensional points x. The overlap between fields, which measures their similarity, replaces the distance. The smearing parameter, σ, can be set by ensuring that the overlap between fields corresponding to structurally distinct landmark points is negligible. Then, with this machinery in place, we can create an algorithm that is analogous to metadynamics (32) and use a history-dependent bias to discourage the system from returning to previously visited configurations. Now, though, this bias† is calculated from the overlap between the instantaneous field, ϕX(x), and a bias field v(x,t), constructed from previously visited configurations:
 |
[3] |
where
 |
[4] |
When χ2 has a well-defined global minimum, this field is strongly concentrated about the minimum, with a shape that is nearly Gaussian. Consequentially, the algorithm described above reduces to well-tempered metadynamics (33) in this limit (SI Text). The pleasing thing though is that, as shown in Fig. 1, when the minimization is not straightforward the probability distribution splits itself between the various degenerate minima in Eq. 1. Therefore, these fields give a better description of the trajectory in regions where the sketch-map projection is poor. Furthermore, the field changes smoothly even when the out-of-sample projection changes discontinuously. A slight problem is that there is no longer a simple mathematical relationship between the final bias and the free-energy surface. However, one can always reconstruct the free-energy surface using on-the-fly reweighting (8–11). In fact, calculating the free energy in this way is advantageous as the converged FES will not be affected if the fields are broader than the features in the free-energy landscape. Hence, a poorly chosen σ will not adversely affect the accuracy of the method.
Results
Model Potential.
To test our algorithm we first examined the model potential shown in Fig. 1. As we have explained here and in our previous paper (16), it is difficult to produce a two-dimensional, geometry-preserving map of the low energy portions of this potential. The sketch-map projection nicely separates the eight basins but only by introducing severe distortions in four of the transition pathways. These four discontinuities make the machinery discussed above absolutely critical. The results in Fig. 2 show that our algorithm performs admirably. We are able to quickly explore the entirety of the space, we see many recrossing events and the bias converges by the end of our simulations (SI Text). Together these factors make it so that we can safely extract the free energies surfaces shown in Fig. 2 by reweighting the histogram of visited configurations. In Fig. 2 we compare the reweighted free energies with those obtained by integrating out explicitly one of the three degrees of freedom in Fig. 2. Also shown is the free energy computed as a function of the sketch-map coordinates, which is perhaps more revealing as in this representation the complex topology of the free-energy surface with its eight identical basins can be clearly seen. This representation also demonstrates that there are six escape routes from each basin and that every pathway that is not broken by the projection (i.e., every pathway that we can examine using these CVs) is energetically equivalent.
Fig. 2.
Free-energy surfaces for the model potential shown in Fig. 1 calculated by reweighting the trajectories obtained from the field-overlap metadynamics simulations. A shows the free energy as a function of the modulus of two of the three degrees of freedom. In this panel we compare the free energies obtained by reweighting the trajectory with those calculated by explicitly integrating the free energy using the known Hamiltonian. B shows the free-energy surface as a function of the sketch-map coordinates, which was calculated by reweighting the metadynamics trajectories and using the out-of-sample extension from ref. 16 to define the instantaneous position in sketch-map space. Insets show the free energy in the vicinity of one of the basins and along a pair of transition pathways.
Polyalanine-12.
Having demonstrated our algorithm on a relatively simple energy landscape we now turn our attention to a more complex system; namely, the landscape of polyalanine-12 in implicit solvent. This system has been extensively studied and it has been shown that the potential energy surface, although very rough, is overall funnel-shaped with an alpha-helical global minimum (5, 34). However, in spite of this structure, local minima in the potential energy surface (35) prevent the system from forming the helix during long, unbiased MD simulations (15), which suggests that MD alone is not a suitable tool for exploring this landscape. In contrast, reconnaissance metadynamics can find the global minimum so we have thus used this technique to collect the data (15) we used to construct sketch-map projections (16).
To asses the quality of our metadynamics simulations we first computed free-energy surfaces for ala12 using parallel tempering (36, 37). In and of themselves these results are interesting as they demonstrate that, when free energies are displayed as a function of a set of simple collective coordinates, the resulting pictures can give a myopic view of the underlying physics. Fig. 3 shows the free-energy surface as a function of the gyration radius and the mean square displacement from an alpha helix configuration. This surface is very smooth, and has just two prominent features, one peaked basin and one very broad basin, which can be associated with the folded and unfolded states. This smoothness is in sharp contrast to the picture in Fig. 4, which shows the free energy in terms of the sketch-map coordinates. In this representation the free-energy surface appears to be very rough with a large number of very well-localized basins. This result is more in line with what one would expect given the results from potential energy methods (34) and given that the system does not fold during a 1-μs MD simulation (15). Nevertheless, the CVs used in Fig. 3 can differentiate between the folded and unfolded states. Hence, they can safely be used to compare the relative populations of these states in unbiased MD simulations so that comparisons can be made with experiment. The problem though is that the description of the free-energy landscape that these CVs provide is incomplete—these simple collective coordinates can distinguish folded configurations from the sea of unfolded states but are unable to detect the sometimes marked differences between the various unfolded configurations.
Fig. 3.
The free-energy landscape for ala12 in implicit solvent calculated from parallel tempering (A) and field-overlap metadynamics (B, using sketch-map coordinates) simulations. Here the free energy is shown as a function of the radius of gyration and the mean square displacement from the native state configuration. These CVs project radically different structures close together and thus many of the energetic barriers between structures disappear. In the highlighted structures the red and blue balls indicate the positions of the N and C termini, respectively.
Fig. 4.
The free-energy landscape for ala12 in implicit solvent calculated from parallel tempering (A) and field-overlap metadynamics (B) simulations. Here the free energy is shown as a function of the sketch-map coordinates and is seen to be very rough. In contrast to Fig. 3 each of the highlighted structures lies in a separate basin in the free-energy surface. In these structures the red and blue balls indicate the positions of the N and C termini, respectively.
When CVs do not discriminate well between states interesting features in the landscape get blurred out. This fact explains why the many basins, which are visible in Fig. 4, become blurred into a broad, featureless valley in which there are no well-defined minima in Fig. 3. A direct consequence of this blurring is that, when these simple collective coordinates are used in a metadynamics simulation, the estimate of the free energy will converge very slowly. In fact, in all probability, it will only be possible to converge the free energy by combining metadynamics with parallel tempering (38) so as to ensure that barriers in the transverse degrees of freedom can be crossed. Using multiple replicas of the system in this way is expensive and limits the sizes of the systems that can be studied. Furthermore, one is left feeling that, were the collective coordinates better able to describe the various barriers to motion, much of this computational expense could be avoided.
In Figs. 3 and 4 we show the free-energy surfaces obtained through on-the-fly reweighting of field-overlap metadynamics simulations performed using sketch-map coordinates in tandem with the field formalism laid out previously. These free-energy surfaces were calculated at 525 K, which is the unfolding temperature for this system. Reproducing the free-energy surface at this temperature is particularly challenging because, unlike at 300 K, there is significant occupancy of the unfolded state. Even so the surfaces obtained from metadynamics are very close to the results from the parallel tempering (see SI Text for a more quantitative analysis). This result is particularly impressive given the complexity of the free-energy surface when it is projected as a function of the sketch-map coordinates (Fig. 4).
Conclusions
Enhanced sampling and free-energy methods have been used to understand a wide variety of chemical and physical phenomena. In producing these successful applications, choosing collective variables that describe the problem of interest is critical. Making this choice is enormously difficult and it is perhaps this problem, more than any other, which prevents these methods being used even more widely. This choice will always be challenging, however, as we perform simulations to understand the properties of complex Hamiltonians that describe enormous numbers of interrelated energetic constraints. We should thus not be surprised when the results cannot be explained using a simple function of the atomic positions.
When using collective variables in any approach, it is critical to remember that low-dimensional descriptions of complex chemical processes are inherently limited. Invariably certain features of the intrinsically high-dimensional process will be left out. Therefore, any decision as to what CV to use should be based on what information can be safely discarded. For example, if one wants to examine the folding equilibrium by analyzing an unbiased MD simulation, the CVs used in Fig. 3 are sufficient as these coordinates ably distinguish between folded and unfolded configurations. In contrast, where more detailed descriptions of the unfolded landscape are required these CVs fail because they cannot describe the subtleties in regions of phase space that are not in the immediate vicinity of the minimum-energy, folded state.
In biased MD choosing CVs is particularly critical as, for these methods, barriers to motion in orthogonal degrees of freedom can prevent free-energy estimates from converging. Furthermore, given that in many applications one is endeavoring to accelerate rare events a thousandfold or even a millionfold times, even barriers that are small compared to that of the rare event represent significant hurdles. Consequentially, using CVs that, like those used in Fig. 3, only distinguish the folded state from the sea of unfolded configurations in algorithms such as umbrella sampling or metadynamics will always be problematic. In these cases sketch-map CVs are a better approach as the data-driven strategy used to derive these coordinates ensures that distinct energetic basins are mapped to different parts of the low-dimensional space. The downside is that the sketch-map representation can contain discontinuities. However, as we have shown herein, this problem can be resolved by using fields to describe the instantaneous state. Admittedly, calculating the overlap integrals in this approach is considerably more computationally expensive than calculating the value of a CV. However, it is straightforward to parallelize these calculations on cheap GPU processors and, more importantly, unlike parallel tempering, the cost of this method is independent of system size. Hence, it can be used to calculate the free energies in very large systems or in ab initio calculations, where multiple replicas are less feasible. Furthermore, because the free energy is extracted by reweighting, it can be calculated as a function of any collective coordinate or examined using the collective variable free approaches that have been applied to the analysis of unbiased MD trajectories (27, 39, 40).
Dimensionality reduction is a generic tool that is used in fields of science ranging from chemistry and physics to social sciences and psychology. In all these fields this technique serves to identify low-dimensional trends in easy-to-measure, high-dimensionality data so that diverse features in the underlying phenomena can be classified. This understanding can then be used to classify points from outside the fitting set so that their likely behavior can be inferred. If these inferences are made by minimizing Eq. 1, one is forced to assume that the fitting set describes every possibility and that the low-dimensional representation is a sensible topological description of the high-dimensionality data. In contrast, representing a configuration by a field like that in Eq. 2 allows one to perform these out-of-sample classifications more tentatively and to identify regions of the high-dimensional space where the low-dimensional representation is perhaps lacking. This approach is generic and builds on the notion that the overlap between normalized fields gives a measure of their similarity. In some cases, where it is natural to represent the high-dimensionality data using a normalized histogram (41), it may even be possible to use the overlap between these probability distributions directly, and to avoid the dimensionality reduction step completely.
Materials and Methods
Reweighting.
All the free-energy surface obtained from metadynamics simulations were calculated using on-the-fly reweighting of multiple trajectories. The free energies as a function of a collective coordinate, s, were calculated based on a single trajectory using
 |
[5] |
where the sum runs over the entirety of the trajectory. The free energies shown in the paper were then calculated by averaging the free energies obtained from a number of statistically uncorrelated simulations.
Model Potential.
The model potential shown in Fig. 1 is given by V(θ,ϕ,ψ) = exp(3[3 - sin4(θ) - sin4(ϕ) - sin4(ψ)]) - 1. We study the thermodynamics of a particle of mass m at temperature T. Hence, if one defines the unit of length as l∗, then the characteristic time unit, t∗, is equal to  . To integrate the equation of motion we used the velocity Verlet algorithm with a timestep of 0.01 t∗. Temperature was kept fixed using a Langevin thermostat that had a relaxation time of 0.1 t∗. The sketch-map projection of this landscape that was described in ref. 16 was used throughout. The integrals in Eq. 3 and the equations for the forces were evaluated numerically on a 250 × 250 grid of points. However, because evaluating the value of Eq. 1 at every one of these points would be prohibitively expensive, we chose instead to only evaluate this function on a 15 × 15 grid of points. The function was then interpolated onto the remaining grid points using a bicubic interpolation algorithm (42). The bias field was augmented with a new function every 100 steps, while the initial height, ω, and the well-tempered factor, ΔT, were set equal to 0.44 kBT and 4 kBT, respectively. To collect adequate statistics the free-energy surfaces shown in Fig. 2 were calculated from sixteen statistically uncorrelated runs, which each ran for a total time of 52,800 t∗.
. To integrate the equation of motion we used the velocity Verlet algorithm with a timestep of 0.01 t∗. Temperature was kept fixed using a Langevin thermostat that had a relaxation time of 0.1 t∗. The sketch-map projection of this landscape that was described in ref. 16 was used throughout. The integrals in Eq. 3 and the equations for the forces were evaluated numerically on a 250 × 250 grid of points. However, because evaluating the value of Eq. 1 at every one of these points would be prohibitively expensive, we chose instead to only evaluate this function on a 15 × 15 grid of points. The function was then interpolated onto the remaining grid points using a bicubic interpolation algorithm (42). The bias field was augmented with a new function every 100 steps, while the initial height, ω, and the well-tempered factor, ΔT, were set equal to 0.44 kBT and 4 kBT, respectively. To collect adequate statistics the free-energy surfaces shown in Fig. 2 were calculated from sixteen statistically uncorrelated runs, which each ran for a total time of 52,800 t∗.
Alanine 12.
All simulations of polyalanine were run using gromacs-4.5.1 (43), the amber96 force field (44), and a distance dependent dielectric (34). A timestep of 1 fs was used throughout, all bonds were kept rigid using the LINCS algorithm, and the van der Waals and electrostatic interactions were calculated without any cutoff. The temperature was maintained using an optimal-sampling, colored noise thermostat (45). Once again the sketch-map projection from ref. 16 was used and integrals were calculated on a 401 × 401 grid of points that was constructed by performing a bicubic interpolation from a sparser 21 × 21 grid of points. The bias field was augmented with a new function every 500 steps, while the initial height, ω, and the well-tempered factor, ΔT, were set equal to 0.5 kJ mol-1 and 2,100 K, respectively. The free-energy surfaces shown in Figs. 3 and 4 were calculated by reweighting from 16 such runs—a total of 800 ns of simulation time. For comparison we also calculated the free-energy surface for this system from a single, 800 ns parallel tempering calculation with five replicas in which swapping moves were attempted every 100 steps. The temperatures of the replicas in this calculation were 525.00 K, 601.86 K, 688.23 K, 785.17 K, and 886.17 K. The radius of gyration and distance from the alpha-helical configuration were calculated using PLUMED (13).
Supplementary Material
Acknowledgments.
The authors would like to thank Davide Branduardi and Giovanni Bussi for useful discussions along with Ali Hassanali and Federico Giberti for reading early drafts of the manuscript and giving suggestions. This work was funded by European Union Grant ERC-2009-AdG-247075, the Royal Society, and the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201152109/-/DCSupplemental.
 |
References
- 1.Garcia AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 2.Amadei A, Linssen ABM, Berendsen HJC. Essential dynamics of proteins. Proteins Struct Funct Genet. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 3.Piana S, Laio A. Advillin folding takes place on a hypersurface of small dimensionality. Phys Rev Lett. 2008;101:208101. doi: 10.1103/PhysRevLett.101.208101. [DOI] [PubMed] [Google Scholar]
- 4.Hegger R, Altis A, Nguyen PH, Stock G. How complex is the dynamics of peptide folding? Phys Rev Lett. 2007;98:028102. doi: 10.1103/PhysRevLett.98.028102. [DOI] [PubMed] [Google Scholar]
- 5.Wales DJ. Energy Landscapes. Cambridge, UK: Cambridge Univ Press; 2003. [Google Scholar]
- 6.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 7.Frenkel D, Smit B. Understanding Molecular Simulation. San Diego: Academic; 2002. [Google Scholar]
- 8.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1012. [Google Scholar]
- 9.Dickson BM, Lelièvre T, Stoltz G, Legoll F, Fleurat-Lessard P. Free energy calculations: An efficient adaptive biasing potential method. J Phys Chem B. 2010;114:5823–5830. doi: 10.1021/jp100926h. [DOI] [PubMed] [Google Scholar]
- 10.Bonomi M, Barducci A, Parrinello M. Reconstructing the equilibrium Boltzmann distribution from well-tempered metadynamics. J Comput Chem. 2009;30:1615–1621. doi: 10.1002/jcc.21305. [DOI] [PubMed] [Google Scholar]
- 11.Dickson BM. Approaching a parameter-free metadynamics. Phys Rev E. 2011;84:037701. doi: 10.1103/PhysRevE.84.037701. [DOI] [PubMed] [Google Scholar]
- 12.Laio A, Gervasio FL. Metadynamics: A method to simulate rare events and reconstruct the free energy in biophysics, chemistry and materials sciences. Rep Prog Phys. 2008;71:126601. [Google Scholar]
- 13.Bonomi M, et al. Plumed: A portable plugin for free-energy calculations with molecular dynamics. Comput Phys Commun. 2009;180:1961–1972. [Google Scholar]
- 14.Barducci A, Bonomi M, Parrinello M. Metadynamics. Wiley Interdiscip Rev: Comput Mol Sci. 2011;1:826–843. [Google Scholar]
- 15.Tribello GA, Ceriotti M, Parrinello M. A self-learning algorithm for biased molecular dynamics. Proc Natl Acad Sci USA. 2010;107:17509–17514. doi: 10.1073/pnas.1011511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceriotti M, Tribello GA, Parrinello M. Simplifying the representation of complex free-energy landscapes using sketch-map. Proc Natl Acad Sci USA. 2011;108:13023–13029. doi: 10.1073/pnas.1108486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geissler PL, Dellago C, Chandler D. Kinetic pathways of ion pair dissociation in water. J Phys Chem B. 1999;103:3706–3710. [Google Scholar]
- 18.Cox TF, Cox MAA. Multidimensional Scaling. London: Chapman & Hall; 1994. [Google Scholar]
- 19.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 20.Das P, Moll M, Stamati H, Kavraki LE, Clementi C. Low-dimensional, free-energy landscapes of protein-folding reactions by nonlinear dimensionality reduction. Proc Natl Acad Sci USA. 2006;103:9885–9890. doi: 10.1073/pnas.0603553103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schölkopf B, Smola A, Müller KR. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998;10:1299–1319. [Google Scholar]
- 22.Schölkopf B, Smola A, Muller KR. Advances in Kernel Methods Support Vector Learning. Cambridge, MA: MIT Press; 1999. pp. 327–352. [Google Scholar]
- 23.Coifman RR, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: Multiscale methods. Proc Natl Acad Sci USA. 2005;102:7432–7437. doi: 10.1073/pnas.0500896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coifman RR, Lafon S. Diffusion maps. (Diffusion Maps).Applied and Computational Harmonic Analysis. 2006;21:5–30. [Google Scholar]
- 25.Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 2003;15:1373–1396. [Google Scholar]
- 26.Ferguson AL, Panagiotopoulos AZ, Debenedetti PG, Kevrekidis IG. Systematic determination of order parameters for chain dynamics using diffusion maps. Proc Natl Acad Sci USA. 2010;107:13597–13602. doi: 10.1073/pnas.1003293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohrdanz MA, Zheng W, Maggioni M, Clementi C. Determination of reaction coordinates via locally scaled diffusion map. J Chem Phys. 2011;134:124116. doi: 10.1063/1.3569857. [DOI] [PubMed] [Google Scholar]
- 28.Donoho DL, Grimes C. When does Isomap recover the natural parameterization of families of articulated images? 2002. (Department of Statistics, Stanford University), Technical Report No. 2002-27.
- 29.Donoho DL, Grimes C. Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data. Proc Natl Acad Sci USA. 2003;100:5591–5596. doi: 10.1073/pnas.1031596100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branduardi D, Gervasio FL, Parrinello M. From a to b in free energy space. J Chem Phys. 2007;126:054103. doi: 10.1063/1.2432340. [DOI] [PubMed] [Google Scholar]
- 31.Spiwok V, Kralova B. Metadynamics in the conformational space nonlinearly dimensionally reduced by isomap. J Chem Phys. 2011;135:224504. doi: 10.1063/1.3660208. [DOI] [PubMed] [Google Scholar]
- 32.Laio A, Parrinello M. Escaping free energy minima. Proc Natl Acad Sci USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barducci A, Bussi G, Parrinello M. Well tempered metadynamics: A smoothly converging and tunable free energy method. Phys Rev Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 34.Mortenson PN, Evans DA, Wales DJ. Energy landscapes of model polyalanines. J Chem Phys. 2002;117:1363–1376. [Google Scholar]
- 35.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 37.Hansmann UE. Parallel tempering algorithm for conformational studies of biological molecules. Chem Phys Lett. 1997;281:140–150. [Google Scholar]
- 38.Bussi G, Gervasio FL, Laio A, Parrinello M. Free-energy landscape for β hairpin folding from combined parallel tempering and metadynamics. J Am Chem Soc. 2006;128:13435–13441. doi: 10.1021/ja062463w. [DOI] [PubMed] [Google Scholar]
- 39.Krivov SV, Karplus M. Free energy disconnectivity graphs: Application to peptide models. J Chem Phys. 2002;117:10894–10903. [Google Scholar]
- 40.Gfeller D, De Los Rios P, Caflisch A, Rao F. Complex network analysis of free-energy landscapes. Proc Natl Acad Sci. 2007;104:1817–1822. doi: 10.1073/pnas.0608099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tribello GA, Cuny J, Eshet H, Parrinello M. Exploring the free energy surfaces of clusters using reconnaissance metadynamics. J Chem Phys. 2011;135:114109. doi: 10.1063/1.3628676. [DOI] [PubMed] [Google Scholar]
- 42.Press WH, Teukolosky SA, Vetterling WT, Flannery BP. Numerical Recipes: The Art of Scientific Computing. Cambridge, UK: Cambridge Univ Press; 2007. [Google Scholar]
- 43.Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: Algorithms for highly efficient, load-balanced and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 44.Kollman PA. Advances and continuing challenges in achieving realistic and predictive simulations of the properties of organic and biological molecules. Acc of Chem Res. 1996;29:461–469. [Google Scholar]
- 45.Ceriotti M, Bussi G, Parrinello M. Colored-noise thermostats à la carte. J Chem Theory Comput. 2010;6:1170–1180. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.