Abstract
1O2 (singlet oxygen) is a reactive O2 species produced from triplet excited chlorophylls in the chloroplasts, especially when plants are exposed to excess light energy. Similarly to other active O2 species, 1O2 has a dual effect: It is toxic, causing oxidation of biomolecules, and it can act as a signal molecule that leads to cell death or to acclimation. Carotenoids are considered to be the main 1O2 quenchers in chloroplasts, and we show here that light stress induces the oxidation of the carotenoid β-carotene in Arabidopsis plants, leading to the accumulation of different volatile derivatives. One such compound, β-cyclocitral, was found to induce changes in the expression of a large set of genes that have been identified as 1O2 responsive genes. In contrast, β-cyclocitral had little effect on the expression of H2O2 gene markers. β-Cyclocitral–induced reprogramming of gene expression was associated with an increased tolerance to photooxidative stress. The results indicate that β-cyclocitral is a stress signal produced in high light that is able to induce defense mechanisms and represents a likely messenger involved in the 1O2 signaling pathway in plants.
Keywords: oxidative stress, reactive electrophile species
Reactive O2 species (ROS) are inevitably produced in chloroplasts during photosynthesis, especially under environmental stress conditions that inhibit the photosynthetic processes and, hence, lead to excessive absorption of light energy (1, 2). Reduced forms of O2 are generated by transfer of electrons from the photosynthetic electron transport chain to molecular O2, whereas triplet excited chlorophylls can transfer excitation energy to O2, resulting in the formation of singlet oxygen (1O2) (3, 4). The latter ROS is a strong electrophile agent that can react with many classes of biological molecules, including lipids, proteins, and DNA (4). Using hydroxy fatty acids as specific reporters of enzymatic and nonenzymatic lipid peroxidation mechanisms, 1O2 was demonstrated to play a major destructive role during the execution of ROS-induced cell death in leaves (5). However, besides its toxic effects, 1O2 can also trigger a signaling cascade, leading to programmed cell death (6) or to acclimation (7). Genetic studies of the conditional Arabidopsis mutant flu that produces massive amounts of 1O2 during a dark-to-light transition showed that 1O2 signaling has specific features in terms of gene induction compared with signaling by other ROS (6, 8, 9). Despite the identification of several components of the 1O2 signaling pathway, it remains unclear how the 1O2 signal is transduced from the chloroplast to the nucleus, leading to changes in gene expression. Because of its high reactivity and short lifetime, the direct involvement of 1O2 as a signaling compound seems unlikely. More probably, signaling finds its origin in the reaction of 1O2 with preferential target molecules that can serve as mediators. Among the antioxidants present in the chloroplasts, carotenoids are considered to be the first line of defense of plants against 1O2 toxicity (4, 10, 11) and, therefore, products resulting from their direct oxidation by 1O2 are potential candidates for this function. This possibility is explored in the present work.
Results
Products Generated by in Vitro and in Vivo 1O2 Oxidation of Carotenoids.
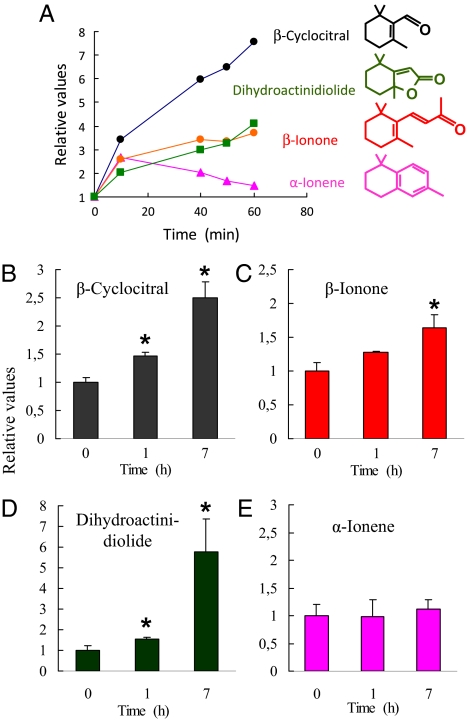
The oxidative breakdown of β-carotene is known to produce a number of volatile short-chain compounds (12, 13), and we looked for those products in a β-carotene solution illuminated for up to 60 min in the presence of the 1O2 generator Rose Bengal. GC-MS analysis indicated that several products rapidly accumulated during 1O2 oxidation of β-carotene: β-cyclocitral (β-CC), β-ionone (β-I), and dihydroactinidiolide (Fig. 1A). This effect was accompanied by a less pronounced, transient production of α-ionene.
Fig. 1.
Volatile oxidation products of β-carotene measured by GC-MS. (A) Relative changes of four major products generated by the in vitro oxidation of β-carotene by 1O2 produced by Rose Bengal in the light: β-I, β-CC, dehydroactinidiolide, and α-ionene. (B–E) Relative changes of β-carotene oxidation products in Arabidopsis leaves during high light stress and low temperature (1,400 μmol photons m−2⋅s−1, 7 °C). Data are normalized values to the value measured at time 0. For β-I and β-CC in leaves, 1 = 40 and 58 ng⋅g−1 fresh weight, respectively. Data are mean values of three independent measurements + SD. *, significantly different from the control (time 0) at P < 0.05 (t test).
The compounds detected in vitro were then studied in vivo in Arabidopsis leaves subjected to high light stress (1,400 μmol photons m−2⋅s−1, 7 °C). All four products shown in Fig. 1A were present in dichloromethane extracts of control, unstressed leaves, indicating chronic oxidation of β-carotene (Fig. 1 B–E). β-CC, dihydroactinidiolide and, to a lesser extent, β-I accumulated in plants exposed to high light stress (Fig. 1 B–D). In contrast, the α-ionene levels did not change with the light treatment (Fig. 1E). We searched for the corresponding molecules derived from the oxidation of xanthophylls, such as 3-hydroxy-β-CC or 3-hydroxy-β-I, but none of those compounds could be detected in Arabidopsis leaves.
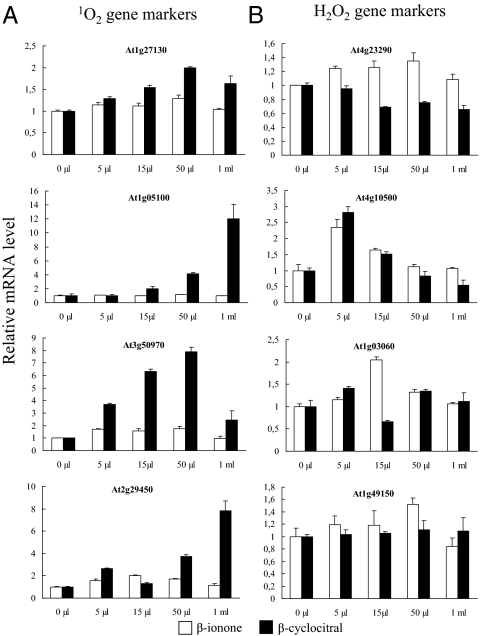
In Arabidopsis cell suspension cultures, 1O2 was found to be the main ROS produced in high light (14). We checked the formation of the latter ROS in Arabidopsis leaves during high light stress by using the transcript levels of several 1O2 marker genes: At1g57630, At1g05100, At3g50970, and At2g29450 (8). Quantitative RT-PCR (qRT-PCR) measurements revealed a strong induction of all 1O2 gene markers after 7 and/or 51 h of illumination (Fig. S1A), indicating that Arabidopsis leaves produced 1O2 during the light treatment. In contrast, because the expression of H2O2 gene markers responded to the light stress in a very complex manner, without a clear trend display, it cannot be concluded that the light stress treatment led to a rapid and massive production of H2O2. Only the At1g49150 gene showed an induction after 7 h of illumination treatment, whereas the two genes At4g23290 and At4g03060 were repressed, instead of induced, and At4g10500 exhibited a late induction that took place only after 51 h of stress (Fig. S1B).
Effects of Oxidized Products of β-Carotene on the Expression of 1O2 and H2O2 Gene Markers.
The volatile oxidized derivatives of β-carotene that accumulated in Arabidopsis leaves during high light stress contain an α,β-unsaturated carbonyl, classifying them as reactive electrophile species (RES) (15, 16). To explore the potential role of those molecules in the signaling of photooxidative stress, Arabidopsis plants placed in an airtight Plexiglas box were exposed to different volumes (0 μL, 5 μL, 15 μL, 50 μL, and 1 mL) of pure β-CC or β-I applied to cotton wicks. As shown in Fig. S2 A and B, these treatments had no impact on leaf stomatal conductance. Similarly, the photochemical activity of chloroplasts, measured in the dark (Fv/Fm) or in the light (ΔF/Fm′), was not affected by the treatments (Fig. S2 C and D). Thus, at the levels used here, volatile carotenoid oxidation products did not appear to be toxic to Arabidopsis plants.
We examined the effects of β-CC and β-I on the expression of the 1O2 and H2O2 gene markers (Fig. 2 and Fig. S3). The transcript analysis revealed contrasted effects of the two molecules: β-CC induced a marked expression of all 1O2 gene markers (Fig. 2A and Fig. S3), and this effect displayed a clear dose dependence. Conversely, none of the H2O2 gene markers were induced by β-CC, with the exception of At4g10500, which exhibited an increased expression at one concentration only (Fig. 2B). In striking contrast with β-CC, only one gene (At2g33380) among five 1O2 gene markers tested by qRT-PCR was slightly induced by β-I. This low bioactivity of β-ionone cannot be explained by its lower volatility relative to β-CC, which did not lead to significant differences in the concentrations of β-I and β-CC reached in the gas phase in the airtight boxes (Fig. S4). Surprisingly and in contrast with β-CC, β-I was able to induce some H2O2 gene markers. However, the dependence of this effect on the β-I level was very complex, so that no clear picture could emerge from the data. Taken together, the results show that β-CC is able to induce noticeable changes in gene expression, and this effect seems to be specific to 1O2 responsive genes. GC-MS analysis of the β-CC concentration in plants treated with volatile β-CC (50 and 500 μL) gave values close to the endogenous concentration measured after high light stress (≈140 ng⋅g−1 fresh weight): 127 ± 24 and 375 ± 59 ng⋅g−1 for the 50-μL and 500-μL treatments, respectively. These data indicate that the internal β-CC levels reached in leaves after the β-CC treatments were in the physiological range.
Fig. 2.
Effects of β-I or β-CC on the expression of 1O2 and H2O2 gene markers in Arabidopsis leaves, as measured by qRT-PCR. Transcript levels of 1O2 gene markers [GST tau3 (GSTU13, At1g27130), Mitogen activated protein kinase kinase kinase 18 (MAPKKK18, At1g05100), Toll-Interleukin-Resistance domain-containing protein (At3g50970) and GST 103–1A (GSTU5, At2g29450)] (A) and H2O2 gene markers [Cysteine-rich receptor-like protein kinase 21 (CRK21, At4g23290), Oxidoreductase (At4g10500), 2-oxoglutarate-dependent dioxygenase (OAP2, At4g03060) and Hypothetical protein (At1g49150)] (B) in Arabidopsis leaves exposed for 4 h to different amounts (0, 5, 15, 50 μL, and 1 mL) of β-I (open bars) or β-CC (filled bars) in an airtight box under a photon flux density of 60 μmol photons m−2⋅s−1. The gene markers were selected from ref. 9 and Genevestigator. Data are expressed in relative values normalized to the value at time 0. Data are mean values of four to five independent measurements + SD.
Gene Expression Reprogramming by β-CC Treatment.
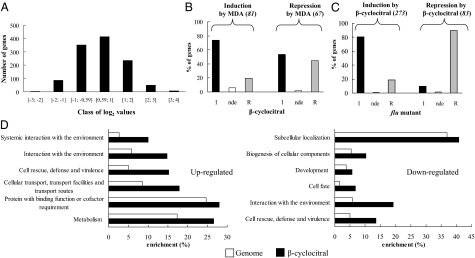
To obtain further insight into the effects of β-CC on the whole genome, a transcriptional analysis was realized with a CATMAv5 array (17) on plants treated for 4 h with 50 μL of β-CC in an airtight box (60 μmol photons m−2⋅s−1, 20 °C) and compared with control plants treated with water. Among the 31,987 gene-specific tags contained on the array corresponding to 31,599 genes, 439 genes were down-regulated, whereas 706 were up-regulated by β-CC compared with control condition (Dataset S1). The distribution of up- and down-regulated genes (Fig. 3A) indicated that most of the gene responses had a log2 value comprised between 2 and −2 (corresponding to fourfold changes). A functional classification of differentially expressed genes by β-CC relative to water was performed by the FunCat annotation scheme (18). The results reported in Fig. 3D contained only the main functional categories with a P value <0.005 (see Tables S1 and S2 for the complete list). Among the up-regulated genes, the “(Systemic) interaction with the environment” and “Cell rescue, defense and virulence” categories were overrepresented; they correspond to genes responding to oxidative stress or participating in cellular sensing, hormone signaling, and detoxification mechanisms, such as receptor-like kinase proteins (CRK3; HAESA, and RLK7), MAP kinases (MKK9, MPK17, and MPK15) and various regulatory proteins (Zinc finger family protein, At1g63840, Heat shock protein binding, At1g65280, MYB4, AP2, At1g71520). We also observed the activation of genes involved in hormone biosynthesis (jasmonate, LOX2, OPR1, AOC1, AOS; ethylene, ATERF-2, CEJ1) and of various defense genes (DHAR2, ATMDAR2, AtGPX6, glutaredoxin, At1g28480, and SAG21). The categories “Metabolism” “Protein with binding function,” and “Cellular transport” were also overrepresented and include genes implicated in detoxification processes, such as cytochrome P450 (At3g28740; At2g121910) and monooxygenases (At4g15760), GST (ATGSTU7; ATGSTU1; and ATGSTF8) and glycosyl transferases (UGT73B4; UGT73B2; and UGT73B1), and a number of membrane transporters (At1g33110; At3g23550; and At1g79410).
Fig. 3.
DNA microarray analysis of changes in gene expression induced by β-CC in Arabidopsis leaves. Plants were exposed for 4 h to 50 μL of β-cyclocitral in an airtight box under a photon flux density of 60 μmol photons m−2⋅s−1. (A) Distribution of the 1,145 genes induced or repressed by β-CC. The plot shows the number of genes in each class of log2 values of the gene expression ratio β-CC/H2O. (B) Comparison of the effects of malondialdehyde (MDA) and β-CC on gene expression. The plot represents the % of the 148 stress- and defense-related genes induced or repressed by MDA that are induced (I), repressed (R), or not differential expressed (nde) by β-CC (compared with the H2O-treated control samples). This comparison is based on the microarray study of MDA-exposed Arabidopsis plants performed in ref. 19. (C) Microarray-based comparison of the effects of β-CC and 1O2 on gene expression. The plot represents the % of genes induced or repressed by β-CC by a factor of 2 or more that were induced (I), repressed (R), or not differentially expressed (nde) by 1O2 in the flu mutant after 2 h of illumination (compared with the illuminated WT). This comparison is based on the microarray study of the flu Arabidopsis mutant performed in ref. 8. The comparison flu (2 h)/WT (2 h) allows the identification of genes induced or repressed by 1O2 while eliminating genes that respond to the dark/light transition. We eliminated also the limited number of false positives (six for the induced genes and two for the repressed one), which occur when the genes are already induced or repressed in flu at time 0 and (flu (2 h)/WT (2 h))/(flu (0 h)/WT (0 h)) is <1 or >1, respectively. The number of genes in the experimental datasets is given in italics in brackets. (D) Overview of the functional categories significantly enriched in genes induced (Left) or repressed (Right) by β-CC in comparison with their relative abundance in the genome.
Among the down-regulated genes, the “Biogenesis of cellular components” “Development,” and “Cell fate” categories were overrepresented, with repression of genes involved in growth and development such as EXPA8, EXPA11, and XTH4 (Fig. 3D).
A comparative analysis of our microarray data with previously published transcriptomes allows us to confront the biological activity of β-CC with other volatile RES (Fig. 3B), such as malondialdehyde (MDA), a secondary end-product of lipid peroxidation. Although ≈70% of the 81 genes induced by MDA were also induced by β-CC, the comparison of the two volatile RES gave a very poor correlation for the repressed genes because >50% of the genes repressed by MDA were induced, instead of repressed, by β-CC. A similar conclusion was reached for the comparison with methyl vinyl ketone (19), a model RES compound. We also compared the transcriptomic data generated by the analysis of β-CC–treated plants with the expression profile induced by 1O2 in the Arabidopsis flu mutant (8). Strikingly, Fig. 3C showed very similar gene expression profiles, with >80% and >90% of similitude for the up-regulated and down-regulated genes, respectively. Among the 219 genes that were induced both in flu and β-CC-treated plants, 22 can be considered as specifically induced by 1O2 (and not by other ROS such as H2O2) according to ref. 9 (Table S3).
Photooxidative Stress Tolerance of Arabidopsis Plants After Exposure to β-CC.
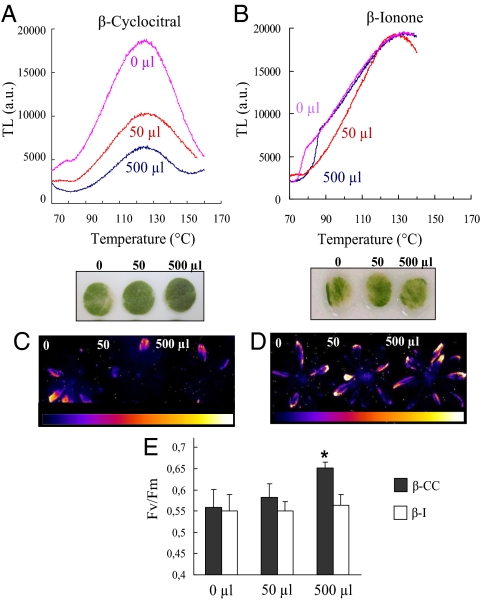
Arabidopsis plants exposed for 4 h to β-CC in an airtight box were subsequently transferred to high light stress conditions. Leaf discs were illuminated for 20 h with white light (1,500 μmol photons m−2⋅s−1) at 10 °C. This treatment induced chlorophyll bleaching and lipid peroxidation (as measured by the amplitude of the 135 °C thermoluminescence band; ref. 20) (Fig. 4A). Interestingly, β-CC protected leaf discs against both phenomena in a dose-dependent manner. We also exposed whole plants to high light stress at low temperature (Fig. 4 C and E). Again, β-CC had a protective effect: PSII photochemical efficiency measured after the light stress was higher in β-CC–treated plants compared with control plants (Fig. 4E), and much less β-CC–treated leaves exhibited an increased autoluminescence, indicative of lipid peroxide accumulation (21) (Fig. 4C). Thus, the changes in gene expression triggered by β-CC were associated with an increased tolerance of Arabidopsis leaves toward photooxidative damage. In contrast, when plants were treated with β-I, no such protection was observed (Fig. 4 B, D, and E), indicating that an inactive carotenoid oxidation product is unable to enhance the tolerance to photooxidative stress.
Fig. 4.
Effects of β-CC or β-I on the tolerance of Arabidopsis plants to photooxidative stress. Plants were exposed for 4 h to β-CC or β-I (50 or 500 μL) or H2O (0 μL) in an airtight box, and then exposed to a photooxidative stress treatment. (A and B) Lipid peroxidation as measured by the 135 °C thermoluminescence (TL) band (Upper) and chlorophyll bleaching (Lower) in leaf discs exposed for 20 h to 1,500 μmol photons m−2⋅s−1 at 10 °C. (C–E) Whole plants exposed for 48 h to high light stress at low temperature (1,400 μmol photons m−2⋅s−1, 7 °C). Lipid peroxidation imaged by measuring plant autoluminescence [color scale indicates signal intensity from 0 (blue) to saturation (white)] (C and D) and PSII photochemical efficiency, as measured by the Fv/Fm ratio, after the light stress treatment (E). *, significantly different from control (0 μL) with P < 0.05 (t test).
Discussion
Plants have been reported to produce volatile organic compounds, such as oxylipins, isoprene, or monoterpenes, when exposed to high light intensities (22, 23). This study shows that exposure of Arabidopsis plants to high light stress also induces the production of short-chain compounds derived from the oxidation of β-carotene, such as β-CC, β-I, and dihydroactinidiolide—a phenomenon previously reported in cyanobacteria and microalgae (24) but not yet in vascular plants. Being electrophilic, the α,β-unsaturated carbonyl group of β-CC and β-I can react with electron donors such as, for example, sulphydryl groups in proteins (15, 16). Because of their reactivity, RES can be cytotoxic and, accordingly, a number of previous studies have reported the deleterious effect of high concentrations of β-carotene oxidation products, including β-I, on mitochondrial and chloroplastic functions, DNA intactness, and cell viability (25–27). In addition, low levels of RES, and particularly those produced during lipid peroxidation, have also effects on plant genome (19, 28, 29). In animals, ROS-induced oxidized carotenoid derivatives have been shown to be biologically active, playing a role in changes in gene expression, transcription activation, and apoptosis (e.g., refs. 30–33). This study demonstrates that carotenoid oxidation products accumulating in light-stressed Arabidopsis plants are bioactive molecules: The expression of a large range of genes (>1,000) was changed in Arabidopsis plants exposed to β-CC. The majority of the induced genes encode proteins involved in the interactions with the environment, in stress responses, and in cellular transport, whereas many repressed genes are related to development, growth, and biogenesis of cellular components. Thus, β-CC appears to function as a stress signal that can reprogram gene expression, shifting plant cells from active growth to cellular defense toward stress.
Interestingly, all 1O2 gene markers examined by qRT-PCR were induced by β-CC, in a dose-dependent manner. β-I was found to be much less efficient in inducing expression of 1O2 gene markers. Many structural factors, such as the presence of a methyl group and its relative position to the terminal aldehyde or the length of the backbone of the carotenoid molecule, determine RES reactivity (34), possibly explaining the differential effects of β-CC and β-I. A more complete analysis of gene expression in β-CC–exposed Arabidopsis plants using DNA microarray technology showed that ≈80% of the induced or repressed genes correspond to genes that are also induced or repressed in the 1O2-overproducing flu Arabidopsis mutant (8). The overlap between gene expression changes induced by β-CC and by other volatile RES, such as MDA and methyl vinyl ketone (19), was smaller. This finding indicates that the β-CC effects on gene expression cannot be merely considered as a general response to RES activity, pointing at a more specific function. Considering that β-CC is generated by 1O2 attack on β-carotene and the gene induction profile of β-CC overlaps strongly with the activation profile of 1O2, an obvious possibility is that β-CC functions as an intermediate in the 1O2 signaling pathway.
Carotene oxidation products can also be formed enzymatically by carotenoid cleavage dioxygenases (CCD) (35). However, the involvement of those enzymes in the production of CC in light-stressed Arabidopsis leaves seems unlikely. An in silico analysis with Genevestigator (www.genevestigator.com) indicated that the 4 CCD genes of Arabidopsis are not induced by light, with CCD4 being strongly repressed under high light and cold. This was confirmed by a separate cDNA microarray-based gene expression analysis of wild-type Arabidopsis exposed to high light (1,500 μmol m−2⋅s−1, 10 °C, 2 d): CCD1, CDD7, and CDD8 were not induced by high light (Log2 = 0.098, −0.11, and −0.042, respectively), whereas CCD4 was strongly repressed (log2 = −3.14) (http://urgv.evry.inra.fr/CATdb; Project: CEA10-02_Light).
A striking feature of the gene expression reprogramming induced by β-CC in Arabidopsis leaves is the induction of various defense mechanisms. Among the most induced genes, we found 10 GST genes and 12 UDP-glycosyltransferase genes. Similar phenomena were observed in a photosensitive Arabidopsis mutant deficient in two xanthophylls (36) and in the green alga Chlamydomonas exposed to 1O2 (7). By catalyzing the conjugation of glutathione or sugar with a variety of substrates, GSTs and glycosyltransferases can detoxify compounds (37, 38). In Chlamydomonas, it was shown that constitutive expression of a GST gene confers tolerance to 1O2 (7). In line with this observation, we found that β-CC led to an increase in the tolerance of Arabidopsis to a subsequent photooxidative stress treatment. Moreover, the β-CC–induced changes in gene expression were not accompanied by visible symptoms of toxicity (e.g., leaf necrosis) or by effects on photosynthetic electron transport or stomatal conductance, although the latter processes are known to be sensitive to RES (27, 39). This observation confirms that the amounts of volatile β-CC that penetrated inside the leaves were low, as also inferred from the gene expression levels, which were less pronounced than expression levels induced by high light stress (compare Figs. S1A and 2A). Taken together, our results strongly support the idea that the 1O2 signaling pathway can lead to stress acclimation in plants, as reported in algae (7). Possibly, the involvement of the signaling pathway in acclimation or in cell death is determined by the stress intensity and the resulting levels of 1O2 production and signal molecules (40). Moreover, β-CC–induced changes in gene expression and in phototolerance were not cancelled in the executer1 Arabidopsis mutant (Fig. S5), in line with the induction of an acclimatory response rather than the cell death response reported in the flu mutant (6). In a recent study (36), it was also shown that 1O2 responsive genes can be induced under conditions that do not lead to cell death.
To sum up, this study has shown that exposing whole Arabidopsis plants to high light stress induced a rapid accumulation of both 1O2 and β-carotene oxidation products within hours. β-CC, one of the β-carotene derivatives produced in high light, is able to induce changes in the expression of a large set of genes, which strongly overlap with the network of genes induced by 1O2. Taken together, this study identifies β-CC as a signal molecule produced during photooxidative stress and indicates that this signal is a likely candidate to be involved in the 1O2 signaling pathway in Arabidopsis. Thus, besides their well-established antioxidant and light-harvesting functions, carotenoids, through their oxidation by ROS, play also a role in the sensing and signaling of oxidative stress conditions.
Materials and Methods
Plant Material, Growth Conditions, and Treatments.
Arabidopsis plants (Arabidopsis thaliana, ecotype Colombia) were grown for 4 wk under controlled conditions, as described (41). Plants were placed for 4 h in a transparent airtight box (≈22 L) installed in a growth chamber under controlled conditions of light and temperature (60 μmol photons m−2⋅s−1, 22 °C). The effects of β-I and β-CC were tested at different concentrations: Known volumes of a pure compound (5 μL, 15 μL, 50 μL, 500 μL, or 1 mL) were deposited on a wick of cotton to increase the contact area with the air and consequently to enhance their volatilization in the airtight box. For the control conditions, the carotenoid oxidation derivatives were replaced by distilled water.
In Vitro and in Vivo Oxidation of β-Carotene.
β-Carotene supplemented with Rose Bengal was dissolved in toluene/methanol (85:15; vol/vol) and kept while bubbled with O2 under illumination (21). Photooxidative stress was imposed on Arabidopsis plants by transferring them to a chamber with the following parameters: 1,400 μmol photons m−2⋅s−1, day/night 8 h/16 h, 7 °C/12 °C day/night. Carotenoid oxidation products were extracted from ≈500 mg of leaves in 4 mL of dichloromethane containing 4-nonanol as an internal reference (10 μg/500 μL final volume). After centrifugation, the supernatant was collected, transferred into a vial, and evaporated to obtain a final volume of 500 μL.
GC-MS.
Analyses were performed by using a GC/MS Shimadzu QP2010 system. The instrument was equipped with a cpsil 8CB LB fused silica capillary column 15 m × 0.1 mm × 0.1 μm (Varian), and the velocity of the carrier gas (He) was at 37 cm/s. Injections of 2 μL of the extracts were carried out with a splitless mode, and the injector temperature was set at 250 °C. Oven temperature was initially set at 60 °C for 1 min and then progressed at a rate of 20 °C⋅min−1 to 250 °C. Oven temperature program was the following: 50 °C (initial temperature); 15 °C⋅min−1 to 160 °C; 3 °C⋅min−1 to 200 °C; and 20 °C⋅min−1 to 250 °C (final temperature). The mass spectra were recorded in electron impact (70 eV). In the first time, acquisition was performed in scan mode to identify the volatiles compounds. Identification was confirmed by injection of standards. Quantification was then done in single ion monitoring on selected ions [177 atomic mass units (amu) for β-I, 152 amu for β-CC, 111 amu for dihydroactinidiolide, 159 amu for α-ionene, and 55 amu for 4-nonanol internal standard].
RNA Isolation.
Total RNA was extracted by using the NucleoSpin RNA Plant kit (Macherey-Nagel) and then treated with the Turbo DNA-free (Ambion) according to the manufacturers’ instructions. Each extraction from leaves of three different plants was performed at least five times.
qRT-PCR.
qRT-PCR experiments were carried out with cDNA synthesized with the SuperScript III Reverse Transcriptase (Invitrogen) from 500 ng of total RNA. Specific primers for each gene selected for analysis were designed by using Primer3plus software (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). qRT-PCR was performed by using LightCycler 480 SYBR Green I Master (Roche) in the qPCR thermal cycler (LightCycler 480 Real-Time PCR System; Roche). Each reaction was prepared by using 2 μL of cDNA diluted 20-fold, 2 μL of SYBR Green I Master and 1 μM forward and reverse primers, in a total volume of 5 μL. The amplification profile consisted of: 95 °C for 10 min and 45 (95 °C for 15 s, 62 °C for 15 s, and 72 °C for 15 s) cycles. All reactions were performed in triplicates. Gene specific primers and oligonucleotide sequences are listed in Table S4.
Transcriptome Studies and Statistical Analysis of Data.
Microarray analysis was performed on CATMAv5 (Complete Arabidopsis Transcriptome MicroArray) arrays containing gene-specific tags from Arabidopsis (42). Three independent biological replicates were produced. The cDNA synthesis, amplification, labeling, and hybridizations and scanning of the slides were performed as described (43). A global intensity-dependent normalization using the loess procedure (44) was performed to correct the dye bias. Then, differential analysis was based on the log ratios averaged on the dye-swap, and these values were used to perform a paired t test. A trimmed variance is calculated from spots that do not display extreme variance (45). The raw P values were adjusted by the Bonferroni method, which controls the Family Wise Error Rate to keep a strong control of the false positives in a multiple-comparison context (46). We considered as being differentially expressed the probes with a Bonferroni P value <0.05.
Photosynthetic Parameters.
The maximal quantum yield of PSII photochemistry (Fv/Fm chlorophyll fluorescence ratio) and the quantum yield of linear electron transport (ΔF/Fm′ fluorescence ratio) were measured in the dark and in the light (200 μmol photons m−2⋅s−1), respectively, as described (41).
Lipid Peroxidation.
Lipid peroxidation was imaged at room temperature by measuring spontaneous photon emission using a high-sensitivity cooled CCD camera, as described (21), using acquisition times of 20 min. Lipid peroxide-related luminescence signal was also measured by thermoluminescence as a band peaking at ≈130 °C, as described in ref. 20. Hydroxy fatty acids (hydroxy octadecatrienoic acids) were analyzed by HPLC using the method described in ref. 47.
Stomatal Conductance.
Stomatal conductance was measured in relative values with a porometer (Delta-T devices, model MKII). Reference values were provided by the ost2 Arabidopsis mutant that keeps its stomata open (48) and wild-type Arabidopsis plants adapted for 2 h in darkness (stomata closed).
Supplementary Material
Acknowledgments
We thank Christophe Laloi (Aix-Marseille University) for useful discussions and the Groupe de Recherches Appliquées en Phytotechnologie (GRAP) platform for help in growing plants. This work is supported by Agence Nationale de la Recherche (ANR) (“Programme Blanc,” Photox Project).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.K.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115982109/-/DCSupplemental.
References
- 1.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 3.Krieger-Liszkay A. Singlet oxygen production in photosynthesis. J Exp Bot. 2005;56:337–346. doi: 10.1093/jxb/erh237. [DOI] [PubMed] [Google Scholar]
- 4.Triantaphylidès C, Havaux M. Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 2009;14:219–228. doi: 10.1016/j.tplants.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Triantaphylidès C, et al. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008;148:960–968. doi: 10.1104/pp.108.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner D, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 7.Ledford HK, Chin BL, Niyogi KK. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot Cell. 2007;6:919–930. doi: 10.1128/EC.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.op den Camp RGL, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadjev I, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogdell RJ, Frank HA. How carotenoids function in photosynthetic bacteria. Biochim Biophys Acta. 1987;895:63–79. doi: 10.1016/s0304-4173(87)80008-3. [DOI] [PubMed] [Google Scholar]
- 11.Edge R, Mcgarvey DJ, Truscott TG. The carotenoids as anti-oxidants – a review. J Photochem Photobiol B. Biol. 1997;41:189–200. doi: 10.1016/s1011-1344(97)00092-4. [DOI] [PubMed] [Google Scholar]
- 12.Stratton SP, Schaefer WH, Liebler DC. Isolation and identification of singlet oxygen oxidation products of β-carotene. Chem Res Toxicol. 1993;6:542–547. doi: 10.1021/tx00034a024. [DOI] [PubMed] [Google Scholar]
- 13.Sommerburg O, et al. β-carotene cleavage products after oxidation mediated by hypochlorous acid—a model for neutrophil-derived degradation. Free Radic Biol Med. 2003;35:1480–1490. doi: 10.1016/j.freeradbiomed.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 14.González-Pérez S, et al. Early transcriptional defense responses in Arabidopsis cell suspension culture under high-light conditions. Plant Physiol. 2011;156:1439–1456. doi: 10.1104/pp.111.177766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmer EE, Davoine C. Reactive electrophile species. Curr Opin Plant Biol. 2007;10:380–386. doi: 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Mueller MJ, Berger S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry. 2009;70:1511–1521. doi: 10.1016/j.phytochem.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 17.Crowe ML, et al. CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 2003;31:156–158. doi: 10.1093/nar/gkg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber H, Chételat A, Reymond P, Farmer EE. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004;37:877–888. doi: 10.1111/j.1365-313x.2003.02013.x. [DOI] [PubMed] [Google Scholar]
- 20.Havaux M. Spontaneous and thermoinduced photon emission: New methods to detect and quantify oxidative stress in plants. Trends Plant Sci. 2003;8:409–413. doi: 10.1016/S1360-1385(03)00185-7. [DOI] [PubMed] [Google Scholar]
- 21.Birtic S, et al. Using spontaneous photon emission to image lipid oxidation patterns in plant tissues. Plant J. 2011;67:1103–1115. doi: 10.1111/j.1365-313X.2011.04646.x. [DOI] [PubMed] [Google Scholar]
- 22.Loreto F, Barta C, Brilli F, Nogues I. On the induction of volatile organic compound emissions by plants as consequence of wounding or fluctuations of light and temperature. Plant Cell Environ. 2006;29:1820–1828. doi: 10.1111/j.1365-3040.2006.01561.x. [DOI] [PubMed] [Google Scholar]
- 23.Bao H, et al. Biogenic volatile organic compound emission potential of forests and paddy fields in the Kinki region of Japan. Environ Res. 2008;106:156–169. doi: 10.1016/j.envres.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Walsh K, Jones GJ, Dunstan RH. Effect of high irradiance and iron on volatile odour compounds in the cyanobacterium Microcystis aeruginosa. Phytochemistry. 1998;49:1227–1239. doi: 10.1016/s0031-9422(97)00943-6. [DOI] [PubMed] [Google Scholar]
- 25.Siems W, et al. β-carotene cleavage products induce oxidative stress in vitro by impairing mitochondrial respiration. FASEB J. 2002;16:1289–1291. doi: 10.1096/fj.01-0765fje. [DOI] [PubMed] [Google Scholar]
- 26.Kalariya NM, Ramana KV, Srivastava SK, van Kuijk FJGM. Carotenoid derived aldehydes-induced oxidative stress causes apoptotic cell death in human retinal pigment epithelial cells. Exp Eye Res. 2008;86:70–80. doi: 10.1016/j.exer.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao J, et al. Elucidating the toxicity targets of β-ionone on photosynthetic system of Microcystis aeruginosa NIES-843 (Cyanobacteria) Aquat Toxicol. 2011;104:48–55. doi: 10.1016/j.aquatox.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Alméras E, et al. Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 2003;34:205–216. doi: 10.1046/j.1365-313x.2003.01718.x. [DOI] [PubMed] [Google Scholar]
- 29.Mueller S, et al. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell. 2008;20:768–785. doi: 10.1105/tpc.107.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharoni Y, Danilenko M, Dubi N, Ben-Dor A, Levy J. Carotenoids and transcription. Arch Biochem Biophys. 2004;430:89–96. doi: 10.1016/j.abb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Siems WG, Sommerburg O, Hurst JS, van Kuijk FJGM. Carotenoid oxidative degradation products inhibit Na+-K+-ATPase. Free Radic Res. 2000;33:427–435. doi: 10.1080/10715760000300961. [DOI] [PubMed] [Google Scholar]
- 32.Kuntz E, et al. Beta-carotene and apocarotenals promote retinoid signaling in BEAS-2B human bronchioepithelial cells. Arch Biochem Biophys. 2006;455:48–60. doi: 10.1016/j.abb.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Liu J-R, et al. β-Ionone suppresses mammary carcinogenesis, proliferative activity and induces apoptosis in the mammary gland of the Sprague-Dawley rat. Int J Cancer. 2008;122:2689–2698. doi: 10.1002/ijc.23453. [DOI] [PubMed] [Google Scholar]
- 34.Linnewiel K, et al. Structure activity relationship of carotenoid derivatives in activation of the electrophile/antioxidant response element transcription system. Free Radic Biol Med. 2009;47:659–667. doi: 10.1016/j.freeradbiomed.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Bouvier F, Isner J-C, Dogbo O, Camara B. Oxidative tailoring of carotenoids: A prospect towards novel functions in plants. Trends Plant Sci. 2005;10:187–194. doi: 10.1016/j.tplants.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Alboresi A, et al. Reactive oxygen species and transcript analysis upon excess light treatment in wild-type Arabidopsis thaliana vs a photosensitive mutant lacking zeaxanthin and lutein. BMC Plant Biol. 2011;11:62. doi: 10.1186/1471-2229-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biol. 2002;3:reviews3004. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowles D, Isayenkova J, Lim EK, Poppenberger B. Glycosyltransferases: Managers of small molecules. Curr Opin Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi T, Ito Y, Okada M, Sakagami Y. Isolation and stomatal opening activity of two oxylipins from Ipomoea tricolor. Bioorg Med Chem Lett. 2005;15:263–265. doi: 10.1016/j.bmcl.2004.10.088. [DOI] [PubMed] [Google Scholar]
- 40.Kim C, Meskauskiene R, Apel K, Laloi C. No single way to understand singlet oxygen signalling in plants. EMBO Rep. 2008;9:435–439. doi: 10.1038/embor.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levesque-Tremblay G, Havaux M, Ouellet F. The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J. 2009;60:691–702. doi: 10.1111/j.1365-313X.2009.03991.x. [DOI] [PubMed] [Google Scholar]
- 42.Hilson P, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 2004;14(10B):2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang YH, et al. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gagnot S, et al. CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 2008;36(Database issue):D986–D990. doi: 10.1093/nar/gkm757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge Y, Dudoit S, Speed TP. Resampling-based multiple testing for microarray data analysis. Test. 2003;12:1–77. [Google Scholar]
- 47.Montillet JL, et al. The upstream oxylipin profile of Arabidopsis thaliana: A tool to scan for oxidative stresses. Plant J. 2004;40:439–451. doi: 10.1111/j.1365-313X.2004.02223.x. [DOI] [PubMed] [Google Scholar]
- 48.Merlot S, et al. Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007;26:3216–3226. doi: 10.1038/sj.emboj.7601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.