Abstract
Adaptive immunity depends on specific recognition by a T-cell receptor (TCR) of an antigenic peptide bound to a major histocompatibility complex (pMHC) molecule on an antigen-presenting cell (APC). In addition, T-cell activation generally requires binding of this same pMHC to a CD4 or CD8 coreceptor. Here, we report the structure of a complete TCR–pMHC–CD4 ternary complex involving a human autoimmune TCR, a myelin-derived self-peptide bound to HLA-DR4, and CD4. The complex resembles a pointed arch in which TCR and CD4 are each tilted ∼65° relative to the T-cell membrane. By precluding direct contacts between TCR and CD4, the structure explains how TCR and CD4 on the T cell can simultaneously, yet independently, engage the same pMHC on the APC. The structure, in conjunction with previous mutagenesis data, places TCR-associated CD3εγ and CD3εδ subunits, which transmit activation signals to the T cell, inside the TCR–pMHC–CD4 arch, facing CD4. By establishing anchor points for TCR and CD4 on the T-cell membrane, the complex provides a basis for understanding how the CD4 coreceptor focuses TCR on MHC to guide TCR docking on pMHC during thymic T-cell selection.
T-cell receptors (TCRs) recognize peptides presented by major histocompatibility complex molecules (pMHC) to discriminate foreign from self-antigens and trigger adaptive immune responses. In addition, T-cell signaling is enhanced by the coreceptors CD4 and CD8, which are expressed on T-helper cells and cytotoxic T lymphocytes, respectively (1). These transmembrane glycoproteins bind MHC class II (CD4) or MHC class I (CD8) molecules on the surface of antigen-presenting cells (APCs). The interaction of CD4 with MHC class II greatly increases cytokine production by helper T cells (1) and substantially reduces the number of antigenic peptides on APCs required for T-cell triggering (2). Similarly, the CD8–MHC class I interaction augments the sensitivity and response of cytotoxic T cells to pMHC ligands (3).
It is generally believed that the main function of the CD4 and CD8 coreceptors is to recruit the Src tyrosine kinase Lck to the TCR–pMHC complex upon coreceptor binding to MHC, leading to formation of a TCR–pMHC–CD4 or TCR–pMHC–CD8 ternary complex (4–8). Recruitment of Lck, which occurs via its association with the cytoplasmic tail of CD4 or CD8, promotes phosphorylation of immunoreceptor tyrosine activation motifs (ITAMs) in the cytoplasmic tails of CD3 subunits associated with the TCR, resulting in signal amplification.
Although numerous (>25) structures of TCR–pMHC binary complexes have been determined (9–11), no structure of a TCR–pMHC–CD4 or TCR–pMHC–CD8 ternary complex has been reported. A hypothetical model of the TCR–pMHC–CD4 complex has been constructed that would rule out direct interactions between CD4 and TCR (12), which seemingly contradicts certain functional and biochemical evidence that CD4 physically associates with TCR (13). One possibility is that CD4 and TCR associate indirectly through other proteins, in particular CD3. Another is that CD4 bridges TCRs interacting with agonist pMHC and endogenous pMHC complexes, creating a TCR “pseudodimer” capable of intracellular signaling (14). A third possibility is that CD4, which consists of four Ig-like domains (D1–D4), possesses a greater degree of segmental flexibility than implied by the CD4 D1–D4 crystal structure (15). In this regard, small angle X-ray scattering (SAXS) has provided evidence that CD4 undergoes a large conformational rearrangement at the D2–D3 junction upon binding the HIV viral entry protein gp120 (16). Clearly, a structure of CD4 D1–D4 bound to TCR–pMHC is critical to resolving this issue.
There is considerable controversy over the mechanism of TCR triggering, and a variety of models have been proposed to explain how the TCR transduces signals across the T-cell membrane after binding pMHC (17). Some of these models evoke dimerization or oligomerization of CD4 (15), MHC (18), or TCR (19) as a means of enhancing phosphorylation of CD3 ITAMs by increasing the proximity of associated tyrosine kinases. However, the plausibility of these models must be assessed in terms of the geometrical constraints that would be imposed by a TCR–pMHC–CD4 structure.
Much effort has been directed at explaining the remarkably conserved diagonal binding mode observed in TCR–pMHC complexes (9, 10). This has been hypothesized to result from a genetically encoded bias of TCRs toward MHC (10, 20–22) and/or the need for the CD4 or CD8 coreceptor for T-cell development and efficient signaling (22–25). Whereas the search for evolutionarily conserved interactions between TCR and MHC molecules has benefited from the wealth of X-ray crystallographic information on TCR–pMHC binary complexes (9–11), a structural framework for understanding how CD4 or CD8 could restrict TCR docking options on pMHC has been lacking. The TCR–pMHC–CD4 ternary complex described here, comprising a human autoimmune TCR, HLA-DR4, and CD4 D1–D4, provides such a framework.
Results and Discussion
Structure Determination.
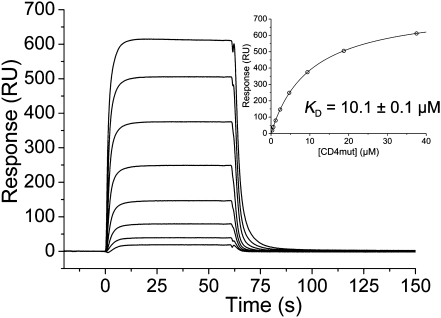
A major obstacle to crystallizing a TCR–pMHC–CD4 ternary complex is its intrinsic instability, attributable to the low affinities of the binary interactions. Indeed, the affinity of the pMHC–CD4 interaction, the dissociation constant (KD) of which has been variously estimated to range from ∼200 μM to >2 mM (13, 26), is even weaker than that of most TCR–pMHC interactions (1–100 μM) (13). To overcome this difficulty, we used in vitro-directed evolution by yeast surface display to increase the affinity of human CD4 for the MHC class II molecule HLA-DR1 (27). We subjected the CD4 D1 domain to in vitro random mutagenesis, and displayed the resulting mutant CD4 library on the yeast surface by fusion to agglutinin protein Aga2p. The library was sorted by flow cytometry with HLA-DR1 to isolate CD4 variants with increased affinity. One of these variants, which contains two substitutions in CD4 D1 (Gln40Tyr/Thr45Trp), was produced as a full-length ectodomain (CD4 D1–D4) by secretion from baculovirus-infected insect cells. This CD4 mutant bound HLA-DR1 with KD = 8.8 μM, compared with no detectable binding for wild-type CD4, even at concentrations up to 400 μM (27). The CD4 mutant exhibited similar affinity for HLA-DR4 (KD = 10.1 μM), as measured by surface plasmon resonance (SPR) (Fig. 1). Therefore, it was used for cocrystallization with HLA-DR4 bearing a self-peptide from myelin basic protein (MBP) and the human autoimmune TCR MS2-3C8. We previously showed that MS2-3C8 engages its ligand via the canonical docking mode of αβ TCRs (28), in which the TCR adopts a central diagonal orientation over pMHC (9, 10). The affinity of MS2-3C8 for MBP–DR4 is 5 μM (28), which is at the high end of the range for TCR–pMHC interactions (13).
Fig. 1.
SPR analysis of the binding of affinity-matured human CD4 mutant to HLA-DR4. SPR sensorgrams for the interaction of the CD4 mutant (0.3, 0.6, 1.2, 2.3, 4.7, 9.4, 18.8, and 37.5 μM) with immobilized HLA-DR4 (1,600 resonance units). Inset shows fitting curve for equilibrium binding that resulted in a KD of 10.1 ± 0.1 μM.
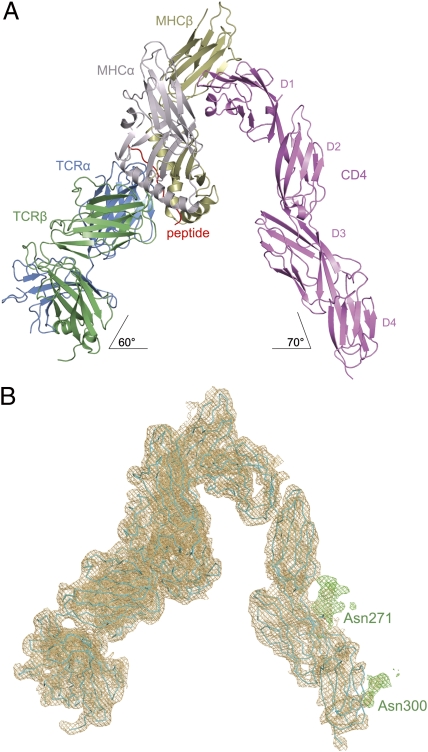
Remarkably, the MS2-3C8–MBP–DR4–CD4 complex crystallized spontaneously from an equimolar mixture of the three proteins, without the aid of precipitants. The structure was determined to 4.0-Å resolution by molecular replacement using the MS2-3C8–MBP–DR4 complex (28) and CD4 (15) as search models (Fig. 2A). The final Rwork and Rfree values are 23.8% and 30.5%, respectively (Table S1). Despite the modest resolution, the electron density was interpretable throughout, and the maps were of sufficient quality to unambiguously assemble the entire TCR–pMHC–CD4 complex (Fig. 2B). Moreover, electron density was visible for carbohydrates attached to CD4 residues Asn271 and Asn300 in domains D3 and D4, respectively, the only two N-linked glycosylation sites in human CD4.
Fig. 2.
Overview of the TCR–pMHC–CD4 ternary complex. (A) Ribbon diagram of the complex oriented as if the TCR MS2-3C8 and CD4 molecules are attached to the T cell at the bottom and the HLA-DR4 MHC class II molecule is attached to an opposing APC at the top. TCR α chain, blue; TCR β chain, green; CD4, pink; MHC α chain, gray; MHC β chain, yellow; MBP peptide, red. (B) σA-weighted 2Fo – Fc electron density map of the complete TCR–pMHC–CD4 complex (contoured at 1σ). The orientation is the same as in A. In green is electron density for carbohydrates attached to CD4 Asn271 and Asn300 in domains D3 and D4, respectively. Protein molecules are shown in cyan as α-carbon traces.
Overview of the TCR–pMHC–CD4 Complex.
The TCR–pMHC–CD4 complex resembles a pointed arch in which both TCR and CD4 are tilted rather than oriented vertically (Fig. 2A). The apex of the arch is formed by the α2 and β2 domains of HLA-DR4 and the D1 domain of CD4. In this view, the complex is oriented as if the TCR and CD4 molecules stand on the T-cell surface at the bottom where they reach up to engage the MHC class II molecule on an opposing APC above. The TCR makes an angle of ∼60° with the T-cell surface and the CD4 molecule an angle of ∼70°; the apical angle between pMHC and CD4 is ∼50° (Fig. 2A).
Importantly, superposition of the pMHC–CD4 D1–D2 portion of the MS2-3C8–MBP–DR4–CD4 structure onto the complex between mouse I-Ak and wild-type human CD4 D1–D2 (12) gave a root mean squared (r.m.s.) difference of 1.6 Å for 465 α-carbon atoms, indicating that affinity maturation of CD4 did not affect its overall binding mode to MHC class II. In both cases, CD4 engages MHC class II through its membrane-distal D1 domain at a concavity formed by the MHC α2 and β2 domains (Fig. 2A). As we showed previously (27), the Gln40Tyr/Thr45Trp substitutions in affinity-matured CD4 improved shape complementarity and increased hydrophobic interactions at the interface with MHC class II through minor adjustments at the mutation sites, without altering overall complex orientation. Moreover, all CD4-contacting residues are invariant across human HLA-DR, -DP, and -DQ alleles and nearly invariant across mouse I-A and I-E alleles (27). Therefore, the topology of the CD4–MHC class II interaction observed in MS2-3C8–MBP–DR4–CD4 structure is likely to be maintained in all TCR–pMHC–CD4 complexes, irrespective of the particular MHC molecule involved.
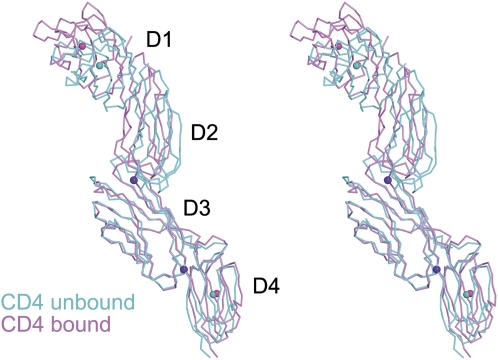
The CD4 molecule bound to TCR–pMHC retains the overall extended conformation observed in different crystal forms of unbound CD4 (15), with some hinge-like variability at the interdomain junctions (Fig. 3). There are no direct contacts between TCR and CD4 in the MS2-3C8–MBP–DR4–CD4 complex (Fig. 2A). An examination of crystal contacts showed no such contacts between TCR and HLA-DR4 in neighboring complex molecules. All crystal contacts are between CD4 and TCR or HLA-DR4: CD4 D2 and TCR Cα/Cβ, CD4 D3 and TCR Cα, and CD4 D3 and HLA-DR4 β2. However, it is the rigidity of CD4, rather than crystal contacts, that is most likely responsible for keeping TCR and CD4 splayed ∼70 Å apart in the crystal lattice. Evidence for this rigidity comes from seven independent views of CD4 D1–D4, all having a similar extended structure. Six of these views come from three different crystal forms of unbound CD4 D1–D4, each containing two molecules per asymmetric unit (15). The seventh is from the MS2-3C8–MBP–DR4–CD4 structure. In the unbound CD4 structures, the angle at the D2–D3 junction displayed a divergence of no more than 6°, whereas the angles at the D1–D2 and D3–D4 junctions were maintained. In bound CD4, the D2–D3 angle varies by no more than 11° in pairwise comparisons with the six unbound CD4 structures, whereas the D3–D4 angle differs by only 4°. The maximum divergence between bound CD4 D1–D4 and any of the unbound structures is shown in Fig. 3. The limited segmental flexibility of CD4 therefore explains the large separation between TCR Cα/Cβ and CD4 D4 observed in the MS2-3C8–MBP–DR4–CD4 complex (Fig. 2A). It also precludes models of T-cell activation calling for large conformational changes in the coreceptor upon assembly of the TCR–pMHC–CD4 complex (29). The relative rigidity of CD4 implies that any significant variations in overall complex architecture must arise from differences in TCR docking on pMHC (see below).
Fig. 3.
Limited segmental flexibility of CD4. Stereoview of CD4 in the TCR–pMHC–CD4 complex overlayed onto CD4 in unbound form (PDB ID code 1WIO) (15). The CD4 molecules were superposed through the D3 domain. Bound CD4 is pink, and unbound CD4 is cyan. The D1–D2 junction shows no variability, whereas the D2–D3 and D3–D4 junctions display angular differences of 11° and 4°, respectively. The pink and cyan spheres mark the centers of mass of the D1 and D4 domains in the bound and unbound CD4 structures, respectively. The purple spheres mark the D2–D3 and D3–D4 junctions.
Implications for the TCR–CD3 Receptor Complex.
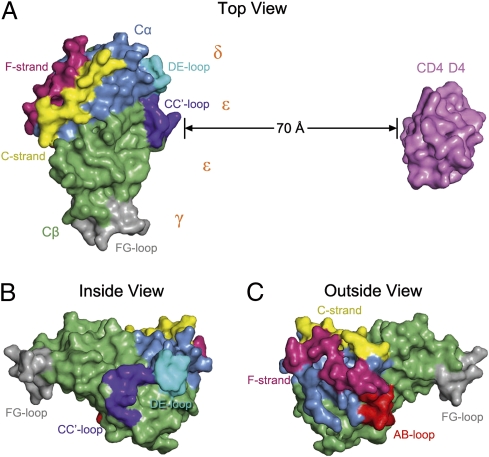
The absence of direct contacts between TCR and CD4 in the TCR–pMHC–CD4 structure (Fig. 2A) is in agreement with an earlier prediction (12) and explains how these molecules can simultaneously, yet independently, bind to pMHC (26, 30). In addition, the wide separation (∼70 Å) between the membrane-proximal TCR Cα/Cβ module and CD4 D4 domain provides ample space for the placement of TCR-associated CD3εγ, -εδ, and -ζζ subunits, which relay activation signals to the T cell via ITAMs (17). Although no crystal structure of a TCR–CD3 complex has been reported, extensive mutational studies have identified docking sites for the ectodomains of CD3εδ and CD3εγ, which interact with the TCR through adjacent Cα DE and Cβ CC′ loops, respectively (31, 32). Based on this information, and on the demonstration that the CD3ε ectodomains are juxtaposed in the TCR–CD3 complex (19), CD3εγ and CD3εδ would be situated under the TCR–pMHC–CD4 arch, wedged between the TCR and T-cell membrane (Fig. 4 A and B). If the angle of the TCR relative to the membrane is greater for the unbound than the bound receptor (∼60°), then engagement of pMHC by CD4 in the ternary complex could displace the CD3 ectodomains into the membrane, resulting in dissociation of CD3ε ITAMs from the membrane and their exposure to Src kinases (33). Of note in this regard, the carbohydrate moiety attached to CD4 Asn300 is located on the outside of the TCR–pMHC–CD4 arch at the base of the D4 domain (Fig. 2B). This glycan may predispose CD4 to adopt a tilted orientation for binding the TCR–pMHC complex in a role analogous to that observed for the adhesion molecule ICAM-2 (34).
Fig. 4.
Location of sites for CD3 docking and TCR dimerization. (A) Top view of the TCR–pMHC–CD4 complex, as if looking down on the T cell. The membrane-proximal TCR C domains and CD4 D4 domain are shown in surface representation; other domains and pMHC are omitted for clarity. The proposed arrangement of the extracellular domains of the CD3εγ and CD3εδ heterodimers (19) is shown in relation to interaction sites identified by mutational studies (31, 32): Cα DE loop (cyan), Cβ CC’ loop (dark blue), and Cβ FG loop (gray). TCR dimerization in the plasma membrane is mediated by the C and F stands of Cα (yellow and magenta, respectively) (19), which are located on the side of the TCR opposite the docking sites for CD3εγ and CD3εδ. (B) View of the TCR C domains from inside the TCR–pMHC–CD4 arch, showing the Cα DE, Cβ CC′, and Cβ FG loops that interact with the CD3 heterodimers (31, 32). (C) View of the TCR C domains from outside the TCR–pMHC–CD4 arch, showing the Cα C and F strands that mediate TCR dimerization. The AB loop of Cα (red), which is not visible in the top view in A, is also involved in TCR dimerization (19).
Implications for TCR Triggering.
A variety of models have been proposed to explain how pMHC binding to TCR initiates signaling across the T-cell membrane (17). The structural constraints imposed by the TCR–pMHC–CD4 ternary complex allow us to discriminate among several models that evoke dimerization (or oligomerization) of CD4 (15), MHC (18), or TCR (19) as triggering mechanisms.
The structure of unbound human CD4 D1–D4 showed that CD4 molecules form dimers through the D4 domain, at least in the crystal (15). Although it has been suggested that D4–D4-associated CD4 dimers may contribute to T-cell activation by interacting with TCR–pMHC complexes (15, 35), the TCR–pMHC–CD4 structure is incompatible with this idea. First, CD4 is monomeric in the TCR–pMHC–CD4 crystal. Second, in a hypothetical model constructed by superposing the TCR–pMHC–CD4 structure onto the D4–D4-associated CD4 dimer, the vertical distance between the C termini of the D4 domains and the hypothetical T-cell surface is ∼40 Å, which is too far to be spanned by the short (eight-residue) stalk region of CD4 (Fig. S1). This assembly is, therefore, unlikely to form on the T-cell membrane.
The observation that some HLA-DR molecules crystallize as dimers (18, 36), as well as the detection of MHC class II dimers on cells (37), suggested a mechanism for T-cell triggering whereby an MHC class II dimer cross-links two TCRs. However, the CD4-binding site on HLA-DR4 is nearly coincident with the putative HLA-DR dimerization site (Fig. S2), which would preclude formation of MHC class II dimers, at least through the interface identified in HLA-DR crystals (18, 36).
According to the pseudodimer model of T-cell triggering, one TCR binds an agonist pMHC ligand, whereas a second TCR binds a self-pMHC ligand (14). Dimerization occurs because the CD4 coreceptor associated with the TCR bound to the agonist pMHC complex also interacts with the self-pMHC complex. However, experimental evidence for CD4-mediated TCR cross-linking is lacking. By contrast, it was recently shown that TCRs can dimerize in the cell membrane independently of CD4 and that mutations which impair TCR dimerization also inhibit activation of CD4+ T cells (19). Notably, these mutations are located in the C and F strands and AB loop of Cα, which form a contiguous surface on the outside of the TCR–pMHC–CD4 arch (Fig. 4C), opposite the site mediating TCR–CD3 interactions (Fig. 4A). As such, CD4 would not interfere sterically with TCR dimerization, nor would it be necessary to displace CD3 subunits to expose the TCR dimerization site in a modified pseudodimer model of T-cell activation (Fig. S3).
CD4 and TCR–pMHC Docking Orientation.
Structural studies of >25 TCR–pMHC complexes have demonstrated remarkable similarities in the overall topology of TCR binding to pMHC, irrespective of MHC class I or class II restriction (9, 10). Typically, the TCR docks on pMHC in a central diagonal orientation, with the Vα domain over the N-terminal half of the antigenic peptide and the Vβ domain over the C-terminal half, although the exact angle and pitch of TCR engagement vary. Two competing (although not mutually exclusive) hypotheses have been put forward to explain this roughly conserved diagonal binding mode. The first holds that coevolution of TCR and MHC genes has led to specific interaction motifs between the germline-encoded CDR1 and CDR2 loops of TCRs and the α-helices of MHC proteins (10, 20–22). However, if the interaction of TCR V regions with MHC is governed solely by evolutionarily selected rules, these are not always apparent, even when a particular V region recognizes the same MHC allele (10). According to the second hypothesis, it is the need for coreceptor function during thymic T-cell selection that restricts the geometry of TCR–pMHC recognition and eliminates from positive selection CD4+CD8+ double-positive thymocytes expressing TCRs unable to engage pMHC in a manner that generates a signal to induce maturation (22–25). In this view, TCR docking topology is guided by the CD4 and CD8 coreceptors during T-cell development to achieve intracellular juxtaposition of coreceptor-bound Lck with CD3 ITAMs.
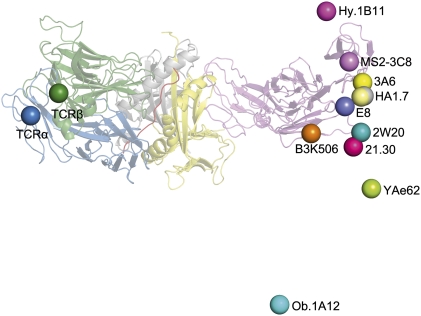
By establishing anchor points for TCR and CD4 on the T-cell membrane, the TCR–pMHC–CD4 structure imposes constraints on the orientation of CD3 relative to Lck associated with CD4 on the cytoplasmic side of the membrane, thereby providing a framework for understanding how CD4 can focus TCR on MHC to restrict TCR docking options on pMHC. Fig. 5 shows the position of the C terminus of CD4 observed in the complex with TCR MS2-3C8 and HLA-DR4, as well as the predicted position of the C terminus of CD4 in hypothetical ternary complexes constructed using nine other TCR–pMHC class II structures, both human and mouse, assuming that CD4 engages human and mouse MHC class II molecules in the same manner (27). With a single exception, the human autoimmune TCR Ob.1A12 (38), the C termini of CD4 in these modeled complexes form a relatively tight cluster that includes the C terminus of CD4 in the MS2-3C8–MBP–DR4–CD4 complex. Other autoimmune TCRs (3A6, Hy.1B11) also fall within the cluster. The observed variability in the position of the CD4 membrane anchor point is the consequence of variations in the overall diagonal docking topology of the TCR–pMHC structures, which places CD3εγ and CD3εδ inside the TCR–pMHC–CD4 arch, opposite CD4 (Fig. 4A). If the TCR–pMHC docking polarity were reversed (i.e., Vα over the C terminus of the peptide and Vβ over the N terminus), we postulate that CD4-bound Lck would be impeded from phosphorylating CD3 ITAMs, thus preventing positive selection of T cells bearing those TCRs.
Fig. 5.
Orientation of TCR and CD4 in TCR–pMHC–CD4 complexes. Bottom view of the TCR–pMHC–CD4 complex, as if looking up from inside the T cell. TCR α chain, blue; TCR β chain, green; CD4, pink; MHC α chain, gray; MHC β chain, yellow; MBP peptide, red. On the left side of the figure, the C termini of the extracellular portions of the α and β chains of TCR MS2-3C8, as defined in the crystal structure, are indicated by blue and green spheres, respectively. On the right side of the figure, the C terminus of the extracellular portion of CD4 in the complex with MS2-3C8 and HLA-DR4 is marked by a pink sphere labeled MS2-3C8. The right side also shows the predicted position of the C terminus of CD4 in hypothetical ternary complexes constructed using other TCR–pMHC class II structures [human: HA1.7 (PDB ID code 1JH8), Ob.1A12 (PDB ID code 1YMM), 3A6 (PDB ID code 1ZGL), E8 (PDB ID code 2IAM), Hy.1B11 (PDB ID code 3PL6); mouse: B3K506 (PDB ID code 3C5Z), 2W20 (PDB ID code 3C6L), YAe62 (PDB ID code 3C60), 21.30 (PDB ID code 3MBE)]. In each case, the C terminus of CD4 is marked by a colored sphere labeled with the name of the corresponding TCR. The TCR–pMHC–CD4 complexes were modeled by superposing each TCR–pMHC class II structure onto the MS2-3C8–MBP–DR4–CD4 complex through the C domains of the TCRs. In addition, CD4 was assumed to engage mouse MHC class II molecules in the same manner as HLA-DR4, based on the structures of human CD4 D1–D2 bound to mouse I-Ak (12) and of human CD4 D1–D2 bound to HLA-DR1 (27). With the exception of TCR Ob.1A12, the C termini of CD4 in the modeled TCR–pMHC–CD4 complexes form a cluster that includes the C terminus of CD4 in the MS2-3C8–MBP–DR4–CD4 complex. Variability in the position of the CD4 membrane anchor point results from differences in the docking topology of the TCR–pMHC structures. Based on mutational analysis (31, 32), CD3εγ and CD3εδ would be located inside the TCR–pMHC–CD4 arch, facing CD4.
Although the TCR–pMHC–CD8 complex is probably not as conformationally constrained as the TCR–pMHC–CD4 complex, there is evidence that the CD8 stalk may not be as flexible as generally believed, because of O-glycosylation at multiple sites in CD8α and CD8β (39). Biophysical studies of mucins have shown that O-glycans stiffen polypeptides through steric interactions between peptide-linked N-acetylgalactosamine residues and adjacent peptide residues (40, 41). Similarly, O-glycans in the CD8 stalk polypeptides were found to reduce the overall extension of the stalk from a theoretical maximum of 3.4 Å per residue to 2.6 Å per residue, suggesting rigidification (39, 42). Hence, O-glycosylation may limit the mobility of the CD8 head group that binds MHC class I, thereby imposing constraints on the orientation of CD3 relative to CD8-bound Lck, as in the TCR–pMHC–CD4 complex.
At the same time, some flexibility must exist within the overall signaling complex to accommodate variations in TCR–pMHC docking geometry that alter the location of anchor points for TCR and CD4 on the T-cell surface (Fig. 5). Given the relative rigidity of the CD4 ectodomain (Fig. 3), this flexibility most likely resides in interactions involving the cytoplasmic tails of CD3 and CD4 with Lck, which can adopt multiple conformations (43). We conclude that the diagonal docking orientation of αβ TCRs reflects not only genetically encoded interactions with MHC but also the requirement to form a ternary complex with CD4 or CD8 that is geometrically competent to deliver a maturation signal to double-positive thymocytes during T-cell selection.
Materials and Methods
Protein Production and Purification.
Soluble TCR MS2-3C8 and MBP–HLA-DR4 were prepared by in vitro folding from inclusion bodies produced in Escherichia coli as described (28). For affinity maturation of CD4 by yeast surface display, 12 residues in the D1 domain (positions 35, 40, 42–48, 59, 60, and 63) were mutated by overlap PCR with degenerate primers to generate a CD4 D1–D2 mutant library of 4 × 107 clones (27). The CD4 library was displayed on the surface of yeast by N-terminal fusion to agglutinin protein Aga2p and sorted by flow cytometry with HLA-DR1 tetramers or monomers to isolate CD4 mutants with increased affinity. One of these mutants, containing two substitutions in CD4 D1 (Gln40Tyr/Thr45Trp), was produced as a full-length ectodomain. Thus, CD4 D1–D4 (residues 1–363) was fused to the gp67 secretion signal sequence of baculovirus expression vector pAcGP67-B (BD Biosciences) with a C-terminal FLAG tag (DYKDDDDK) and expressed in Sf9 insect cells (Invitrogen). In a typical preparation, 1 L of Sf9 cells at 1.6 × 106 cells/mL in Sf-900 II SFM medium (Invitrogen) was inoculated with 12 mL of recombinant baculovirus at 4 × 108 pfu/cell. Supernatants were harvested 4 d postinfection and loaded onto an anti-FLAG M2 column (Sigma) for affinity purification. Recombinant CD4 D1–D4 was eluted with 0.1 mg/mL FLAG peptide and further purified using sequential Superdex S-75 and MonoQ columns (GE Healthcare).
SPR Analysis.
The interaction of affinity-matured CD4 with HLA-DR4 was assessed by SPR using a BIAcore T100 biosensor at 25 °C. Biotin-tagged MBP–HLA-DR4 was prepared as described (28) and directionally coupled to a streptavidin-coated BIAcore SA chip. Solutions containing different concentrations of CD4 D1–D4 were injected sequentially over flow cells immobilized with HLA-DR4 or buffer alone as a control. Injections were stopped at 60 s after SPR signals reached a plateau. Equilibrium data were fitted with a 1:1 binding model using BIAevaluation 4.1 software (BIAcore) to obtain the dissociation constant (KD).
Crystallization and Data Collection.
Purified TCR MS2-3C8, MBP–HLA-DR4, and CD4 D1–D4 were mixed in equimolar amounts and concentrated to 13 mg/mL in 10 mM Tris-HCl (pH 8.0), 10 mM sodium acetate (pH 5.2), and 5 mM NaCl. The ternary complex crystallized spontaneously from this protein solution. For data collection, crystals with dimensions up to 0.2 × 0.1 × 0.1 mm were transferred to a cryoprotectant solution containing 25% (wt/vol) polyethylene glycol 4000 and 20% (vol/vol) glycerol, before flash-cooling in liquid nitrogen. X-ray diffraction data were collected to 4.0-Å resolution at beamline 23ID-B of the Advanced Photon Source. The data were indexed, integrated, and scaled with the program CrystalClear (Rigaku). The crystals belong to space group P43212 with unit cell dimensions a = b = 146.2 Å, c = 231.4 Å and one ternary complex molecule per asymmetric unit. Data collection statistics are presented in Table S1.
Structure Determination and Refinement.
The structure of the TCR–MHC–CD4 complex was solved by molecular replacement with the program Phaser (44) using MS2-3C8–MBP–DR4 [Protein Data Bank (PDB) ID code 3O6F] (28) and the first three domains (D1–D3) of CD4 (PDB ID code 1WIO) (15) as search models. The D4 domain of CD4 was positioned into the electron density after rigid body refinement of the solution found with CD4 D1–D3. Initial refinement was performed using the deformable elastic network (DEN)-assisted refinement module in CNS v1.3 (45, 46). The model was then manually built in COOT (47) and further refined using PHENIX (48). Refinement statistics are summarized in Table S1. The final Rwork and Rfree values (23.8% and 30.5%, respectively) at 4.0-Å resolution compare favorably with statistics for 51 structures deposited in the PDB within the last 3 y with resolutions in the range of 3.9–4.1 Å. The average Rwork for these structures is 27.8% and average Rfree is 31.2%.
Supplementary Material
Acknowledgments
We are grateful to S. T. Walsh (University of Maryland) for help with X-ray data collection and to M. Mo for assistance with protein expression. We thank Y. Li and L. Deng for comments on the manuscript and R. Martin (University of Zürich) for TCR clones. This work was supported by National Institutes of Health Grants AI036900 and AI073654 and National Multiple Sclerosis Society Grant RG2747 (to R.A.M.). X.X.W. was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3T0E).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118801109/-/DCSupplemental.
References
- 1.Janeway CA., Jr The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu Rev Immunol. 1992;10:645–674. doi: 10.1146/annurev.iy.10.040192.003241. [DOI] [PubMed] [Google Scholar]
- 2.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 3.Holler PD, Kranz DM. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 4.Xu H, Littman DR. A kinase-independent function of Lck in potentiating antigen-specific T cell activation. Cell. 1993;74:633–643. doi: 10.1016/0092-8674(93)90511-n. [DOI] [PubMed] [Google Scholar]
- 5.Li QJ, et al. CD4 enhances T cell sensitivity to antigen by coordinating Lck accumulation at the immunological synapse. Nat Immunol. 2004;5:791–799. doi: 10.1038/ni1095. [DOI] [PubMed] [Google Scholar]
- 6.Artyomov MN, Lis M, Devadas S, Davis MM, Chakraborty AK. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc Natl Acad Sci USA. 2010;107:16916–16921. doi: 10.1073/pnas.1010568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang N, et al. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity. 2011;34:13–23. doi: 10.1016/j.immuni.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Merwe PA, Cordoba SP. Late arrival: Recruiting coreceptors to the T cell receptor complex. Immunity. 2011;34:1–3. doi: 10.1016/j.immuni.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 10.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wucherpfennig KW, Call MJ, Deng L, Mariuzza RA. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol. 2009;21:590–595. doi: 10.1016/j.coi.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JH, et al. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. Proc Natl Acad Sci USA. 2001;98:10799–10804. doi: 10.1073/pnas.191124098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis SJ, et al. The nature of molecular recognition by T cells. Nat Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- 14.Krogsgaard M, et al. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 15.Wu H, Kwong PD, Hendrickson WA. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 16.Ashish , et al. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J Biol Chem. 2008;283:2761–2772. doi: 10.1074/jbc.M708325200. [DOI] [PubMed] [Google Scholar]
- 17.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 18.Brown JH, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 19.Kuhns MS, et al. Evidence for a functional sidedness to the alphabetaTCR. Proc Natl Acad Sci USA. 2010;107:5094–5099. doi: 10.1073/pnas.1000925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 21.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazza C, Malissen B. What guides MHC-restricted TCR recognition? Semin Immunol. 2007;19:225–235. doi: 10.1016/j.smim.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. A correlation between TCR Valpha docking on MHC and CD8 dependence: Implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 25.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Xiong Y, Kern P, Chang H, Reinherz E. T cell receptor binding to a pMHCII ligand is kinetically distinct from and independent of CD4. J Biol Chem. 2001;276:5659–5667. doi: 10.1074/jbc.M009580200. [DOI] [PubMed] [Google Scholar]
- 27.Wang XX, et al. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex. Proc Natl Acad Sci USA. 2011;108:15960–15965. doi: 10.1073/pnas.1109438108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Li Y, Kerzic MC, Martin R, Mariuzza RA. Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection. EMBO J. 2011;30:1137–1148. doi: 10.1038/emboj.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maekawa A, Schmidt B, Fazekas de St Groth B, Sanejouand YH, Hogg PJ. Evidence for a domain-swapped CD4 dimer as the coreceptor for binding to class II MHC. J Immunol. 2006;176:6873–6878. doi: 10.4049/jimmunol.176.11.6873. [DOI] [PubMed] [Google Scholar]
- 30.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26:357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes RA, et al. The T-cell receptor is a structure capable of initiating signalling in the absence of large conformational rearrangements. J Biol Chem. 2012 doi: 10.1074/jbc.M111.332783. 2012 Jan 19 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casasnovas JM, Springer TA, Liu JH, Harrison SC, Wang JH. Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface. Nature. 1997;387:312–315. doi: 10.1038/387312a0. [DOI] [PubMed] [Google Scholar]
- 35.Moldovan MC, et al. CD4 dimers constitute the functional component required for T cell activation. J Immunol. 2002;169:6261–6268. doi: 10.4049/jimmunol.169.11.6261. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 37.Cherry RJ, et al. Detection of dimers of dimers of human leukocyte antigen (HLA)-DR on the surface of living cells by single-particle fluorescence imaging. J Cell Biol. 1998;140:71–79. doi: 10.1083/jcb.140.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shore DA, Wilson IA, Dwek RA, Rudd PM. Glycosylation and the function of the T cell co-receptor CD8. Adv Exp Med Biol. 2005;564:71–84. doi: 10.1007/0-387-25515-X_12. [DOI] [PubMed] [Google Scholar]
- 40.Shogren R, Gerken TA, Jentoft N. Role of glycosylation on the conformation and chain dimensions of O-linked glycoproteins: Light-scattering studies of ovine submaxillary mucin. Biochemistry. 1989;28:5525–5536. doi: 10.1021/bi00439a029. [DOI] [PubMed] [Google Scholar]
- 41.Gerken TA, Butenhof KJ, Shogren R. Effects of glycosylation on the conformation and dynamics of O-linked glycoproteins: Carbon-13 NMR studies of ovine submaxillary mucin. Biochemistry. 1989;28:5536–5543. doi: 10.1021/bi00439a030. [DOI] [PubMed] [Google Scholar]
- 42.Merry AH, et al. O-glycan sialylation and the structure of the stalk-like region of the T cell co-receptor CD8. J Biol Chem. 2003;278:27119–27128. doi: 10.1074/jbc.M213056200. [DOI] [PubMed] [Google Scholar]
- 43.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 44.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 45.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Schröder GF, Levitt M, Brünger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.