Abstract
The malaria parasite, Plasmodium falciparum, and the human immune system have coevolved to ensure that the parasite is not eliminated and reinfection is not resisted. This relationship is likely mediated through a myriad of host–parasite interactions, although surprisingly few such interactions have been identified. Here we show that the 33-kDa fragment of P. falciparum merozoite surface protein 1 (MSP133), an abundant protein that is shed during red blood cell invasion, binds to the proinflammatory protein, S100P. MSP133 blocks S100P-induced NFκB activation in monocytes and chemotaxis in neutrophils. Remarkably, S100P binds to both dimorphic alleles of MSP1, estimated to have diverged >27 Mya, suggesting an ancient, conserved relationship between these parasite and host proteins that may serve to attenuate potentially damaging inflammatory responses.
Keywords: placental, yeast two-hybrid, surface plasmon resonance, high-throughput screen
Malaria is an infectious disease, the most deadly form of which is caused by the parasite Plasmodium falciparum. P. falciparum infections result in nearly 1 million deaths each year in Africa alone, almost exclusively among young children and pregnant women (1). Immunity to malaria is a complex phenomenon that appears to be acquired in at least two phases. Children living in malaria endemic areas with high P. falciparum transmission have been reported to relatively rapidly acquire the ability to control severe, life-threatening malaria after only a small number of infections (2), a resistance that may be short lived in the absence of continued exposure to P. falciparum. Severe malaria, including severe anemia, cerebral malaria, and respiratory distress, has long been recognized to have many features that are similar to the uncontrolled inflammatory immune responses in sepsis (3). Thus, learning to control severe malaria may be a process of learning to control the innate immune system's inflammatory responses through mechanisms that are relatively short lived. Placental malaria is another example of severe malaria in which acquisition of immunity is rapid, occurring after one or two infections (4). Resistance has been correlated with antibodies (Abs) to a particular parasite adhesion molecule expressed by infected red blood cells (iRBCs), P. falciparum erythrocyte membrane protein 1, VAR2CSA, which promotes sequestration of the iRBCs in the placenta (5). However, a role for the control of inflammation in resistance to placental malaria has not been extensively explored. Immunity to uncomplicated malaria, fever and parasitemia, is not acquired until early adolescence despite intense exposure to P. falciparum from birth (6). Immunity that would protect against P. falciparum infections is rarely acquired such that adults living in malaria endemic areas are seldom sick with malaria but often have asymptomatic P. falciparum infections. This slowly acquired resistance to uncomplicated malaria appears to reflect the gradual acquisition of adaptive immunity over years that allows control of parasitemia. Indeed, passive transfer studies carried out in the early 1960s demonstrated that Abs from adults living in endemic areas were sufficient to control malaria fever and high parasite levels in sick children (7).
This complex interplay between the human host and P. falciparum presumably involves a myriad of molecular interactions between the human immune system and the parasite that have coevolved together. However, remarkably, to date the only interactions that have been described are those between the parasite's hemozoin (8) or hemozoin–DNA complexes (9) and the host's toll-like receptor 9 (TLR9) or the NLRP3 inflammasome (10, 11) and the parasite's DNA and an unknown host receptor through a STING-dependent pathway (12). To identify interactions between P. falciparum and human host immune system proteins, we carried out a screen for interactions between 145 P. falciparum protein fragments and approximately half of the human ORFeome, using a yeast two-hybrid (Y2H) approach (13). One of the interactions identified was between P. falciparum merozoite surface protein 1 (MSP1) and human S100P.
MSP1 is the most abundant protein on the surface of P. falciparum merozoites, the RBC invasive form of the malaria parasite (14). Although a considerable amount has been learned about the structure of MSP1 and its immunogenicity as an antigen in both human and nonhuman primates and in rodents, little is known about its function in malaria. During the blood stage of infection merozoites undergo rounds of invasion of RBCs, replication in RBCs, and RBC rupture and release of new infective merozoites. MSP1 is synthesized and expressed as a GPI-linked protein in complex with two additional MSPs (MSP6 and MSP7) on the surface of the merozoite replicating inside the iRBC (15). Before lysis of the RBC and release of merozoites, MSP1 is proteolytically processed by the P. falciparum protease SUB1 into four fragments named on the basis of their apparent molecular weights (from the N to the C terminus, MSP183, MSP130, MSP138, and MSP142) (16). These fragments are held together through noncovalent interactions and anchored on the merozoites’ surface through a GPI moiety on MSP142. At the point of the merozoite's invasion of a RBC, MSP142 is cleaved by a second P. falciparum protease, SUB2, into a C-terminal fragment, MSP119, which contains the GPI moiety and remains on the invading merozoite's surface, and MSP133, which is shed from the merozoite's surface as part of the larger MSP complex (17). Abs specific for MSP119 block merozoite invasion of RBCs and/or development in the infected RBC in an in vitro assay, suggesting a role for MSP119 in these processes (15). The function of MSP133 or the larger shed MSP complex is not known.
S100s are members of a multigene family of EF hand Ca2+-binding proteins composed of at least 21 members that have been studied in a variety of biological processes (18). Several of the S100 family members mediate inflammatory responses, and because they are released by activated or damaged cells under conditions of stress, they have been referred to as damage-associated molecular pattern (DAMP) proteins (19). S100 proteins have been described to have both proinflammatory and chemotactic activities mediated by their binding to TLRs or receptor of advanced glycation end products (RAGE), a multiligand receptor of the Ig superfamily (20). S100 proteins exhibit tissue- and cell-specific expression patterns. S100P is expressed most highly in human placenta but it is also expressed at significant levels in blood cells and in bone marrow (21). S100P has been reported to activate the proinflammatory NFκB pathway through RAGE in NIH 3T3 cells (22).
Results
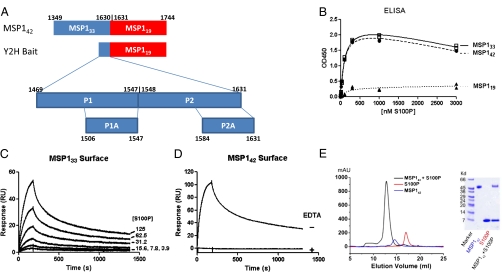
To identify interactions between P. falciparum proteins and proteins functioning in the immune system we carried out a Y2H screen between 10,000 human ORFs used as prey and used as bait 145 P. falciparum protein fragments of the P. falciparum 3D7 strain, predicted to be exposed to the immune system either as secreted proteins or on the parasite or iRBC surface (Dataset S1). One interaction identified was that between S100P and a fragment of P. falciparum MSP1 that contained MSP119 and the 162-residue C terminus of MSP133 (Fig. 1A). By ELISA, using recombinant MSP142, MSP133, or MSP119 from the P. falciparum 3D7 strain to coat the assay plates, S100P bound strongly to both MSP142 and MSP133 but only weakly to MSP119 (Fig. 1B). By surface plasmon resonance (SPR) S100P bound to both MSP133 (Fig. 1C) and MSP142 (Fig. S1A) immobilized on the chip surface in a dose-dependent fashion, but not to MSP119. Together these data indicate that S100P binds to the 162-aa C-terminal fragment of MSP133 that is shed from the merozoite's surface during RBC invasion. The binding of S100P to both MSP142 and MSP133 was of a similar high affinity with an equilibrium constant, KD = 20.0 ± 16.7 nM (calculated from the binding data in Fig. 1C and Fig. S1A). S100P is a calcium-binding protein and its binding to MSP142 was calcium dependent (Fig. 1D).
Fig. 1.
S100P binds with high affinity, in a calcium-dependent fashion, to MSP133. (A) Schematic representation of MSP142 indicating the cleavage products MSP133 (blue) and MSP119 (red). Also shown is the bait used for the Y2H screens that identified S100P as a binding partner for the MSP142 fragment and the synthetic peptides used for characterizing the interaction of S100P with MSP133. Numbers represent residues relative to the MSP1 protein sequence as described previously (30). (B) The binding of His-tagged S100P in ELISAs to MSP133(□), MSP142 (●), or MSP119 (▲) of the 3D7 P. falciparum strain coated onto assay plates and detected using polyclonal rabbit Abs specific for S100P followed by HRP-labeled goat antibodies specific for rabbit Ig. Data were background subtracted and analyzed with a one-site total binding model. (C) SPR sensograms of the binding of S100P at increasing concentrations (3.9–125 nM) to immobilized MSP133, analyzed with the two-state reaction model for curve fitting. Association data were collected for 3 min and dissociation data for 30 min. (D) SPR sensograms of the binding of S100P to immobilized MSP142 in the presence or absence of EDTA. Association data were collected for 3 min and dissociation data for 30 min. (E) Elution profiles of size-exclusion chromatography of 3D7 MSP142 (blue), S100P (red), and a mixture of 3D7 MSP142 (160 μM) and S100P (360 μM) (black). SDS/PAGE analysis of elution peaks was performed by size-exclusion chromatography.
The S100 family members exist as dimers (18) and to characterize the S100P and MSP142 complex in solution, purified S100P expressed in Escherichia coli and MSP142 expressed in insect cells were mixed and analyzed by gel filtration followed by SDS/PAGE of column fractions (Fig. 1E). S100P alone ran as a dimer of ∼18 kDa on the column that resolved as S100P monomers by SDS/PAGE. MSP142 alone ran as a dimer on the column and resolved on SDS/PAGE as a 50-kDa protein, a glycosylated form of MSP142. The MSP142–S100P mixture ran as a major higher molecular weight peak on gel filtration that resolved into 50-kDa and 7-kDa bands by SDS/PAGE. This behavior is consistent with MSP142 and S100P forming a heterodimer of MSP142 and S100P dimers in solution.
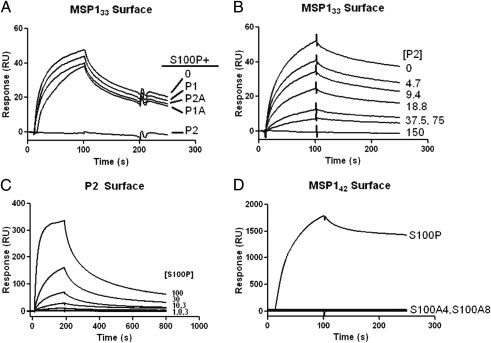
To determine which residues of the C-terminal fragment of MSP133 were sufficient for S100P binding, peptides spanning the 162 residues of the C terminus of MSP133 (Fig. 1A) were synthesized and tested for their ability to block the binding of S100P to MSP133 by SPR. Recombinant MSP133 in solution blocked completely the binding of S100P to MSP133 on the chip surface in a concentration-dependent fashion (Fig. S1B). Of the four peptides synthesized only peptide 2 (P2) blocked S100P's binding to MSP133 (Fig. 2A) and did so in a concentration-dependent fashion (Fig. 2B), indicating that the binding site on MSP1 for S100P is within this 83-residue peptide. The inability of P2A to block the binding of S100P to MSP133 does not necessarily exclude this region from the binding site as P2A alone may not take on the proper conformation for binding to S100P, for example. S100P binds to P2 on the chip surface in a concentration-dependent fashion (Fig. 2C) with a calculated KD = 12.7 ± 13.5 nM, nearly equivalent to the KD of the full-length MSP133 binding to S100P.
Fig. 2.
S100P binding selectivity to MSP133. (A) SPR sensograms of S100P (200 nM) binding to immobilized 3D7 MSP133 as in Fig. 1 in the presence of 200-nM solutions of P1, P1A, P2, or P2A. (B) SPR sensogram of the binding of S100P to 3D7 MSP133 in the presence of increasing concentrations of P2 (0–150 nM). (C) SPR sensogram of the binding of S100P at increasing concentrations (0.3–100 nM) to immobilized P2. (D) SPR sensogram of the binding of S100P, S100A8, and S100A4 to immobilized 3D7 MSP142.
S100P is a member of a family of proteins that show cell-type–specific expression and for some S100s unique receptor-binding specificities. To evaluate the specificity of the interaction between MSP1 and other S100 family members, we tested the binding of two highly related S100 proteins, S100A4 and S100A8 to MSP142 (Fig. 2D) or P2 (Fig. S1C) on the chip surface by SPR. Only S100P bound to MSP142 or P2. The fact that S100A4 and S100A8 do not bind to MSP142 is striking because these proteins are not only highly similar in amino acid sequence to S100P (ranging from 59% to 64% similarly) but also have been shown to bind to RAGE, as has S100P (23).
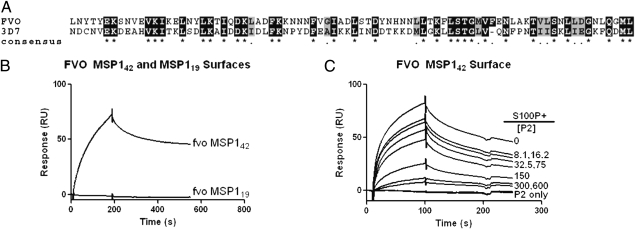
P. falciparum-MSP1 has two major allelic dimorphisms that are estimated to have diverged >27 Mya (Fig. 3A) (24). Both allelic forms are maintained in Africa; however, as yet no function has been assigned to the dimorphic sequences. The binding studies described thus far were with MSP1 from the 3D7 strain, the so-called MAD20 allele. The MSP1 of the FVO strain is a representative of the alternative, K1 allele. By SPR S100P bound to FVO MSP142, but not to FVO MSP119 (Fig. 3B), with a KD = 14.0 ± 7.8 nM equivalent to the KD of 3D7 MSP142 binding to S100P. Moreover, the 3D7 P2 competed for the binding of S100P to FVO MSP142 (Fig. 3C). These results indicate that the ancient dimorphic alleles of P. falciparum have maintained the ability to bind to S100P.
Fig. 3.
S100 binds equally well to the dimorphic forms of MSP142. (A) Alignment of the sequences encompassing the P2 regions of FVO and 3D7 MSP133. P2 has one additional asparagine residue at the C terminus, which is not shown. (B) SPR sensogram of S100P (125 nM) binding to immobilized FVO MSP142 or immobilized FVO MSP119. (C) SPR sensograms of S100P binding to FVO MSP142 in the presence of increasing concentrations of P2 (0–600 nM).
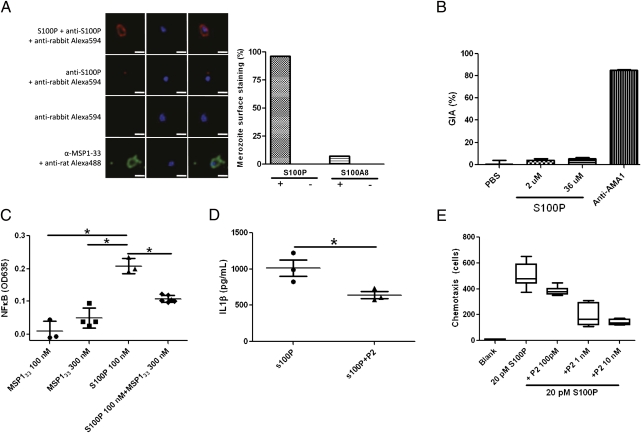
To determine whether S100P bound to MSP1 on the surface of merozoites we incubated free P. falciparum merozoites with S100P detected using S100P-specific Abs and fluorescently labeled secondary Abs and found that S100P bound to >98% of merozoites (Fig. 4A). In contrast, the binding of the highly related S100A8 to merozoites was nearly undetectable. Thus, the selectivity of binding of MSP1 to S100P vs. S100A8 observed in SPR was recapitulated in the binding to merozoites. Because binding of S100P to the surface of merozoites could potentially block RBC invasion, we tested the effect of S100P in an assay that measures the RBC invasion and intracellular development of P. falciparum merozoites in vitro. Even at the highest concentration tested, 36 μM, S100P had no effect on parasite invasion or development (Fig. 4B) whereas in controls Abs specific for merozoite apical membrane antigen 1 (AMA1) blocked parasite growth by >80% (Fig. 4B).
Fig. 4.
MSP133 blocks proinflammatory and chemotaxis responses in vitro. (A) Confocal microscopy images and quantification of the number of free merozoites stained with S100P, S100A8, or Ab specific for MSP133. Shown are representative images of merozoites stained with S100P detected with S100P-specific rabbit Abs followed by Alexa594-conjugated rabbit Ig-specific Abs or with the detecting Abs alone in the absence of S100P or MSP133-specific rat Abs detected using Alexa 488-conjugated rat Ig-specific Abs. Approximately 100 merozoites were counted for each condition and the percentage that bound S100P or S100A8 using the appropriate primary and secondary Abs is given. In controls, the percentage of stained merozoites in the presence of the primary and secondary Abs alone is also given. (B) S100P (2–36 μM) was analyzed in a growth inhibition assay as previously described. Results shown are the mean ± SEM from pooled data from three independent experiments. Rabbit IgG specific for a combination of 3D7 and FVO AMA1 and PBS were used as positive and negative controls, respectively. (C) MSP133, S100P, and a mixture of MSP133 and S100P were tested for their ability to activate NFκB in the monocyte THP1-XBlue reporter cell line. NFκB activation was determined by SEAP reporter gene assay, using the colorimetric detection method. Bars represent mean ± SEM. (D) S100P and S100P in the presence of P2 were tested for their ability to stimulate IL-1β secretion from human PBMCs in vitro. Results shown are the mean ± SEM of three donors (n = 3). (E) The ability of S100P to stimulate chemotaxis of HL-60 cells in vitro was tested in the presence of increasing concentrations of P2.
Many members of the S100 family have been shown to induce inflammatory responses in cells of the innate immune system, including monocytes, macrophages, and neutrophils, through TLRs and RAGE (23). S100P has been reported to activate NFκB through RAGE in NIH 3T3 cells (22). S100P induced NFκB activation in the THP-1 NFκB reporter cell line (Fig. 4C) in a dose-dependent fashion (Fig. S1D). MSP133 itself had no effect on NFκB expression in the THP-1 cells but significantly blocked the induction of NFκB by S100P. In human peripheral blood mononuclear cells (PBMCs), S100P at 100 nM induced the expression of the inflammatory cytokine IL-1β and this effect was partially blocked by MSP133 P2 (Fig. 4D). We also characterized the chemotactic effects of S100P on neutrophils, using a transwell assay. S100P increased the number of migrating cells significantly (Fig. 4E). The effect was blocked by MSP133 P2 in a dose-dependent fashion. Thus, S100P has the ability to induce both inflammatory responses and chemotaxis and these functions are blocked by P. falciparum MSP133.
Discussion
Collectively our data provide evidence that MSP133 that is shed from the merozoite surface as it invades RBCs binds to S100P, a proinflammatory cytokine, and blocks its inflammatory and chemotactic activity. MSP133 bound to S100P in a calcium-dependent, high-affinity, and selective fashion and blocked the ability of S100P to induce NFκB activation and chemotaxis in vitro. These findings suggest that one function of the shed MSP133 is to block inflammatory responses or chemotaxis presumably for the benefit of both the host and the parasite. Although inflammation is necessary for the initiation of adaptive immune responses, hyperinflammation is detrimental to the host and at least indirectly to the parasite. The uncontrolled inflammatory response in severe malaria, which can result in death, is an undesirable outcome for the parasite. It is of interest that the NFκB pathway is also compromised by Plasmodium in the liver stage of the disease (25), although by a very different mechanism. The circumsporozoite protein's nuclear localization signal was shown to block the import of hepatocyte proteins into the nucleus, including NFκB, thus shutting down the NFκB pathway.
One important question raised by the finding presented here is, Where in the host does MSP133 function to attenuate inflammation? The observations that MSP133 binds in a highly selective fashion to S100P, produced in greatest amounts in the placenta, and not to S100A4 or S100A8 produced primarily by macrophages and neutrophils in the blood, suggests that MSP133 may have evolved to selectively block the function of S100P in the placenta. Placental malaria is a particularly lethal disease that can result in the death of the mother and the fetus. Placental malaria is caused by parasites that express one particular PfEMP1, VAR2CSA, at the surface of the iRBC that allows the iRBCs to sequester in the placenta (5). It may be an advantage for the parasite to attenuate inflammation at this site, driving the evolution of the selective binding of MSP133 to S100P. Because S100P, S100A4, and S100A8 are all expressed to some level in leukocytes and in the placenta, the selectivity of MSP1 for S100P may also suggest a nonredundant role for S100P in inducing inflammation. It is also remarkable that S100P has maintained its ability to bind to both P. falciparum MSP1 dimorphic alleles that diverged >27 Mya, suggesting that the interaction of MSP1 with S100P is critical in the P. falciparum–host interaction. The interaction between P. falciparum MSP1 and S100P is one of very few interactions described between the human host's immune system and P. falciparum products and a unique protein–protein interaction, the only other characterized interactions with the human immune system being that with the P. falciparum product of heme detoxification, hemozoin, hemozoin-P. falciparum DNA, or P. falciparum DNA. The interaction described here identified by a Y2H screen suggests that such unbiased screens may prove useful in identifying additional interactions between P. falciparum and the human host's immune system.
Materials and Methods
Materials.
P. falciparum 3D7 MSP133, 3D7 MSP142, FVO MSP142, 3D7 MSP119, and FVO MSP119 and rabbit 3D7 MSP142-specific Abs were obtained from the National Institute of Allergy and Infectious Diseases (NIAID) Laboratory of Malaria Immunology and Vaccinology. Synthetic peptides were made by the NIAID Research Technologies Branch. Human S100P, S100A4, and S100A8 and antibodies were obtained from Abcam.
Yeast Two-Hybrid Screening.
ELISA.
ELISA was performed as previously described (26) with some modifications. In brief, plates were coated at room temperature for 2 h with 50 μL of 3D7 MSP119, 3D7 MSP133, or 3D7 MSP142 protein solution at 5 μg/mL concentration in 1× TBS. Wells were blocked with 5% (wt/vol) BSA in 1× PBS for 2 h and then washed three times with wash buffer (1× TBS, 0.05% Tween 20, 1 mM CaCl2). S100P diluted in dilution buffer [2% (wt/vol) BSA in 1× TBS, pH 7.4, 0.05% Tween 20, and 1 mM CaCl2] was added. After 2 h, plates were washed three times and incubated with 50 μL of polyclonal rabbit anti-S100P antibody (Abcam; ab42288) at 1 μg/mL in dilution buffer. After 1 h, wells were washed three times and then incubated with goat anti-rabbit HRP (Jackson Immunoresearch; 111-036-047) at 1:3,000 diluted in dilution buffer. After 1 h, wells were washed four times with wash buffer and 100 μL of peroxidase substrate (KPL; 5076-11) was added to each well. After 5–10 min the reaction was stopped by adding 100 μL of 1 M phosphoric acid. Colorimetric analysis was performed by measuring the absorbance values at 450 nm.
Growth Inhibition Assay.
The growth inhibition assay (GIA) was performed as described previously (27) with some modification; i.e., S100P was used as a test material, instead of antibody. In brief, the GIA was performed using the late trophozoite and schizont stage of P. falciparum prepared by Percoll gradient and 5% (wt/vol) sorbitol treatment. S100P was tested at 2 and 36 μM in the presence of 1 mM calcium. Forty hours after incubation, parasite growth was determined by a parasite-specific lactate dehydrogenase assay.
Confocal Microscopy.
P. falciparum FVO strain merozoites were isolated from purified schizonts as previously described (31). They were air dried onto coverslips at room temperature (RT) and fixed with ice-cold fixative [2.5% (vol/vol) paraformaldehyde and 0.05% glutaraldehyde in 1× PBS] for 10 min at RT. After washing in PBS (pH 7.5), the cells were incubated with blocking buffer [3% (wt/vol) BSA in PBS, pH 7.5] for 1 h at room temperature. Samples were then incubated with S100P recombinant protein (10 μg/mL in blocking buffer) at RT for 1 h. After washing, S100P was detected with an anti-S100P antibody (1:100) and visualized using anti-rabbit Alexa594 secondary antibody. S100P antibody and anti-rabbit Alexa594 secondary antibody in the absence of S100P protein were used as negative controls. Anti-MSP133 antibody (1:500) was used as a positive control and was visualized using anti-rat Alexa488. Scale bars represent 1 μM. The samples were analyzed on a Leica SP2 confocal microscope and images generated using Bitplane Imaris software. For quantitation, purified merozoites in solution were incubated with recombinant S100P and S100A8 (10 μg/mL in blocking buffer) for 1 h at RT. Samples were washed in PBS to remove any unbound protein and fixed [2.5% (vol/vol) paraformaldehyde and 0.05% glutaraldehyde in 1× PBS] for 10 min at RT and labeled as described above. Mouse anti-S100P and sheep anti-S100A8 antibodies (1:100) along with their respective Alexa488 and Alexa594 secondary antibodies were used to quantify binding efficiency.
NFκB Assay.
NFκB activation was assayed in THP1-XBlue reporter cells (InvivoGen) according to the company's instructions. In brief, cells were grown in RPMI 1640 with 10% (vol/vol) heat-inactivated FBS, 100 μg/mL Normocin, 50 units/mL penicillin, 50 μg/mL streptomycin, and 200 μg/mL of Zeocin. Cells were passaged every 3 d to maintain concentration <2 × 106 cells/mL and used with <20 passages after thawing. Cells were centrifuged at 300 × g for 5 min and washed with fresh medium, and then 5 × 105 cells were plated into 96-well wells and incubated with MSP133, S100P, or a mixture of the two. HKLM cells (107) were used as a positive control and endotoxin-free water as a negative control. Plates were incubated for 24 h and activity was determined in 20 μL of supernatant by using QUANTI-Blue substrate after 8 h of incubation. Secreted Alkaline Phosphatase (SEAP) levels were determined using a spectrophotometer (Molecular Devices SpectraMax Plus 384) at 625 nm.
SPR Analysis.
All SPR experiments were performed using a T100 instrument (Biacore) according to manufacturer's instructions. The Biacore T100 evaluation software was used for kinetic analysis. Sensor CM5, amine coupling reagents, and buffers were purchased from Biacore. Fresh HBS-P (10 mM Hepes, pH 7.4, 150 mM NaCl, 0.005% vol/vol P20 surfactant) amended with 1 mM CaCl2 was used as the running buffer for all SPR experiments.
For kinetic analysis soluble 3D7 MSP133 (20 μg/mL) and 3D7 MSP142 (20 μg/mL) in acetate buffer, pH 5.0, and 3D7 MSP119 in acetate buffer, pH 4.0, were immobilized on a CM5 sensor via amine coupling aiming at 400 Resonance Units (RU) (for MSP142 and MSP119) or 700 RU (MSP133), resulting in final immobilization of 444.5 RU, 412.3 RU, and 668.4 RU, respectively. For end-point analysis a target of 3,000 RU was used, resulting in final immobilization of 3,026.4 RU (MSP142) and 2,947.4 RU (MSP119). Peptides (30 μg/mL) were immobilized on a CM5 sensor via amine coupling as recommended by Biacore. The final immobilized levels were as follows: peptide 1, 356 RU; peptide 2A, 230 RU; and peptide 2, 474 RU. Blank flow cells were used to subtract the buffer effect on sensograms. Kinetic experiments were performed for a contact time of 180 s at a flow rate of 30 μL⋅min−1 at 25 °C. S100P-MSP133-S100P and S100P-peptide complex dissociation was monitored for 1200 s, and the sensor surface was regenerated by a 20 s pulse of 10 mM HCl at 30 μL⋅min−1. After subtraction of the contribution of the bulk refractive index and nonspecific interactions with the CM5 chip surface, the individual association (Ka) and dissociation (Kd) rate constants were obtained by global fitting of the data, using the two-state reaction (conformational change) interaction model in BIAevaluation (Biacore):
These values were then used to calculate the equilibrium constant (KD).
The values of the mean square residual obtained were not significantly improved by fitting data to models that assumed other interactions. Conditions were chosen so that the contribution of mass transport to the observed values of KD was negligible. In addition, the models in the T100 evaluation software fitted for the mass transfer coefficient to mathematically extrapolate the true Ka and Kd.
For endpoint competition experiments, S100P at 100 nM or 200 nM concentration was mixed with MSP133, synthetic peptides encompassing multiple regions of MSP133, or various proteins (S100A4, S100A8, S100A12, Ixolaris, or PACAP) at concentrations varying from 75 nM to 300 nM. Blank flow cells were used to subtract the buffer effect on sensograms.
Chemotaxis Assay.
HL-60 cells (ATCC) were maintained in an undifferentiated state in RPMI 1640 media containing 10% (vol/vol) FBS and 25 mM Hepes at 37 °C, 5% CO2. Cells were differentiated at a density of 4.5 × 105 cells/mL for 6 d in 1.3% (vol/vol) DMSO RPMI complete media, and the status of differentiation was monitored by CD11b staining. Differentiated HL-60 chemotaxis was analyzed in 48-well chemotaxis chambers (Neuroprobe). Lower wells were loaded with 26 μL of test compound at the indicated concentration and RPMI containing 0.1% BSA as a negative control. Differentiated HL-60 cells in RPMI containing 0.1% BSA were added to upper wells and allowed to migrate through a 3.0 μm pore membrane filter for 90 min at 37 °C and 5% CO2. After incubation, the nonmigrated cells were removed from top side of the filter and the filter was fixed and stained with a three-step staining kit (Thermo Scientific; 3313). The total number of migrated cells beyond the lower surface of membrane was counted in a microscopic field.
PBMC Stimulation Assay.
Blood is from anonymized healthy donors and was drawn for research purposes at the National Institutes of Health (NIH) Blood Bank under an NIH institutional review board approved protocol with informed consent. The PBMCs were obtained by density-gradient centrifugation. In separate tubes we prepared solutions of S100P or S100P and Peptide 2 in RPMI 1640 containing 10% heat-inactivated FBS and 1 mM CaCl2 (amended RPMI 1640). The solutions were preincubated for 10 min to allow S100P and the synthetic peptide to bind. Cells were then diluted at 2.5 × 106 cells/mL in amended RPMI. Two hundred microliters of cell suspension, containing 5 × 105 cells, was dispensed on each well of a flat bottom 96-well culture plate according to the experimental design. Cells were incubated at 37 °C and 5% CO2 for 24 h, and supernatants were collected and frozen at −80 °C until further analysis. Cytokines (IL-1α, IL-1β, IL-4, IL-6, IL-8, IL-10, IL-12p70, IFN-γ, TNF-α, TNF-β, MCP-1, and RANTES) were analyzed using a Q-Plex Human Cytokine Kit according to manufacturer's instructions (Quansys Biosciences).
Analytical Size Exclusion Chromatography.
MSP142 was expressed in insect cells as previously described (28). Human S100P was cloned into pET30a vector and expressed and purified as described previously (29). Analytical size-exclusion chromatography of MSP1-S100P complexes was done using a Superdex 200 10/30 column (GE Healthcare). Briefly, S100P and MSP142 were mixed in chromatography buffer [20 mM Hepes (pH7.4), 0.15 M NaCl, 2 mM CaCl2] for complex formation using starting concentrations of 360 μM and 160 μM, respectively. The purity and size-exclusion profile of single components of S100P and MSP142 were analyzed in the same buffer as described above.
Supplementary Material
Acknowledgments
The authors thank Dr. George Makhatadze for sharing reagents, Ms. Julie Kim for logistic support, and Ms. Terri Fantasia for editorial support. This study was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202689109/-/DCSupplemental.
References
- 1.WHO . WHO World Malaria Report. Geneva: WHO; 2008. [Google Scholar]
- 2.Gupta S, Snow RW, Donnelly CA, Marsh K, Newbold C. Immunity to non-cerebral severe malaria is acquired after one or two infections. Nat Med. 1999;5:340–343. doi: 10.1038/6560. [DOI] [PubMed] [Google Scholar]
- 3.Clark IA. Does endotoxin cause both the disease and parasite death in acute malaria and babesiosis? Lancet. 1978;2:75–77. doi: 10.1016/s0140-6736(78)91386-7. [DOI] [PubMed] [Google Scholar]
- 4.Fried M, Nosten F, Brockman A, Brabin BJ, Duffy PE. Maternal antibodies block malaria. Nature. 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- 5.Duffy PE. Plasmodium in the placenta: Parasites, parity, protection, prevention and possibly preeclampsia. Parasitology. 2007;134:1877–1881. doi: 10.1017/S0031182007000170. [DOI] [PubMed] [Google Scholar]
- 6.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: More questions than answers. Nat Immunol. 2008;9:725–732. doi: 10.1038/ni.f.205. [DOI] [PubMed] [Google Scholar]
- 7.Cohen S, McGregor IA, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 8.Coban C, et al. Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J Exp Med. 2005;201:19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parroche P, et al. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc Natl Acad Sci USA. 2007;104:1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shio MT, et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 2009;5:e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostert C, et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS ONE. 2009;4:e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Kadekoppala M, Holder AA. Merozoite surface proteins of the malaria parasite: The MSP1 complex and the MSP7 family. Int J Parasitol. 2010;40:1155–1161. doi: 10.1016/j.ijpara.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Holder AA. The carboxy-terminus of merozoite surface protein 1: Structure, specific antibodies and immunity to malaria. Parasitology. 2009;136:1445–1456. doi: 10.1017/S0031182009990515. [DOI] [PubMed] [Google Scholar]
- 16.Koussis K, et al. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009;28:725–735. doi: 10.1038/emboj.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heizmann CW, Fritz G, Schäfer BW. S100 proteins: Structure, functions and pathology. Front Biosci. 2002;7:d1356–d1368. doi: 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- 19.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: A novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 20.Ebihara T, et al. Differential gene expression of S100 protein family in leukocytes from patients with Kawasaki disease. Eur J Pediatr. 2005;164:427–431. doi: 10.1007/s00431-005-1664-5. [DOI] [PubMed] [Google Scholar]
- 21.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 23.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: An update. Biochim Biophys Acta. 2009;1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Polley SD, Weedall GD, Thomas AW, Golightly LM, Conway DJ. Orthologous gene sequences of merozoite surface protein 1 (MSP1) from Plasmodium reichenowi and P. gallinaceum confirm an ancient divergence of P. falciparum alleles. Mol Biochem Parasitol. 2005;142:25–31. doi: 10.1016/j.molbiopara.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Singh AP, et al. Plasmodium circumsporozoite protein promotes the development of the liver stages of the parasite. Cell. 2007;131:492–504. doi: 10.1016/j.cell.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Calvo E, et al. Aegyptin displays high-affinity for the von Willebrand factor binding site (RGQOGVMGF) in collagen and inhibits carotid thrombus formation in vivo. FEBS J. 2010;277:413–427. doi: 10.1111/j.1742-4658.2009.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malkin EM, et al. Phase 1 clinical trial of apical membrane antigen 1: An asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect Immun. 2005;73:3677–3685. doi: 10.1128/IAI.73.6.3677-3685.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stowers AW, et al. A recombinant vaccine expressed in the milk of transgenic mice protects Aotus monkeys from a lethal challenge with Plasmodium falciparum. Proc Natl Acad Sci USA. 2002;99:339–344. doi: 10.1073/pnas.012590199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, et al. Purification, crystallization and preliminary X-ray diffraction studies of a Ca2+-binding protein, human S100P. Acta Crystallogr D Biol Crystallogr. 2002;58:694–696. doi: 10.1107/s0907444902002342. [DOI] [PubMed] [Google Scholar]
- 30.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1) Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan , et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci USA. 2011;108:13275–13280. doi: 10.1073/pnas.1110303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.