Abstract
The hydrophobin EAS from the fungus Neurospora crassa forms functional amyloid fibrils called rodlets that facilitate spore formation and dispersal. Self-assembly of EAS into fibrillar rodlets occurs spontaneously at hydrophobic:hydrophilic interfaces and the rodlets further associate laterally to form amphipathic monolayers. We have used site-directed mutagenesis and peptide experiments to identify the region of EAS that drives intermolecular association and formation of the cross-β rodlet structure. Transplanting this region into a nonamyloidogenic hydrophobin enables it to form rodlets. We have also determined the structure and dynamics of an EAS variant with reduced rodlet-forming ability. Taken together, these data allow us to pinpoint the conformational changes that take place when hydrophobins self-assemble at an interface and to propose a model for the amphipathic EAS rodlet structure.
Amyloid fibrils were first identified in association with human diseases, but recent discoveries show that the amyloid ultrastructure also contributes to important functions in normal biology (1, 2). In bacteria, fungi, insects, fish, and mammals, amyloid structures perform a wide variety of roles (3). Functional amyloids in the form of fibrillar rodlets composed of class I hydrophobin proteins are found in filamentous fungi. These hydrophobins are small proteins that are secreted as monomers and self-assemble into rodlets that pack to form amphipathic monolayers at hydrophilic:hydrophobic boundaries, such as the surface of the growth medium (4). These proteins are extremely surface active and lower the surface tension of the aqueous growth medium, allowing hyphae to break through the surface and to produce aerial structures (5, 6). Many of these aerial structures subsequently become coated with amyloid rodlets, creating a hydrophobic layer that serves multiple purposes, including conferring water resistance to spores for easier dispersal in air (7), preventing wetting or collapse of gas transfer channels (8), enhancing adherence to waxy surfaces such as leaves during infection of rice plants by Magnaporthe grisea (9), and mediating evasion of the immune system as is observed in Aspergillus fumigatus infections (10).
Hydrophobins are characterized by the presence of eight cysteine residues that form four disulphide bonds, but the hydrophobin family can be further divided into two classes based on the spacing of the conserved cysteine residues and the nature of the amphipathic monolayers that they form (11). Class I, but not class II, hydrophobins form amyloid-like rodlets that are extremely robust and require treatment with strong acid to induce depolymerization. The amphipathic monolayers formed by class II hydrophobins are not fibrillar and can be dissociated by treatment with detergent and alcohol solutions. The soluble, monomeric forms of hydrophobins share a unique β-barrel topology, and all have a relatively large exposed hydrophobic area on the protein monomer surface (4). The diversity in sequence and chain length between members of the hydrophobin family is accommodated in the regions between the cysteines.
The ability of class I hydrophobins to spontaneously self-assemble into amphipathic amyloid monolayers at hydrophobic:hydrophilic interfaces structures has sparked interest in these unique proteins. Hydrophobin coatings adhere tightly to surfaces and reverse their wettability, making them attractive for coating hydrophobic solids such as carbon nanotubes (12, 13) and increasing the biocompatibility of medical implants (14). In order to understand the multiple roles played by class I hydrophobin monolayers in fungal biology and to exploit potential biotechnological applications, we have focused on delineating the molecular structure of the polymerized amyloid form of the hydrophobin EAS from Neurospora crassa. We have used site-directed mutagenesis and peptide inhibition assays to identify the region of EAS that forms the spine of the amyloid structure in the polymerized form. Surprisingly, this region does not lie on the edge of the β-barrel or in the long, unstructured Cys3–Cys4 loop, two possibilities suggested by our initial determination of the structure of the EAS monomer (15). Instead, the key region driving intermolecular association lies between cysteines 7 and 8, and this region can be transplanted into a class II hydrophobin to render it amyloidogenic. The structure and solution dynamics of a mutant form of EAS give insight into the structural rearrangement that accompanies rodlet formation. We have applied these experimental data to generate a molecular model for the EAS rodlet structure, demonstrating how the cross-β amyloid structure can arise through a simple conformational change in the Cys7–Cys8 region of EAS. In this model the bulk of the protein is accommodated on the periphery of the cross β-spine in a repeating arrangement that gives rise to the amphipathic nature of the functional amyloid monolayer.
Results
Prediction of Aggregation-Prone Regions in EAS.
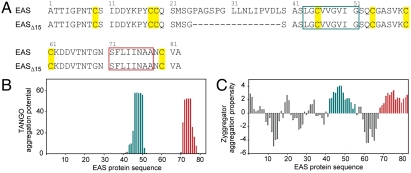
Several algorithms have been developed with the goal of predicting regions of protein sequence that have a high propensity for forming amyloid-like β-sheet polymeric structures. Given the evidence from X-ray fiber diffraction, electron microscopy, and Congo red and Thioflavin-T (ThT) binding that rodlets formed by class I hydrophobins are amyloid-like (15, 16), we analyzed the amino acid sequence of EAS using several of these algorithms. TANGO (17) identified two potential aggregation-prone regions in EAS: residues L43–G51 and S71–A78 (Fig. 1 A and B). Submitting the EAS sequence for Zyggregator (18) analysis returned two regions that overlapped with the Tango prediction: residues S42–G55 and T68–A82 (Fig. 1C). The Waltz algorithm (19) predicted either both of these regions (using the high specificity setting) or only the N70–N76 region (using the best overall performance setting). Use of the 3D profile method implemented by ZipperDB (20) identified the regions G44–V49 and F72–A77 as having the lowest energies and composite scores, which is indicative of high amyloid-forming potential.
Fig. 1.
(A) Sequence of EAS and EASΔ15. Numbering is according to residues in mature, full-length EAS. Regions common to both TANGO and Zyggregator predictions are boxed in green (V43–I51) and red (N71–N78). Cys residues are highlighted in yellow. (B and C) Aggregation potential within the EAS sequence, as predicted by TANGO and Zyggregator algorithms, respectively, and colored as in A.
Identification of Residues Critical for Rodlet Formation.
In order to test the validity of the predictions, 12 single-point mutants were produced by site-directed mutagenesis. In each mutant, a single residue in the V46–I50 or N70–N76 region was mutated to glycine, because this residue has a low propensity for β-sheet formation (21). The mass of each mutant was verified by mass spectrometry and folding was investigated by one-dimensional 1H NMR spectroscopy. Most mutants can be refolded to a native wild-type-like monomer form (see Fig. S1), although several appear to contain a larger proportion of unstructured chain, judging from the increase in intensities of 1H signals in regions of the spectra that typically correspond to disordered polypeptide. The 1H NMR spectra of the V46G and I50G mutants indicated that these proteins did not fold correctly. Residues V46 and I50 lie very close to Cys4 and Cys5, so it is possible these mutations have interfered with disulfide bond formation, which is critical for the core hydrophobin β-barrel. These proteins were not investigated further, but additional mutants, V46I and I50A, were produced and these were able to refold correctly in vitro (Fig. S1).
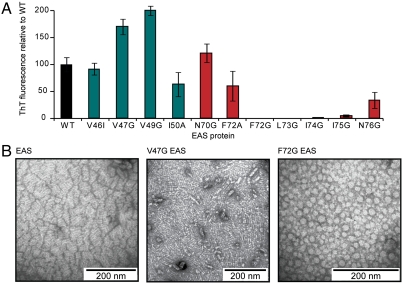
The ability of the mutants to self-associate and to form ordered amyloid rodlets was investigated using a ThT fluorescence assay. Self-assembly of EAS monomers into the β-sheet-rich rodlet form that binds ThT was induced by agitation of the solution with a vortex mixer. This process promotes self-assembly by continuously creating new air:water interfaces that trigger intermolecular association and amyloid formation. As shown in Fig. 2A, substitution of the wild-type residues at any one of the positions between and including F72 and I75 by glycine had a dramatic effect on EAS self-assembly. Under normal assay conditions, wild-type EAS is fully assembled after 2 min, but no rodlets were detected for F72G, L73G, I74G, and I75G after 15 min of agitation. This effect is specific to the glycine substitution, because an additional F72A mutant was indistinguishable from wild type in this assay. In contrast, when the mutant proteins could refold to a WT-like conformation, glycine substitutions in the V46–I50 region did not reduce rodlet formation. The ThT assay result is supported by negative stain transmission electron microscopy (TEM): The morphology of V47G rodlets was very similar to that of wild-type EAS rodlets, although some disturbance of lateral assembly is observed. However, only amorphous aggregates could be observed in the F72G EAS sample after 10 min of agitation (Fig. 2B).
Fig. 2.
Glycine substitutions within the S72–N76 region of EAS, but not the V46-I50 region, affect rodlet formation. (A) The extent of rodlet formation, as measured by Thioflavin-T fluorescence after 15 min agitation of the hydrophobin solution, is unaffected by mutations in the V46–I50 region but glycine substitutions of residues F72–I75 reduce the extent of EAS assembly into rodlets. Replacement of F72 with alanine did not affect rodlet assembly. (B) Negatively stained transmission electron micrographs show that the morphology of wild-type EAS rodlets and those formed by V47G EAS are very similar, while samples of F72G EAS taken from the time period before ThT fluorescence develops do not show fibrillar rodlets; instead, amorphous aggregates are visible.
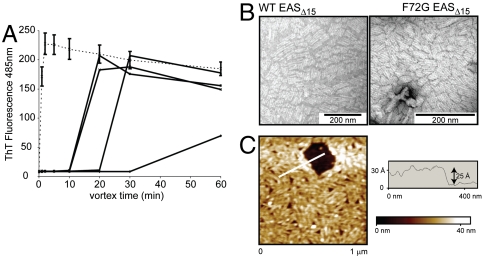
The effect of the F72G mutation was further probed in full-length EAS and also in the context of EASΔ15, a truncated version of EAS that lacks 15 residues from the disordered Cys3–Cys4 loop. EASΔ15 has much improved expression and purification properties compared with wild-type EAS, but it is otherwise indistinguishable in terms of monomer core structure, assembly kinetics, rodlet stability, and morphology (22). We observed that with much extended agitation times, both EAS-F72G and EASΔ15-F72G were able to form rodlets (Fig. 3A). Interestingly, large variations in the length of the lag phase before rodlet detection were observed for F72G on both backgrounds, even between samples from the same protein preparation. Samples removed for analysis by negative stain TEM during the lag phase revealed only amorphous protein aggregates. However, when F72G rodlets do eventually form, as judged by a plateau in ThT fluorescence, they are indistinguishable from those formed by wild-type EAS or EASΔ15 in terms of morphology (Figs. 2B and 3B) and stability (Fig. S2 A and B).
Fig. 3.
Rodlet formation is slowed by the F72G mutation but not prevented entirely; rodlets form after extended periods of vortexing. (A) A comparison of the assembly kinetics for WT EASΔ15 (dotted line, average of four replicates) and F72G EASΔ15 (solid lines, four individual samples illustrated) shows that assembly of the EAS mutant into rodlets is much slower than WT EAS, and there are large differences in the length of the lag phase between the F72G EASΔ15 samples. (B) When the F72G EASΔ15 protein does form rodlets they have the same morphology as WT EASΔ15 rodlets, as judged by TEM. (C) Atomic force microscopy image of EASΔ15 rodlets formed on a mica surface. The height of the monolayer is approximately 25 Å, as determined from the cross-section profile, corresponding to the white line on the monolayer image.
The F72G Substitution Has Effects on Dynamics but Not on Structure.
The F72G mutant was chosen for further study because large aromatic residues have been suggested to be particularly important in stabilization of the β-sheet core of amyloid fibrils (23). In order to determine the effect of the mutation on the solution structure of monomeric EAS and gain insights into the reason for the delay in initiation of rodlet assembly, we determined the solution structure of monomeric EASΔ15-F72G by nuclear magnetic resonance (NMR) spectroscopy.
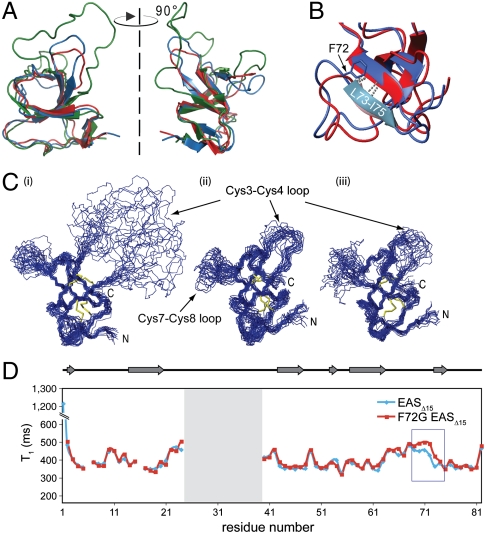
Overall, the structure was of good quality (see Table S1) and demonstrates that the EASΔ15-F72G monomer (Fig. 4 A and B) adopts a fold similar to that of the parent EAS and EASΔ15 (15, 22). The β-barrel core that is constrained by the four disulfide bonds is conserved in all three proteins. Comparison of the 20 lowest-energy conformers from EAS, EASΔ15, and EASΔ15-F72G reveals there are two poorly defined regions: the loop between Cys3 and Cys4 and the loop comprising residues V65–F72 (throughout the manuscript, residues from EASΔ15 are numbered according to their corresponding position in full-length EAS; Fig. 4C). F72 lies at the C terminus of the V65–F72 loop, on the periphery of the β-barrel (Fig. 4B). The conformation of the adjacent L73–I75 β-strand in the EASΔ15-F72G structure is not perturbed by the nearby substitution; in particular, the two backbone hydrogen bonds that connect the L73–I75 β-strand to the β-barrel are still present (Fig. 4B).
Fig. 4.
The F72G mutation does not disrupt the hydrophobin fold but has local effects on the backbone dynamics around the site of the mutation. (A) Overlay of the structures of WT EAS (green), EASΔ15 (blue), and F72G EASΔ15 (red) indicate that the β-barrel core structures are very similar. In WT EAS the Cys3–Cys4 loop is extended and highly mobile. (B) There are minor differences in the structures around the site of the mutation but the L73–I75 βstrand remains hydrogen bonded to the core of the F72G protein. (C) A comparison of the 20 lowest-energy structures calculated for (i) WT EAS, (ii) EASΔ15, and (iii) F72G EASΔ15 shows that the mutant protein adopts a fold similar to that of the parent structures. In all three structures, two disordered regions are observed, namely the segments between Cys3–Cys4 and Cys7–Cys8. (D) 15N relaxation experiments carried out on the EASΔ15-F72G mutant protein demonstrate a small but significant increase in the T1 relaxation time constant for residues around the site of mutation (boxed in blue), indicating an increase in flexibility as a result of the glycine substitution.
Given that the glycine substitution was introduced with the aim of increasing chain flexibility and disfavoring incorporation into a cross-β structure, 15N relaxation experiments were conducted to investigate backbone dynamics of the EASΔ15-F72G mutant. There was a small but significant increase in the T1 relaxation time constant around the site of mutation (Fig. 4D), such that, on average, the T1 values for residues N70–L73 in the mutant are approximately 13% higher than in the parent EASΔ15 protein. While any glycine substitution would be expected to increase local backbone flexibility, it is significant that only substitutions in the region F72–N76 of EAS have an effect on rodlet assembly. Circular dichroism studies show that the secondary structure profile and thermal stability of the WT and F72G EASΔ15 proteins are very similar (Fig. S2 D–F). Together, these data suggest that the most likely reason for the longer lag phase is the delayed formation of a stable nucleus by the F72G mutant, arising from the increased flexibility due to the glycine substitution.
Investigating Intermolecular Contacts with EAS Peptides.
Our results indicate that, of the two regions in EAS predicted to have a high aggregation potential, only residues in the F72–N76 region are critical for rodlet formation in the context of the surface-driven assembly that characterizes hydrophobins. In order to assess the true amyloidogenic potential of these sequences, we synthesized peptides corresponding to the two putative aggregation-prone regions (pep46–52 and pep70–76) and a third from a non-aggregation-prone region (pep10–16) and tested them for amyloid-forming potential (Fig. 5A). When dissolved in water, pep70–76 spontaneously assembled into amyloid fibrils that had typical amyloid morphology when viewed under TEM and gave rise to the characteristic cross-β X-ray fiber diffraction pattern (Fig. 5 B and C). The other two peptides did not assemble into fibrillar structures, even with extended incubation. Surprisingly, even though pep70–76 fibrils have a cross-β substructure, they do not induce ThT fluorescence (Fig. 5D). This may be due to the short length, amino acid composition or individual morphology of the peptide. Similar observations for other peptide fibrils have been made by several laboratories (24, 25).
Fig. 5.
A peptide corresponding to residues N70–N76 in EAS forms amyloid fibrils and interferes with EAS rodlet formation. (A) The regions of EAS corresponding to pep10–16, pep46–52, and pep70–76 are illustrated on the structure of EAS in yellow, turquoise and red, respectively. (B) TEM of fibrils formed by pep70–76 when dissolved in water. (C) X-ray fiber diffraction pattern collected from pep70–76 fibrils shows the reflections at 4.8 Å on the meridian and approximately 10 Å on the equator of the pattern that are characteristic of the cross-β structure of amyloid fibrils. (D) When pep10–16, pep46–52, and pep70–76 were added individually to solutions of EAS to final molar ratios of 1∶1 or 1∶5 [EAS∶peptide] and the solutions were agitated to initiate rodlet formation, only pep70–76 inhibited assembly of EAS into rodlets. (E) Addition of pep70–76 (ratio of EASΔ15∶pep 5∶1) affected the morphology of the EASΔ15 rodlets, such that they were shorter than seen in EASΔ15-only preparations and lacked the ordered lateral assembly of rodlets. (F) A peptide derived from the corresponding Cys7–Cys8 region of the hydrophobin SC3 also spontaneously forms fibrillar structures.
We also carried out ThT assays on EAS in the presence of these short EAS-derived peptides. Because none of the peptides, in either the monomeric or fibrillar form, induces ThT fluorescence, any ThT fluorescence induced in agitated EAS:peptide mixtures must arise solely from EAS rodlets. When pep10–16 and pep46–52 were vortexed with EAS, no measureable change in either the rate or extent of the development of ThT fluorescence was observed (Fig. 5D). However, a mixture containing a 1∶1 molar ratio of pep70–76 and EAS gave significantly less ThT fluorescence under rodlet assembly conditions than EAS alone (30% reduction; p < 0.05; Student’s t test), and an even greater reduction was observed with a fivefold molar excess (60% reduction; p < 0.01). A similar reduction in ThT signal was observed for a pep70–76:EASΔ15 mixture.
The morphology of the rodlets formed in the presence of pep70–76 was examined by TEM. Fibrillar structures could be observed in the vortexed pep70–76:EASΔ15 (5∶1) mixture, but these were generally shorter than pure EAS rodlets and lacked the ordered lateral packing (Fig. 5E). No long fibrils (of the type observed in Fig. 5B) were present in the pep70–76:EASΔ15 mixtures; confirming that pep70–76 interacts directly with EAS in these mixtures and is not available to self-assemble into peptide fibrils.
In order to probe the generality of our finding, we used Waltz (19) to analyze the amyloid-forming potential of the sequence of SC3, a class I hydrophobin from Schizophyllum commune that has been demonstrated to form rodlets (26). This indicated that, as found for EAS, a segment between Cys7–Cys8 had a high aggregation potential. We synthesized a peptide corresponding to this region of SC3 (FNGLINI). This peptide formed fibrils spontaneously in solution (Fig. 5F), suggesting that the presence of an amyloidogenic segment is likely to be a necessary feature of all Class I hydrophobins.
Converting a Class II Hydrophobin into a Class I Hydrophobin.
A key difference between class I and class II hydrophobins is that only class I hydrophobins form amyloid-like rodlets. To test the hypothesis that the presence of an accessible amyloidogenic segment gives rise to rodlet-forming ability, we produced two chimeric hydrophobins: NChi2, in which the Cys7-Cys8 loop from EAS was grafted into the corresponding location in NC2 (a class II hydrophobin from N. crassa), and EChi2, in which the Cys7-Cys8 segment from NC2 replaced residues N70–N79 in EAS (Fig. 6A). The presence of the EAS sequence transformed NC2 into a rodlet-forming hydrophobin, whereas the presence of the NC2 sequence rendered EAS non-rodlet-forming (Fig. 6B). Rodlet formation by NChi2 was surface-dependent and required conditions of extended agitation at low pH and raised temperatures, conditions where wild-type EAS forms rodlets rapidly but NC2 and EChi2 do not. Variation in the length of the lag phase was observed in the NChi2 samples, but once formed, the chimera assemblies display the characteristic hydrophobin rodlet morphology (Fig. 6C).
Fig. 6.
The chimera that has the EAS sequence between Cys7 and Cys8 forms ThT-positive rodlets. (A) Sequences of NC2, EAS, EChi2, and NChi2. (B) Agitation of WT EAS, NC2, NChi2, and EChi2 at pH 2.5 and 45 °C results in formation of ThT-positive rodlets by WT EAS (solid black line) and NChi2 (dotted black line) but not by the class II hydrophobin NC2 nor the EChi2 chimera (solid gray lines). Variation in the length of the lag phase was observed among replicate samples of NChi2. Three replicates each are presented. (C) TEM of the NChi2 chimera sample produced in the ThT assembly assay shows that the assemblies formed by this chimera display the characteristic hydrophobin rodlet morphology.
A Model for the EAS Hydrophobin Rodlet Structure.
Hydrophobin rodlet formation is accompanied by an increase in β-sheet structure (11, 16) (Fig. S2G). The results presented here suggest the S71–I75 region directly forms part of the cross-β core that is generated upon rodlet formation. In the monomer structure of EAS, this region is hydrogen bonded to the β-barrel and does not appear readily accessible for intermolecular β-sheet formation (Fig. 4B). In order to determine whether the formation of such a cross-β core could be compatible with the monomer structure of EAS, we carried out molecular docking simulations using the data-driven docking program HADDOCK (27, 28). Different sets of intermolecular hydrogen bond restraints between neighboring monomers over residues S71–I75 in both parallel and antiparallel arrangements were tested. Residues A1–T2 and those between Cys3 and Cys4, as well as those between Cys7 and Cys8, were defined as flexible in the docking procedure because of their observed flexibility in the NMR structures or involvement in, or proximity to, the identified assembly critical region.
These calculations indicate that the monomer structure of EAS is able to undergo a relatively straightforward conformational change that exposes the amyloidogenic region for intermolecular assembly while maintaining all four disulfide bonds (Fig. 7B). An antiparallel arrangement of monomers is favored because this minimizes the steric clashes between the remaining bulk of the protein (Fig. 7 A and C and Fig. S2C). Although hydrogen bond restraints were only applied to residues in the “trailing” part of the loop (residues S71–I75) in early models, the “leading” part of the loop (residues D64–T68) appeared to be positioned such that hydrogen bonds between adjacent monomers are possible. In addition, two Thr residues are located at positions 66 and 68. Notably, threonine is a residue that is commonly found in β-sheets. In subsequent runs hydrogen bond and dihedral angle restraints for this leading part of the loop were added. This generated a β-arch, with residues D64–T68 forming the leading strand and residues S71–I75 making up the trailing strand (Fig. 7A, Right). In the docked model the side-chains of residues T66, N67, and T68 align and stack very well across adjacent monomers, such that N67 could potentially participate in interstrand side-chain hydrogen bond interactions (Fig. 7D). Asparagine and glutamine ladders are believed to add extra stability to certain amyloids, in particular to some other functional amyloids (29, 30). They are found in parallel β-helix proteins (31) and in the β-solenoid structure of the fungal prion amyloid formed by Het-S (32). Addition of restraints to incorporate an Asn ladder along the rodlet length reduced the total energy of the system by approximately 10%, suggesting that this interaction may indeed be present in EAS rodlets (SI Text).
Fig. 7.
EASΔ15 rodlet model from HADDOCK calculations showing packing of EAS monomers into the amphipathic cross-β structure. (A) Five lowest-energy structures of EASΔ15 hexamer. (B) Overlay of the NMR structure of EASΔ15 (red) with the middle structure in the final HADDOCK model (blue). Conformational change of the Cys7–Cys8 loop is indicated by arrow. Disulphide bonds are shown as yellow sticks. (C) Lowest-energy structure of EASΔ15 hexamer shown as ribbon in the same orientation as middle panel of A. Monomers are coloured and black spheres denote positions of Cα atoms in the Cys3–Cys4 loop. (D) Close up view of the β-spine showing the possible Asn ladder. N67 side-chain oxygen and HNδ atoms are denoted as red and blue spheres, respectively. Hydrogen bonds are indicated by dotted blue lines. (E) Ribbon diagram of EASΔ15 hexamer in gray showing packing of side chains in the β-spine core and location of charged residues. Side chains of F72, L73, and I74 are shown as purple sticks, N67 as green sticks and T66 and T68 as orange sticks. Red and blue spheres denote positions of Cγ and Cε atoms of Asp and Lys side chains, respectively. Horizontal line illustrates that the majority of charges are located on the “bottom” half of the structure (corresponding to the hydrophilic face). Orientation of structure is a 180° rotation through center of the page with respect to right panel of A. (F) Illustration showing how the hexamer model may be stacked end-to-end to give rodlets of varying lengths.
In this hypothetical rodlet model, the spine of the cross-β structure is a pair of antiparallel β-sheets formed by S71–I75 and D64–T68 and the structure has a depth of approximately 2.7 nm. This dimension is consistent with direct measurement of the EAS monolayer thickness by AFM. When EAS is allowed to self-assemble from a dilute solution onto an atomically flat, fresh mica surface and imaged by AFM, the distinct rodlet morphology is visible and the height profile clearly shows that the rodlet structure has a thickness of approximately 2.5 nm (Fig. 3C). We conclude therefore that the EAS rodlet layer is one-monomer thick. Apart from the conformational change in the Cys7–Cys8 loop, the hydrophobin fold is essentially maintained (including preservation of the four disulphide bonds). The bulk of the protein is accommodated along both sides of the β-spine (Fig. 7). The width of the rodlet generated in this way is approximately 65 Å, which agrees very well with the measured width of approximately 61 Å for EASΔ15 rodlets (22). The amphipathic nature of the rodlets is also maintained in this model: In particular, T66 and T68 are located on one side of the structure, together with most of the charged residues from the remainder of the protein, whereas F72, I74, and other mostly hydrophobic residues are located on the diametrically opposite face (Fig. 7E).
To test whether N67 is important for rodlet assembly and/or stability, a possibility suggested by this model, we generated the EASΔ15-N67G mutant and followed its rodlet assembly by ThT fluorescence assay. However, this mutation did not affect the rate of rodlet assembly nor rodlet stability. If this Asn ladder is present in EAS rodlets, it is possible that its contribution is small in the context of the extended network of hydrogen bonding that exists along both faces of the cross-β spine.
Discussion
While the amyloid characteristics of rodlets give a starting point for understanding the formation of fungal hydrophobin amyloid, there is much to learn about the triggers for the conformational change involved in rodlet formation, the sequence segments that are critical for assembly and the molecular structure of the functional monolayers.
Our mutagenesis studies indicate that glycine substitutions only within the F72–N76 segment of EAS and not elsewhere in the protein have a significant effect on rodlet assembly, identifying this region as the critical amyloidogenic segment. Our results demonstrate that the F72–I75 region directly forms the core of the cross-β structure in EAS hydrophobin functional amyloid rodlets. Not only can a peptide encompassing the sequence of this region form amyloid fibrils on its own, but when this sequence is transplanted into a class II hydrophobin, it confers rodlet-forming ability. Significantly, inclusion of pep70–76 in EAS assembly assays reduces the extent of EAS rodlet assembly and disrupts the fibrillar morphology and packing within the monolayer. This finding provides a starting point for the design of inhibitors that can reduce rodlet polymerization, which is an important step in fungal pathogenesis.
We previously proposed that EAS rodlet assembly might involve the monomeric β-barrels stacked head-to-tail at the interface, with or without involvement of the Cys3–Cys4 loop (15). However, we have subsequently demonstrated that the Cys3–Cys4 loop is not required for assembly (22). In addition, although several different amyloid prediction algorithms identified two regions of high fibril-forming potential in EAS, our results demonstrate that in the context of the intact protein and surface-driven functional amyloid assembly, only the region F72–I75 within the Cys7–Cys8 loop is central to EAS rodlet formation.
Goldschmidt et al. highlight the importance of conformational freedom for the amyloidogenic segment to interact with other molecules (20). In the solution structure of EAS, the Cys7–Cys8 bond tethers this loop at its base and the L73–I75 region (a short two-residue β-strand) links it to the edge of the β-barrel by two hydrogen bonds. Therefore, the only way in which the F72–I75 sequence can form the β-spine of an amyloid-structured rodlet is for a conformational change to take place. This does not happen spontaneously in solution, even at the high protein concentrations used for NMR structure determination, but requires association with a hydrophobic:hydrophilic interface. Surface tension is critical for triggering this conformational change, which may be analogous to the phenomenon of surface-induced denaturation of proteins (33) but is a functional requirement for Class I hydrophobins (34).
The structures of EAS, EASΔ15 and EASΔ15-F72G monomers all suggest that residues T66–S71, in the N-terminal half of the Cys7–Cys8 loop, are highly mobile in solution on the nanosecond time scale (15, 16). Therefore, it is likely that a conformational change involving a larger proportion of the Cys7–Cys8 loop can take place upon contact with a suitable interface. In the crystal structures of the class II hydrophobins HFBI and HFBII solved in the presence of detergent, the Cys7–Cys8 loop in some of the monomers is observed in an extended conformation (35, 36). While these class II hydrophobins do not form rodlets and the sequence of this corresponding region in the class II hydrophobins is not predicted to be amyloidogenic, the observation does indicate that this region of the protein is inherently mobile in response to a hydrophobic environment. In contrast, the region V46–I50 in the EAS protein, while predicted by four algorithms to be amyloidogenic, is flanked by Cys4 and Cys5, which form disulfide bonds with Cys3 and Cys2, respectively. The network of disulfide bonds imposes tight structural constraints on this segment. In addition, we did not find that the peptide corresponding to this region formed fibrils in vitro when dissolved in water, in spite of its predicted amyloid-forming potential.
Substitution of residues in the amyloidogenic F72–I75 region by glycines dramatically increases the lag phase for rodlet assembly. Studies on other proteins and polypeptides have indicated that amyloid formation generally follows a nucleation-dependent polymerization mechanism, with the length of the lag phase dependent on the characteristics of the association equilibria that lead to formation of the nucleus (37, 38). Our data indicate that this is also true for EAS rodlet formation. Glycine residues can sample a much wider range of ϕ and ψ angles than other residues, allowing increased local dynamics, and this is indeed observed around the mutation site in EASΔ15-F72G. In the glycine mutants with an extended lag phase, the formation of a stable nucleus is slowed but not prevented and, when rodlets do form, they are indistinguishable from wild-type rodlets in terms of morphology and stability.
While most amyloid structures determined to date have demonstrated a parallel arrangement of monomers (39–41), important characteristics of these functional hydrophobin rodlets support the antiparallel arrangement presented in this model. Critically, the antiparallel arrangement of monomers maintains the dimensions and amphipathic nature of the rodlet monolayer that is characteristic of these fungal assemblies and fundamental to their biological functions. A model with a parallel arrangement of the Cys7–Cys8 loop was generated, but the barrel of the protein could not be accommodated without severe steric clashes from overlapping adjacent monomers, and the docked structures were not consistent with the observed straight rodlet morphology (Fig. S2C). Antiparallel arrangements have been predicted to form amyloid zipper structures (39) but to be rare because of sequence constraints (42). An antiparallel arrangement has been seen in fibrils formed by the Iowa mutant of Aβ1–40 under certain conditions (43), and recent models for RNase A, cystatin, and β2-microglobulin amyloid fibrils that involve domain swapping suggest that in the more complex situation of amyloid formation by segments within a mostly folded protein, antiparallel sheets may form steric zippers (44–46). The model we present for EAS rodlets is similar to the antiparallel, face-to-face steric zipper, as proposed by Sawaya and colleagues (39), but the sheets are composed of different sequences.
Furthermore, this model is compatible with the effects we observed in vortexed EAS:pep70-76 mixtures—namely, reduction in fibril length and disturbance of lateral packing compared with EAS rodlets alone. Lateral packing of the rodlets is required to form a monolayer, and the model predicts that the lateral assembly of rodlets involves interactions between the remaining bulk of the protein, contacts that the peptide cannot provide. This model would also explain the observation that rodlets composed of EASΔ15 are narrower than those composed of wild-type EAS, given the positioning of the Cys3–Cys4 loop at the outer edges (22) (Fig. 7C).
While it has long been postulated that a conformational change is associated with hydrophobin assembly from solution to rodlet states (11, 26), the critical rodlet assembly region has not been pinpointed before. As proposed for other amyloid-forming proteins (20, 47), a conformational change in the protein monomer is thought to expose a previously buried segment that is prone to cross-β stacking, leaving the bulk of the native fold of the protein largely unchanged. These conformational change models were designed to describe the behavior of proteins that “switch” to an amyloidogenic state due to a localized conformational change under appropriate conditions. The parallels with the functional amyloid formed by hydrophobins are apparent: Hydrophobins also have a nonamyloidogenic state (i.e., the monomeric solution conformation) and an amyloidogenic state (i.e., the conformation adopted at hydrophobic–hydrophilic interfaces).
Materials and Methods
All reagents and chemicals were purchased from Sigma-Aldrich, Astral Scientific, Amyl Media, or Ajax Finechem apart from chromatography grade methanol, which was purchased from Burdick & Jackson. Peptides were synthesized by standard FMOC chemistry using a PS3 Peptide Synthesizer (Protein Technologies) in the School of Chemistry, University of Sydney (pep70–76) or purchased from Genscript (pep46–52, pep10–16, and SC3–pep).
Prediction of Aggregation-Prone Regions and the Effect of Glycine Substitutions on Aggregation.
Four different aggregation prediction programs, Tango (17), Zyggregator (18), Waltz (19), and Profile3D (20) were used to identify putative regions responsible for the self-assembly of EAS into rodlets. Predictions of amyloidogenic regions using Tango were not affected by alterations in pH (3–7), temperature (283–310 K), or salt concentration (0–1 M).
Production of Mutants.
The pHUE-EAS plasmid (15, 48) was used as a template to generate the required EAS mutants via either standard or two-step overlap extension PCR. Synthetic genes encoding NC2 and the chimera were purchased from Genscript and subcloned into the pHUE vector (15). All proteins were expressed and purified as described in Kwan et al. (22). Variant proteins were characterized by mass spectrometry and 1D 1H NMR to verify introduction of desired mutation and correct refolding to native structure.
ThT Fluorescence Assays for Rodlet Assembly.
Lyophilized EAS mutants (25 μg/mL) were incubated with Thioflavin-T (30 μM) in 37 mM Tris, pH 8. Rodlet formation was induced by agitating in 1.5-mL Eppendorf tubes for 15 min at 3,000 rpm. Unvortexed solutions were used as negative controls. Fluorescence measurements were carried out as described previously (34) except the excitation and emission slit widths were set to 5 nm. Intensity at 485 nm was used, and the results are the mean of seven replicates.
For peptide experiments, lyophilized peptides were dissolved in MilliQ™ water (MQW) immediately before use, except for pep70–76, which required 15 min of incubation in a sonicator bath to dissolve. EAS (12 μM) was vortexed for 15 min in the absence or presence of each peptide in a 1∶1 or 1∶5 protein-to-peptide molar ratio. After vortexing, each sample was briefly centrifuged, Thioflavin-T and Tris-HCl at pH 8 were added to 40 μM and 50 mM, respectively, and fluorescence measurements were carried out as described for EAS mutants. Rodlet assembly assays with EAS, NC2, and NC2-chimera were performed in a BMG Polarstar fluorescence plate reader, maintained at 45 °C. Protein concentration was 25 μg/mL in Thioflavin-T (30 μM) and 37.5 mM glycine, pH 2.5.
Transmission Electron Microscopy (TEM).
Fresh samples of proteins were prepared at 0.1 mg/mL in 20% ethanol, or samples from ThT assembly assays were analyzed. Sample preparation and imaging were carried out as described previously (34).
X-Ray Fiber Diffraction.
Droplets of a solution containing peptide fibrils were suspended between the ends of two wax-filled capillaries and allowed to air dry at room temperature to promote fibre alignment. X-ray diffraction patterns were collected using a Cu Kα Rigaku rotating anode source (wavelength 1.5418 Å) and MAR-Research image plate detector.
Atomic Force Microscopy (AFM).
The surface morphologies of the protein films were characterized with a PicoSPM II AFM (Molecular Imaging) in tapping mode at ambient atmosphere. The probes used were NT-MDT NSG10 rectangular cantilevers with a resonant frequency of 190–325 kHz and nominal spring constant of 5.5–22.5 N/m. Analysis of the AFM images was performed using PicoScan Version 5.3.3. A 100 or 200 μL drop of 10 μM hydrophobin solution in water was placed on fresh Mica and rinsed gently with MQW. Fresh mica surfaces from Ted Pella were generated by cleaving layers with adhesive tape.
Structure Determination for EASΔ15 F72G.
All NMR spectra were acquired, processed as described previously (34). Spectra were analyzed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). Sequence specific 1H resonance assignment was obtained using standard COSY, TOCSY, and NOESY spectra (49). 15N chemical shifts were measured directly from the 15N-HSQC spectrum, with reference to a previously assigned EASΔ15 15N-HSQC (15) and confirmed with a HNHA.
Structure calculations were carried out in ARIA1.2 using standard parameters (50) unless otherwise stated. NOESY cross-peak volumes were estimated using the Guassian fit mode in Sparky and ϕ angle constraints derived from a HNHA (51). A total of 100, 20, and 1,000 structures were calculated in iterations 0, 2–7, and 8, respectively. Finally, the 50 lowest-energy structures were subjected to water refinement, and the 20 conformers with the lowest value of Etot were visualized and analyzed as described. Procheck statistics (over residues 2–20, 27–47, and 58–65): 66.7% of residues in most favored regions, 31.2% in allowed regions, 1.2% in generously allowed regions, and 0.9% in disallowed regions (52).
15N T1 Relaxation Measurements.
15N T1 (with T1 delay values of 0.012, 0.08, 0.2, 0.32, 0.4, 0.56, 0.8, 1, 1.5, and 2 sec) values were measured on 15N-labeled EASΔ15 and 15N-labeled EASΔ15 F72G using hsqct1etf3gpsi (T1) with recycle delays of 4 sec. Peak heights were fitted to a single exponential curve using the “rh” function within the Sparky program. 15N T2 and 15N-1H heteronuclear NOE measurements could not be carried out due to low sample concentration and rapid degradation experienced with 15N- EASΔ15 F72G.
Construction of a Model of the Rodlet Structure.
Full details are given in SI Text. In brief, a rodlet segment consisting of six EASΔ15 monomers was modeled using HADDOCK2.1 (27, 28) starting with the lowest-energy structure (PDB ID code 2K6A) in the first round of docking. A total of six rounds of docking were performed. In rounds 2–6, the middle molecule in the lowest-energy structure from the previous round was used as the starting structures. During each round of calculation, residues A1–T2, Q20-G44Δ15 and K62–N79 were defined as flexible residues. F72 was defined as “active” and S71–I75 as “passive.” The following restraints were included: (i) hydrogen bond restraints between neighboring monomers; (ii) β-sheet dihedral angle restraints; (iii) intermolecular Asn ladder restraints between N67 residues; and (iv) dihedral angle restraints to prevent β-sheet twisting over the length of the rodlet segment. The five conformers from round 6 with the lowest Etot were analyzed.
Supplementary Material
Acknowledgments.
We thank Dr. J. Font and Dr. B. Crossett for mass spectrometry at the Sydney University Proteome Research Unit and the staff at the Australian Centre for Microscopy and Microanalysis. We acknowledge the Australian Research Council for funding (LP0776672, DP0879121 and DP1093949). M.S. was supported by a National Health and Medical Research Council RD Wright Career Development Fellowship, V.K.M. was supported by a University of Sydney Vice-Chancellor’s Research Scholarship, and I.M. was supported by an Australian Postgraduate Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 5152 (volume 109, number 14).
Data deposition: The lowest 20 energy structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2LFN), and the full set of restraints in the BioMagResBank, www.bmrb.wisc.edu (accession no. 17765).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114052109/-/DCSupplemental.
References
- 1.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Otzen D, Nielsen PH. We find them here, we find them there: Functional bacterial amyloid. Cell Mol Life Sci. 2008;65:910–927. doi: 10.1007/s00018-007-7404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenwald J, Riek R. Biology of amyloid: Structure, function, and regulation. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Linder MB, Szilvay GR, Nakari-Setala T, Penttila ME. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29:877–896. doi: 10.1016/j.femsre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Lugones LG, Wosten HAB, Wessels JGH. A hydrophobin (ABH3) specifically secreted by vegetatively growing hyphae of Agaricus bisporus (common white button mushroom) Microbiology. 1998;144:2345–2353. doi: 10.1099/00221287-144-8-2345. [DOI] [PubMed] [Google Scholar]
- 6.Wosten HA, Richter M, Willey JM. Structural proteins involved in emergence of microbial aerial hyphae. Fungal Genet Biol. 1999;27:153–160. doi: 10.1006/fgbi.1999.1130. [DOI] [PubMed] [Google Scholar]
- 7.Beever RE, Dempsey G. Function of rodlets on the hyphae of fungal spores. Nature. 1978;272:608–610. doi: 10.1038/272608a0. [DOI] [PubMed] [Google Scholar]
- 8.Whiteford JR, Spanu PD. Hydrophobins and the interactions between fungi and plants. Mol Plant Pathol. 2002;3:391–400. doi: 10.1046/j.1364-3703.2002.00129.x. [DOI] [PubMed] [Google Scholar]
- 9.Talbot NJ, et al. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell. 1996;8:985–999. doi: 10.1105/tpc.8.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aimanianda V, et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 11.Sunde M, Kwan AH, Templeton MD, Beever RE, Mackay JP. Structural analysis of hydrophobins. Micron. 2008;39:773–784. doi: 10.1016/j.micron.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Kurppa K, et al. Controlled hybrid nanostructures through protein-mediated noncovalent functionalization of carbon nanotubes. Angew Chem Int Ed Engl. 2007;46:6446–6449. doi: 10.1002/anie.200702298. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Noncovalently functionalized multi-wall carbon nanotubes in aqueous solution using the hydrophobin HFBI and their electroanalytical application. Biosens Bioelectron. 2010;26:1104–1108. doi: 10.1016/j.bios.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Scholtmeijer K, et al. The use of hydrophobins to functionalize surfaces. Biomed Mater Eng. 2004;14:447–454. [PubMed] [Google Scholar]
- 15.Kwan AH, et al. Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci USA. 2006;103:3621–3626. doi: 10.1073/pnas.0505704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay JP, et al. The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures. Structure. 2001;9:83–91. doi: 10.1016/s0969-2126(00)00559-1. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 18.Tartaglia GG, et al. Prediction of aggregation-prone regions in structured proteins. J Mol Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Maurer-Stroh S, et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 20.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parrini C, et al. Glycine residues appear to be evolutionarily conserved for their ability to inhibit aggregation. Structure. 2005;13:1143–1151. doi: 10.1016/j.str.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 22.Kwan AH, et al. The Cys3–Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity. J Mol Biol. 2008;382:708–720. doi: 10.1016/j.jmb.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 23.Gazit E. Self assembly of short aromatic peptides into amyloid fibrils and related nanostructures. Prion. 2007;1:32–35. doi: 10.4161/pri.1.1.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsbury C, et al. Amyloid fibril formation from full-length and fragments of amylin. J Struct Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 26.de Vocht ML, et al. Structural characterization of the hydrophobin SC3, as a monomer and after self-assembly at hydrophobic/hydrophilic interfaces. Biophys J. 1998;74:2059–2068. doi: 10.1016/s0006-3495(98)77912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 28.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 29.Dueholm MS, et al. Functional amyloid in Pseudomonas. Mol Microbiol. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- 30.Tsai HH, et al. Energy landscape of amyloidogenic peptide oligomerization by parallel-tempering molecular dynamics simulation: significant role of Asn ladder. Proc Natl Acad Sci USA. 2005;102:8174–8179. doi: 10.1073/pnas.0408653102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins J, Pickersgill R. The architecture of parallel beta-helices and related folds. Prog Biophys Mol Biol. 2001;77:111–175. doi: 10.1016/s0079-6107(01)00013-x. [DOI] [PubMed] [Google Scholar]
- 32.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 33.Gidalevitz D, Huang Z, Rice SA. Protein folding at the air-water interface studied with X-ray reflectivity. Proc Natl Acad Sci USA. 1999;96:2608–2611. doi: 10.1073/pnas.96.6.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris VK, et al. Recruitment of class I hydrophobins to the air:water interface initiates a multi-step process of functional amyloid formation. J Biol Chem. 2011;286:15955–15963. doi: 10.1074/jbc.M110.214197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakanpaa J, et al. Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A. Acta Crystallogr D Biol Crystallogr. 2006;62:356–367. doi: 10.1107/S0907444906000862. [DOI] [PubMed] [Google Scholar]
- 36.Kallio JM, Linder MB, Rouvinen J. Crystal structures of hydrophobin HFBII in the presence of detergent implicate the formation of fibrils and monolayer films. J Biol Chem. 2007;282:28733–28739. doi: 10.1074/jbc.M704238200. [DOI] [PubMed] [Google Scholar]
- 37.Jarrett JT, Lansbury PT., Jr Amyloid fibril formation requires a chemically discriminating nucleation event: Studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry. 1992;31:12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- 38.Xue WF, Homans SW, Radford SE. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc Natl Acad Sci USA. 2008;105:8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 40.Maji SK, Wang L, Greenwald J, Riek R. Structure-activity relationship of amyloid fibrils. FEBS Lett. 2009;583:2610–2617. doi: 10.1016/j.febslet.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Shewmaker F, McGlinchey RP, Wickner RB. Structural insights into functional and pathological amyloid. J Biol Chem. 2011;286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kajava AV, Baxa U, Steven AC. Beta arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. Faseb J. 2009;24:1311–1319. doi: 10.1096/fj.09-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiang W, Yau WM, Tycko R. Structural evolution of Iowa mutant beta-amyloid fibrils from polymorphic to homogeneous states under repeated seeded growth. J Am Chem Soc. 2011;133:4018–4029. doi: 10.1021/ja109679q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domanska K, et al. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic beta2-microglobulin variant. Proc Natl Acad Sci USA. 2011;108:1314–1319. doi: 10.1073/pnas.1008560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, Sawaya MR, Eisenberg D. beta-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2011;18:49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 47.Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43:8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wuthrich K, Wider G, Wagner G, Braun W. Sequential resonance assignments as a basis for the determination of spatial protein structures by high resolution proton nuclear magnetic resonance. J Mol Biol. 1982;155 doi: 10.1016/0022-2836(82)90007-9. [DOI] [PubMed] [Google Scholar]
- 50.Linge JP, O’Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 51.Liew CK, et al. Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions. Structure. 2000;8:1157–1166. doi: 10.1016/s0969-2126(00)00527-x. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]