Abstract
The lantibiotic nisin has been used as an effective food preservative to combat food-borne pathogens for over 40 y. Despite this successful use, nisin’s stability at pH 7 is limited. Herein, we describe a nisin analog encoded on the genome of the thermophilic bacterium Geobacillus thermodenitrificans NG80-2. This analog termed geobacillin I was obtained by heterologous expression in Escherichia coli and subsequent purification. Extensive NMR characterization demonstrated that geobacillin I contains seven thioether cross-links, two more than the five cross-links found in nisin and the most cross-links found in any lantibiotic to date. The antimicrobial spectrum of geobacillin I was generally similar to that of nisin A, with increased activity against Streptococcus dysgalactiae, one of the causative agents of bovine mastitis. Geobacillin I demonstrated increased stability compared to nisin A. In addition to geobacillin I, the genome of G. thermodenitrificans NG80-2 also contains a class II lantibiotic biosynthetic gene cluster. The corresponding compound was produced in E. coli, and has a ring topology different than that of any known lantibiotic as determined by tandem mass spectrometry. Interestingly, geobacillin II only demonstrated antimicrobial activity against Bacillus strains. Seven Geobacillus strains were screened for production of the geobacillins using whole-cell MALDI-MS and five were shown to produce geobacillin I, but none produced geobacillin II.
Keywords: posttranslational modification, secondary metabolism, thermophile, antibiotic
Lantibiotics are ribosomally synthesized and posttranslationally modified polycyclic peptides containing thioether bridges (1). The cross-links are made in a two-step process of first dehydration of Ser and Thr residues to the corresponding dehydro amino acids dehydroalanine (Dha) and dehydrobutyrine, and subsequent conjugate addition of the thiol of cysteine (Cys) to the dehydro amino acids. The N terminus of the precursor peptide is termed the leader peptide and is removed in the final step of maturation, whereas the C terminus is designated the core peptide and is converted into the lantibiotic (2). For class I lantibiotics, dehydration and cyclization is carried out by two different enzymes, generically called LanB and LanC, whereas for class II lantibiotics, both reactions are performed by a bifunctional enzyme (LanM). The class I lantibiotic nisin, the most extensively studied member of the lantibiotic family, was first approved for use as a food preservative to combat food-borne pathogens in 1969 and is currently used in over 50 countries (3). Despite this widespread use, very little resistance against nisin has been reported in this field, possibly owing to its mode of action (4). Nisin binds to the pyrophosphate group of lipid II, thereby preventing its use as an essential intermediate in bacterial cell wall biosynthesis (5–7). In addition, the lipid II–nisin complex forms long-lived pores resulting in depolarization of the membrane (8, 9). In comparison to other modes of action, it may be more challenging for a target organism to change the structure of an advanced intermediate such as lipid II that is biosynthesized in 10 steps (10, 11), than to acquire other resistance mechanisms such as efflux pumps and enzyme mutations. These latter mechanisms will not affect nisin because it acts on the outside of the bacterial cell and has a small molecule as target.
In addition to its use as a food preservative, the Center for Veterinary Medicine of the US Food and Drug Administration recently ruled positively on application of nisin for intramammary treatment of subclinical mastitis in dairy cattle (12, 13). After approval of the pending New Animal Drug Application, a nisin-containing product would allow treatment of bovine mastitis with a zero milk discard time and zero meat withdrawal period (i.e., milk and/or meat from treated cattle would not have to be discarded). One drawback that has been noted for nisin is its limited stability at pH 7 (14–18). Hence, more stable analogs may prove more effective. We report here the structure of a lantibiotic from Geobacillus thermodenitrificans NG80-2 termed geobacillin I that is an analog of nisin with two additional cross-links. The compound was produced heterologously in Escherichia coli, its ring topology was determined by NMR spectroscopy, and its activity against various bacteria was assessed. The compound is threefold more active than nisin against Streptococcus dysgalactiae, one of the main contagious causative agents of clinical bovine mastitis (19).
In addition to geobacillin I, we also report the structure of a second lantibiotic encoded on the genome of G. thermodenitrificans NG80-2. The compound was heterologously produced in E. coli, its ring topology was determined by tandem mass spectrometry, and its spectrum of activity was evaluated. Interestingly, geobacillin II, which has no obvious structural homologs among lantibiotics of known structure, was only active against bacilli.
Results
Description of Two Gene Clusters from G. thermodenitrificans NG80-2.
G. thermodenitrificans sp. are thermophilic Gram positive bacteria that have been found in diverse niches such as oil reservoirs, geothermal areas, and marine environments. These bacteria have gained recent interest as a potential source for industrially useful thermostable enzymes. G. thermodenitrificans NG80-2 was isolated from Dagang oil fields in China. Its genome was recently sequenced (20) and two groups of genes were annotated in the deposition to the National Center for Biotechnology Information database as lantibiotic biosynthetic genes. The enzymes encoded in one cluster were annotated as homologs of the biosynthetic enzymes of the class I lantibiotic subtilin produced by Bacillus subtilis 6633 (GenBank accession numbers YB_001124395–Y_001124403), whereas the second cluster was only partially annotated as having homology with the enzymes involved in biosynthesis of the class II lantibiotic mersacidin produced by Bacillus sp. HIL Y85/54728 (accession numbers YP_001126158 and YP_001126159). Both clusters have also been noted in bioinformatic genome mining studies (21, 22), but the product of neither cluster is known. Each gene cluster contains short open reading frames for the precursor peptides, which we designate geoAI for the class I cluster and geoAII for the class II cluster (Fig. 1 A and B). The core peptide of the GeoAI precursor peptide has homology with nisin and subtilin, in particular in the N-terminal region. However, GeoAI is predicted to have two more Cys residues than the precursor peptides for these known lantibiotics (Fig. 1C). The 23-residue leader peptide of GeoAI has high homology to the leader peptides for the lantibiotics ericin S and subtilin (Fig. 1D). On the basis of these sequence alignments, the site for proteolytic removal of the leader peptide was predicted as ProAsn↓Val (Fig. 1 C and D); this prediction was confirmed by detection of the lantibiotic by colony mass spectrometry of other Geobacillus strains (see below).
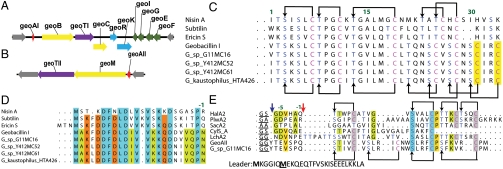
Fig. 1.
Gene clusters and precursor peptides of geobacillin I and geobacillin II. (A and B) Gene clusters for the biosynthesis of geobacillin I and geobacillin II. The gene for the precursor peptide is shown in red, modification enzymes in yellow, transporter/protease in purple, regulatory proteins in blue, and immunity proteins in green. (C) Sequence alignment of the core peptides of selected class I lantibiotics with that of GeoAI. Serines and threonines undergoing dehydration are in blue, cysteines involved in ring formation are in red. The additional two cysteines not found in nisin are highlighted in yellow. (D) Sequence alignment of the leader peptides of select class I lantibiotics with the leader peptide of geobacillin I. (E) Sequence alignment of the core peptide of GeoAII with type II lantibiotics that undergo a second proteolytic processing step after removal of the leader peptide at the double Gly cleavage site (indicated by a dark-blue arrow). The second cleavage site is indicated with a red arrow. The ring topology, when known, is shown at the top and the ring topology deduced in this study for geobacillin is shown at the bottom. The two possible Met residues that could be the start of the leader peptide are in bold font.
Genes typically found in lantibiotic biosynthetic gene clusters are present downstream of geoAI including genes encoding a dehydratase (geoB), ATP-binding cassette (ABC) transporter (geoT), cyclase (geoC), a two-component transcriptional regulator (geoR and geoK), and self-immunity proteins (geoI, geoG, geoE, and geoF) (Fig. 1A). Similar gene clusters with nearly identical precursor peptides (the only difference being a Val for Ile change) were also found in other genomes of Geobacillus (Fig. 1 C and D), but some of these clusters have frameshifted biosynthetic genes (21). No gene was identified that encodes for a typical class I lantibiotic protease to remove the leader peptide.
A search of the nonredundant protein database for homologs of the precursor peptide GeoAII did not find homologous precursor peptides of known lantibiotics suggesting it may be a unique class II lantibiotic. A gene encoding a precursor peptide differing in only one amino acid was found in the genome of Geobacillus sp. G11MC16 (Fig. 1E). Two start codons are present in frame at the N terminus of the leader peptide preventing prediction of the initiation codon and resulting in two possible precursor peptides (Fig. 1E). GeoAII contains a typical double-glycine motif for proteolytic removal of the leader peptide in class II lantibiotics (23, 24). Leader sequence removal after the double Gly site is supported by the presence of a gene product (GeoTII) that shows homology with the family of ABC transporter maturation and secretion proteins whose peptide substrates share the double-glycine type cleavage site (24). Like other family members, GeoTII contains an N-terminal Cys protease domain for leader peptide removal. The class II lanthionine synthetase GeoM is encoded by a gene located next to geoAII and has about 35% sequence identity to known LanM enzymes such as MrsM, HalM1, and HalM2.
Screening of Geobacillus Strains for Lantibiotic Production.
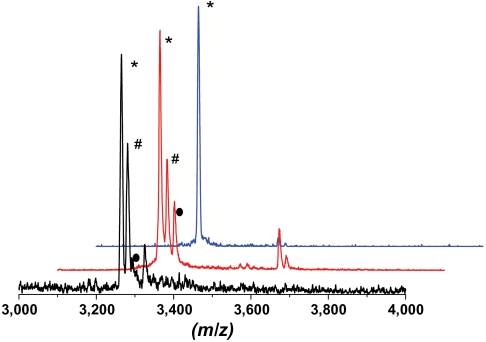
Because G. thermodenitrificans NG80-2 was not available to us, we screened seven different Geobacillus strains (25) for production of the predicted geobacillins (G. thermodenitrificans DSM465, G. thermodenitrificans OHT-1, Geobacillus sp. M10EXG, G. thermodenitrificans OH2-1, G. thermodenitrificans OH5-2, G. thermodenitrificans NM16-2, and Geobacillus kaustophilus DSM7263). The bacteria were grown at 55 °C on Tryptose Blood Agar Base (TBAB) or modified Luria-Bertani (mLB) media for 48 h. Whole-cell MALDI-MS (26) was used to investigate the production of geobacillin I and geobacillin II after 12, 24, and 48 h. A mass corresponding to GeoAI after nine dehydrations and removal of the leader peptide at the predicted site was observed in both media for five of the strains after 24 h. Only G. thermodenitrificans NM16-2 and G. kaustophilus DSM7263 did not produce geobacillin I under the conditions used. A representative MALDI mass spectrum is shown in Fig. 2. All five organisms produced peptides of the same mass, consistent with the near-identical sequences of GeoAI observed in the currently sequenced genomes (Fig. 1C). All five observed peptides had masses consistent with a Val at position 15 rather than an Ile. These observations along with the genome data suggest very high conservation of the sequence of geobacillin I in the Geobacillus genus. The mass spectrum in Fig. 2 also shows peaks corresponding to eight- and sevenfold dehydrated peptides, reminiscent of incomplete dehydration of nisin in Lactococcus lactis 6F3 (27).
Fig. 2.
Geobacillin I production in native and heterologous producers. MALDI-MS spectra showing geobacillin I production by a native producer in black and by E. coli in red. Purified ninefold dehydrated and cyclized core peptide is shown in blue. Nine-, eight-, and sevenfold dehydrated peptides are denoted by *, #, and ●, respectively.
Whole-cell MALDI-MS analysis did not show production of geobacillin II by any of the seven Geobacillus strains. In silico analysis revealed the presence of its class II gene cluster only in Geobacillus thermodenitrificans NG80-2 and Geobacillus sp. G11MC16 and not in any other sequenced Geobacillus genome (21). Indeed, attempts to detect the geoM biosynthetic gene in the genomic DNA of the seven strains used in this study by PCR with various degenerate primers were unsuccessful.
Biosynthesis of Geobacillin I and Geobacillin II in E. coli.
Given our recent success in producing nisin by coexpression in E. coli (28), synthetic genes encoding the precursor peptide GeoAI and the dehydratase GeoB were inserted into the multiple cloning site 1 (MCS-1) and MCS-2 of a pRSFDuet-1 vector, respectively. This construct results in a hexa-histidine-tagged precursor peptide (His6-GeoAI) and an untagged GeoB protein. A synthetic gene encoding the cyclase GeoC was inserted into MCS-2 of a pACYCDuet-1 vector, resulting in an untagged GeoC enzyme. Site-directed mutagenesis was performed to mutate Asn at the -1 position of the GeoAI peptide (Fig. 1D) to Lys to incorporate a trypsin cleavage site in the precursor peptide to be used for leader peptide removal. E. coli BL21 (DE3) cells were transformed with both plasmids and after induction of protein expression, His6-GeoAI was purified from the cell lysate using nickel affinity chromatography followed by RP-HPLC. The modified precursor peptide was treated with trypsin and MALDI-MS analysis of the resultant peptide demonstrated a ninefold dehydrated core peptide (Fig. 2, red line). As seen with the native producers, peaks corresponding to eight- and sevenfold dehydrated peptides were also observed.
A similar approach was taken to produce geobacillin II. Synthetic genes encoding GeoAII and GeoM were inserted into MCS-1 and MCS-2 of a pRSFDuet-1 vector, respectively. Site-directed mutagenesis was performed to mutate the second Gly in the double Gly motif of the GeoAII peptide to Lys to incorporate a trypsin cleavage site for leader peptide removal. Coexpression of GeoM with its substrate precursor peptide in E. coli at 37 °C and subsequent purification as described for geobacillin I resulted in fivefold dehydrated peptide after trypsin removal of the leader peptide (Fig. S1).
Bioactivity of Geobacillin I and Geobacillin II.
The antimicrobial spectrum of geobacillin I and geobacillin II was investigated with a range of Gram positive bacteria. Modified GeoAI was produced in E. coli as described above, treated with trypsin, purified by RP-HPLC (Fig. 2, blue spectrum), and used for bioactivity determinations in liquid medium (Table 1). The compound was active against a wide range of Gram positive bacteria but it was not active against Gram negative E. coli. Interestingly, in parallel experiments, geobacillin I demonstrated threefold higher activity against Streptococcus dysgalactiae ATCC 27957 than nisin. This bacterium is one of the causative agents of bovine mastitis (19). Geobacillin I displayed similar activity as nisin against vancomycin-resistant enterococci and Bacillus anthracis Sterne 7702, and had fivefold lower activity against methicillin-resistant Staphylococcus aureus and Bacillus subtilis ATCC 6633. Its antimicrobial activity against Lactococcus lactis HP is about sevenfold lower than nisin. As anticipated given the larger number of conformation-restraining cross-links and the thermophilic producing strain, geobacillin I was more stable than nisin at pH 7 and 8 at 37 and at 60 °C (Figs. S2–S6).
Table 1.
Specific activity of geobacillin I and nisin in liquid growth inhibition assay
| Strain | Source | Geobacillin I IC50, μM | Geobacillin I IC90, μM | Nisin IC50, μM | Nisin IC90, μM |
| Streptococcus dysgalatiae subsp dysgalactiae | ATCC 27957* | 0.69 ± 0.05 | 0.87 | 2.12 ± 0.04 | 3.87 |
| Vancomycin-resistant Enterococcusfaecium | CNRZ 481† | 0.84 ± 0.05 | 1.1 | 0.39 ± 0.03 | 0.57 |
| Methicillin-resistant Staphylococcus aureus | C5‡ | 2.23 ± 0.03 | 3.39 | 0.42 ± 0.01 | 0.77 |
| Bacillus anthracis Sterne 7702 | Gut et al.§ | 0.49 ± 0.02 | 0.71 | 0.21 ± 0.014 | 0.52 |
| Bacillus subtilis | ATCC 6633 | 0.55 ± 0.01 | 0.81 | 0.11 ± 0.01 | 0.16 |
| Lactococcus lactis HP | ATCC 11602 | 0.12 ± 0.09 | 0.13 | 0.017 ± 0.005 | 0.019 |
*ATCC, American Type Culture Collection.
†CNRZ, National Centre for Zootechnical Research.
‡Clinical isolate from Carle Foundation Hospital.
§Ref. 48.
To determine the bioactivity of geobacillin II, purified fivefold dehydrated GeoAII in which the second Gly of the double-glycine motif was mutated to Lys was treated with trypsin and checked for bioactivity. No antimicrobial activity was observed against any of the strains in Table S1. Although it is not unprecedented that lanthionine-containing peptides do not demonstrate any antimicrobial activity (29–32), we wondered whether perhaps a second proteolytic cleavage event might be required. Removal of six additional N-terminal residues after leader peptide cleavage at a double-glycine site has been reported previously for several class II lantibiotics (33–36). In some examples, such as cytolysin from Enterococcus faecalis, removal of these additional residues from the N terminus of the modified core peptide is necessary for bioactivity (34), but in other examples (e.g., haloduracin; ref. 37) such removal is not required. A sequence alignment of the predicted core peptide of GeoAII with the core peptides of lantibiotics that are currently known to undergo a second proteolytic step is shown in Fig. 1E. The N-terminal amino acids of GeoAII display only low-level sequence homology with the sequences that are removed in the other peptides, but the presence of a Pro-Gln sequence and the observation that the GeoAI leader peptide is removed at a Pro-Asn site prompted us to investigate whether removal of additional amino acids might result in bioactivity. First, a trypsin cleavage site was introduced into wild-type GeoAII by mutation of Gln-1 to Lys (this numbering assumes cleavage after Pro-Gln, Fig. 1E). Unfortunately, this mutant was not processed well in E. coli (predominantly four dehydrations, Fig. S7), presumably because introduction of a positively charged residue is detrimental for GeoM activity. A different mutant was constructed next (GeoAII-Gln-1Glu) and coexpressed with GeoM in E. coli resulting in the desired fivefold dehydrated peptide. Proteolysis with endoproteinase GluC did not result in efficient cleavage after the introduced Glu-1 (Fig. 3A), presumably because the posttranslationally modified Ser at position 1 deactivates the engineered cleavage site. Instead GluC cleaved predominantly after Glu-5 (Fig. 3A) resulting in the removal of only two residues (Tyr and Thr) at the N terminus compared to cleavage at the double Gly site. Interestingly, this GluC-treated core peptide induced zones of growth inhibition for Bacillus strains in agar well diffusion assays (Fig. 3B). Thus, removal of Tyr or Thr (or both) may be required for bioactivity. Geobacillin II only showed bioactivity against Bacillus species and no activity against any other tested bacteria (Table S1). Attempts to increase the observed bioactivity by treatment with aminopeptidase (38) to remove additional amino acids (for MS data, see Fig. S8) did not result in significantly increased zones of growth inhibition (compare spots 1 and 2, Fig. 3B).
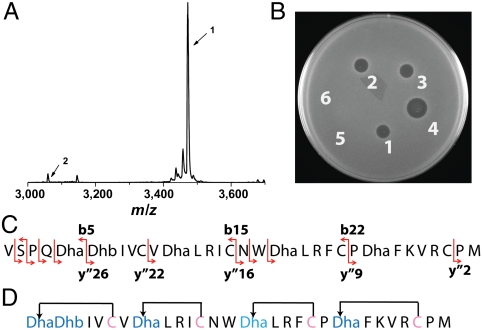
Fig. 3.
Biosynthesis of geobacillin II and growth inhibition assays. (A) MALDI-MS spectrum of GeoAII-Gln-1Glu modified by GeoM in E. coli and treated with GluC. Peptide 1 arises from cleavage at Glu-5; peptide 2 arises from cleavage at Glu-1. (B) Agar well diffusion assay with B. subtilis ATCC 6633. Zone 1: 10 μL of 100 μM GeoAII-Q-1E modified by GeoM and treated with GluC. This treatment results predominantly in peptide 1 (A). Zones 2 and 3: the material used for zone 1 was treated with aminopeptidase for 24 and 48 h, respectively. For MS data, see Fig. S8. Zone 4: 10 μL of 300 μM GeoAII-Q-1K modified by GeoM and treated with trypsin. This sample contains a mixture of four- and fivefold dehydrated peptide (see text and Fig. S7). Spot 5: trypsin-cleaved GeoAII-G-8K. Spot 6: negative control containing GluC and aminopeptidase. (C) ESI fragmentation pattern of GeoAII modified by GeoM and treated with GluC. The b5 and y″26 ions appear to indicate that the A ring is formed from Cys5 and dehydrobutyrine 2 (Dhb2), but three observations argue against this interpretation. First, if the ring were formed between Cys5 and Dhb2, the N-terminal Dha1 would hydrolyze to a lactate group (49), decreasing the mass by 1 Da; the masses in Figs. S7 and S8 do not support this model. Second, as shown in Fig. 1E, A rings formed from Cys5 and a dehydro amino acid at position 1 is well conserved among various lantibiotics, and third, acid hydrolysis followed by derivatization and amino acid analysis by GC-MS only showed lanthionines for geobacillin II (SI Text). Thus, we attribute the b5 and y″26 ions to fragmentation in the A ring. Similar fragmentation is also observed in the A ring of nisin (see figure S7 of ref. 28). (D) Ring topology of geobacillin II derived from the tandem MS results.
Because authentic geobacillin II is not produced by the seven Geobacillus strains we evaluated, at present it is not clear where the core peptide of geobacillin II starts. We favor removal of the entire heptapeptide sequence YTEVSPQ after initial removal of the leader peptide at the double Gly site for the following reasons. First, removal of this peptide would result from cleavage after Gln similar to the cleavage site for haloduracin from Bacillus halodurans and similar to lantibiotic proteases such as NisP involved in nisin biosynthesis (39). A LanP-type protease is not present in the gene cluster or elsewhere in the genome and therefore, like for haloduracin, the identity of the protease is not known, but the same protease may remove the leader of GeoAI at its Pro-Asn cleavage site. Secondly, cleavage after the Pro-Gln sequence would result in an N-terminal structure very similar to that of haloduracin β, plantaricin β, and cytolysin CylLS (Fig. 1E).
Structure Determination of Geobacillin I and Geobacillin II.
Tandem mass spectrometric analysis has been used previously to determine the ring topology of lanthionine-containing peptides that do not contain overlapping rings (32, 36), and for some of these peptides, the topology determined by tandem MS studies has been confirmed by NMR spectroscopy (40). In this work, tandem MS was used to determine the ring pattern of geobacillin II because the amounts of material generated were insufficient for NMR studies. The precursor peptide GeoAII was coexpressed with GeoM in E. coli, purified by immobilized metal ion affinity chromatography, and treated with GluC to remove the leader peptide. The resulting fivefold dehydrated peptide was fragmented by collision-induced dissociation resulting in the fragment ions indicated in Fig. 3C (see also Fig. S9). The observed ions are inconsistent with overlapping rings and agree very well with four nonoverlapping rings. Based on these data, and the discussion in the previous section on the likely N-terminal amino acid, the structure shown in Fig. 3D is proposed for geobacillin II.
It was not possible to use tandem MS to determine the structure of geobacillin I because of overlapping rings. Therefore extensive NMR characterization was carried out on the compound produced in E. coli. First, nearly all resonances were assigned by total correlation spectroscopy (Fig. S10) and gradient double-quantum-filtered COSY experiments. The assignments were further verified by determining the connectivities of dNN(i,i + 1) (Fig. S11) as well as dαN(i,i + 1) from a water-suppressed NOESY spectrum (mixing time, 0.15 s) and taking into consideration the known linear amino acid sequence. Finally, a NOESY spectrum was acquired in D2O to determine intrabridge NOEs, which facilitated the assignment of thioether links. A longer mixing time of 0.40 s was employed in this experiment to obtain stronger NOE signals. The observed cross-peaks arising from correlations of 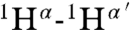 ,
,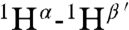 ,
, 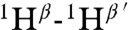 ,
, 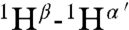 , and
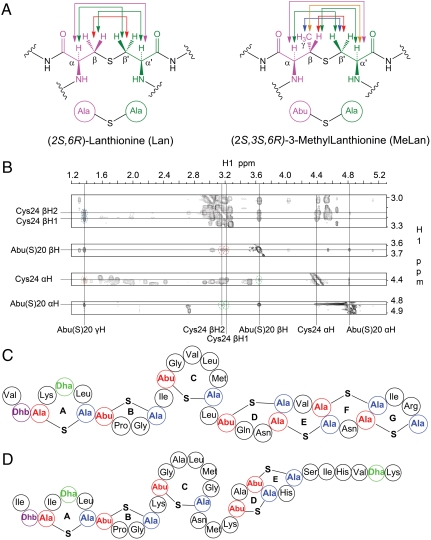
, and 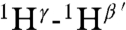 (the prime ′ indicates proton across the thioether bridge; Fig. 4A and Fig. S12) were used primarily to identify the ring patterns of six of the seven lanthionine and methyllanthionine residues (for details, see SI Text). The final lanthionine ring involving Cys30 and Dha26 could not be assigned based on the NMR data because of identical chemical shifts of the β-protons, but given that all other rings were assigned, only one possibility exists for this last ring (the F ring). The final structure determined by NMR spectroscopy is shown in Fig. 4C (for stereochemistry, seeFig. S13).
(the prime ′ indicates proton across the thioether bridge; Fig. 4A and Fig. S12) were used primarily to identify the ring patterns of six of the seven lanthionine and methyllanthionine residues (for details, see SI Text). The final lanthionine ring involving Cys30 and Dha26 could not be assigned based on the NMR data because of identical chemical shifts of the β-protons, but given that all other rings were assigned, only one possibility exists for this last ring (the F ring). The final structure determined by NMR spectroscopy is shown in Fig. 4C (for stereochemistry, seeFig. S13).
Fig. 4.
Structure determination of geobacillin I. (A) Illustration of the NOE correlations used to establish thioether connectivities. (B) Representative NOESY data showing some of the correlations used for D-ring assignment (see Fig. S12 for the other rings). (C) Ring topology of geobacillin I. (D) Ring topology of nisin A.
Discussion
The availability of a rapidly increasing number of bacterial genomes is starting to shift the search for new gene-encoded peptide natural products from activity-based discovery platforms to gene-based discovery approaches (41–43). In the lantibiotic group of compounds, genome mining has resulted in the discovery of several family members (22, 31, 44), including a lantibiotic from an alkaliphilic organism (35, 45). In addition, a large number of biosynthetic gene clusters have been identified that remain to be explored experimentally (21, 46). Our attention was drawn to genomes of thermophiles because only one lantibiotic from a moderately thermophilic organism has been reported (47). The lantibiotics produced by thermophiles could find potential applications as they may be more stable than lantibiotics produced by mesophilic bacteria. Particularly interesting was the discovery of nisin precursor genes in the genomes of Geobacillus because their sequence suggested that the posttranslationally modified products would contain additional rings. Indeed, the masses of the compounds produced by five Geobacillus strains and the corresponding lantibiotic heterologously produced in E. coli showed nine dehydrations and the NMR structure determined for the latter compound demonstrated seven thioether cross-links, including conservation of the A and B rings found in nisin that are important for lipid II binding (5–7). In all, 16 out of the 33 residues in the core peptide of GeoAI are posttranslationally modified, and the final product geobacillin I was shown to have high antimicrobial activity against several pathogens. Furthermore, the compound is more stable than nisin A at pH 7 and 8 and at high temperatures.
This study demonstrates that lantibiotics may be produced in nature at temperatures as high as 50–80 °C, the temperature of the deep subsurface Dagang oilfield where G. thermodenitrificans NG80-2 was isolated. The successful production of geobacillin I and II in E. coli was therefore somewhat surprising because many enzymes from thermophilic organisms are nonfunctional at 37 °C. However, GeoB, GeoC, and GeoM carried out dehydrations and cyclizations with apparently high efficiency at this temperature. Although, at present we cannot determine the precise location of the start of geobacillin II, the demonstration that removal of additional amino acids past the double Gly protease cleavage site resulted in bioactivity along with the sequence homology with other lantibiotics that undergo a second step of proteolytic processing support the lantibiotic structure shown in Fig. 3D.
Methods
Materials.
DNA polymerases, restriction endonucleases, and T4 DNA ligase were purchased from New England Biolabs. All oligonucleotides were purchased from Integrated DNA Technologies. Media components for bacterial cultures were purchased from Difco laboratories. Chemicals were purchased from Sigma-Aldrich unless noted otherwise. Endoproteinase GluC (sequencing grade) was purchased from Roche Biosciences. Trypsin (modified, sequencing grade) was purchased from Worthington Biosciences. Aminopeptidase was purchased from Sigma-Aldrich (ammonium sulfate suspension, L5006).
General Methods.
All PCRs were carried out on a C1000™ thermal cycler (Bio-Rad). E. coli DH5α was used as host for cloning and plasmid propagation, and E. coli BL21 (DE3) was used as a host for coexpression. DNA sequencing was performed by ACGT, Inc. MALDI-TOF MS was carried out on a Voyager-DE-STR (Applied Biosystems) or Bruker Ultraflex TOF/TOF. Liquid chromatography electrospray ionization (ESI) tandem mass spectrometry was carried out and processed using a Synapt ESI quadrupole TOF Mass Spectrometry System (Waters) equipped with an Acquity Ultra Performance Liquid Chromatography system (Waters).
Detection of Geobacillin I and Geobacillin II Production.
Seven Geobacillus strains were purchased from the Bacillus Genetic Stock Center. The strains were streaked on TBAB and mLB plates. A small amount of cells from a colony was picked from the plate with a pipette tip and was spotted on a MALDI plate. The sample was overlaid with 2 μL of a 9∶1 mixture of 2,5-dihydroxybenzoic acid and 2-hydroxy-5-methoxybenzoic acid matrix prepared in 60% acetonitrile (MeCN)/0.1% TFA in water (80 mg/mL). A heat gun was used to dry the sample, and the samples were analyzed by MALDI-MS.
Cloning, Production, and Purification of Geobacillins.
Codon-optimized synthetic genes for GeoAI, GeoB, GeoC, GeoAII, and GeoM were synthesized by GeneArt (Invitrogen). For geobacillin I production, geoAI was inserted in MCS-1 of a pRSFDuet-1 vector using BamHI and HindIII restriction sites, geoB in MCS-2 using NdeI and XhoI sites, and geoC in MCS-2 of pACYCDuet-1 using NdeI and XhoI sites. Site-directed mutagenesis was performed to mutate Asn at the -1 position to Lys (for primers see Table S2). Electrocompetent BL21(DE3) cells were cotransformed with the plasmids. A single colony was picked to inoculate a culture used to overexpress modified GeoAI as described for other lantibiotics (28). The cells were lysed, and modified GeoAI was purified using nickel affinity chromatography and reversed phase (RP) HPLC (48). A similar procedure was followed for production of modified GeoAII. A detailed procedure is described in the SI Text.
Preparation of Bioactive Geobacillin I and Geobacillin II.
A solution of 500 μM posttranslationally modified GeoAI-N-1K in 50 mM Hepes buffer (pH 8.0) was treated with 0.5 μM trypsin for 2 h. The product was analyzed using MALDI-MS. The ninefold dehydrated and cyclized core peptide was purified using RP-HPLC in order to remove incompletely dehydrated peptide and leader peptide using a Phenomenex Luna C18 column (250 × 10 mm, 10 μm) operating at a flow rate of 10 mL/ min. The program used was 3 min of solvent A (2% MeCN/0.1% TFA) followed by a gradient of 2–100% solvent B (80% MeCN/0.1% TFA) over 50 min. For preparation of geobacillin II, GeoM-modified and HPLC-purified GeoAII-G-8K/Q-1E (100 μM) was treated with either 0.1 μM trypsin (20 min) or 0.5 μM GluC (2 h). GeoAII-Q-1K (100 μM) was also treated with 0.1 μM trypsin. The GluC-cleaved GeoAII-G-8K/Q-1E peptide (25 μL, 100 μM) was further treated with 8 μL of Leu aminopeptidase (97 units/mL) for 24 h at 37 °C. The resulting products were analyzed by MALDI-MS (Fig. S8) and the observed and calculated molecular weights are summarized in Table S3.
Agar Diffusion Growth Inhibition Assay.
For IC50 calculations, a 48-well plate (300 μL well size) was used with shaking and a 96-well plate (200 μL well size) was used for nonshaking conditions, depending on the indicator strain. Serial dilutions of nisin and geobacillin I were prepared in sterile deionized water. The 48/96 plate wells contained 50/75 μL of diluted peptide at defined concentrations and 150/225 μL of a 1∶10 dilution (final concentration approximately 1 × 108 cfu mL-1) of a culture of indicator strain prepared in fresh growth medium. Growth medium blank and negative controls lacking the lantibiotics were also prepared. Plates were incubated under appropriate growth conditions (Table S4), and IC50 calculations were performed by monitoring the OD600. For agar diffusion bioactivity assays, the appropriate agar media (Table S4) was melted in a microwave, kept at 45 °C for 10 min, and mixed with 150 μL of overnight culture of indicator organisms (108–109 cfu). Then, 10 μL of a solution of 100 μM peptide was spotted on the plate. For trypsin-cleaved GeoAII Q-1K, 10 μL of 300 μM peptide was spotted.
Supplementary Material
Acknowledgments.
The authors are grateful to Dr. Dan Zeigler, Director, Bacillus Genetic Stock Center, for providing information on Geobacillus strains, and Dr. Dean Olson for NMR training. This work was supported by the National Institutes of Health (GM58822).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116815109/-/DCSupplemental.
References
- 1.Willey JM, van der Donk WA. Lantibiotics: Peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 2.Oman TJ, van der Donk WA. Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 4.de Kruijff B, van Dam V, Breukink E. Lipid II: A central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot Essent Fatty Acids. 2008;79:117–121. doi: 10.1016/j.plefa.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Brötz H, et al. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 6.Breukink E, et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 7.Hasper HE, et al. A new mechanism of antibiotic action. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 8.Breukink E, et al. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J Biol Chem. 2003;278:19898–19903. doi: 10.1074/jbc.M301463200. [DOI] [PubMed] [Google Scholar]
- 9.Wiedemann I, Benz R, Sahl HG. Lipid II-mediated pore formation by the peptide antibiotic nisin: A black lipid membrane study. J Bacteriol. 2004;186:3259–3261. doi: 10.1128/JB.186.10.3259-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 11.Schneider T, Sahl HG. Lipid II and other bactoprenol-bound cell wall precursors as drug targets. Curr Opin Investig Drugs. 2010;11:157–164. [PubMed] [Google Scholar]
- 12.Brigham MF. ImmuCell comments on status of zero milk discard claim for mast out(R) 2011. Jun 9, Available at http://www.reuters.com/article/2011/06/09/idUS120388+09-Jun-2011+MW20110609. Accessed January 27, 2012.
- 13.Brigham MF. ImmuCell presents mast out product offering at World Animal Health Congress. World Animal Health Congress 2011. 2011. Available at http://finance.yahoo.com/news/ImmuCell-Presents-Mast-Out-R-iw-3929215384.html.
- 14.Chan WC, Bycroft BW, Lian LY, Roberts GCK. Isolation and characterization of two degradation products derived from the peptide antibiotic nisin. FEBS Lett. 1989;252:29–36. [Google Scholar]
- 15.Rollema HS, Kuipers OP, Both P, de Vos WM, Siezen RJ. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl Environ Microbiol. 1995;61:2873–2878. doi: 10.1128/aem.61.8.2873-2878.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rollema HS, Metzger JW, Both P, Kuipers OP, Siezen RJ. Structure and biological activity of chemically modified nisin A species. Eur J Biochem. 1996;241:716–722. doi: 10.1111/j.1432-1033.1996.00716.x. [DOI] [PubMed] [Google Scholar]
- 17.Lian LY, et al. Solution structures of nisin A and its two major degradation products determined by n.m.r. Biochem J. 1992;283:413–420. doi: 10.1042/bj2830413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz L, Garden RW, Kaiser HJ, Sweedler JV. Studies of the degradation products of nisin, a peptide antibiotic, using capillary electrophoresis with off-line mass spectrometry. J Chromatogr A. 1996;735:375–385. doi: 10.1016/0021-9673(95)01144-7. [DOI] [PubMed] [Google Scholar]
- 19.Olde Riekerink RG, Barkema HW, Stryhn H. The effect of season on somatic cell count and the incidence of clinical mastitis. J Dairy Sci. 2007;90:1704–1715. doi: 10.3168/jds.2006-567. [DOI] [PubMed] [Google Scholar]
- 20.Feng L, et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci USA. 2007;104:5602–5607. doi: 10.1073/pnas.0609650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh AJ, O’Sullivan O, Ross RP, Cotter PD, Hill C. In silico analysis highlights the frequency and diversity of type 1 lantibiotic gene clusters in genome sequenced bacteria. BMC Genomics. 2010;11:679. doi: 10.1186/1471-2164-11-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begley M, Cotter PD, Hill C, Ross RP. Rational genome mining for LanM proteins leads to the identification of a novel two peptide lantibiotic, lichenicidin. Appl Environ Microbiol. 2009;75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nes IF, Tagg JR. Novel lantibiotics and their pre-peptides. Antonie Van Leeuwenhoek. 1996;69:89–97. doi: 10.1007/BF00399414. [DOI] [PubMed] [Google Scholar]
- 24.Håvarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 25.Zeigler DR. Application of a recN sequence similarity analysis to the identification of species within the bacterial genus Geobacillus. Int J Syst Evol Microbiol. 2005;55:1171–1179. doi: 10.1099/ijs.0.63452-0. [DOI] [PubMed] [Google Scholar]
- 26.Stein T. Whole-cell matrix-assisted laser desorption/ionization mass spectrometry for rapid identification of bacteriocin/lantibiotic-producing bacteria. Rapid Commun Mass Spectrom. 2008;22:1146–1152. doi: 10.1002/rcm.3481. [DOI] [PubMed] [Google Scholar]
- 27.Engelke G, Gutowski-Eckel Z, Hammelmann M, Entian KD. Biosynthesis of the lantibiotic nisin: Genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992;58:3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Yang X, Garg N, van der Donk WA. Production of lantipeptides in Escherichia coli. J Am Chem Soc. 2011;133:2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kodani S, et al. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc Natl Acad Sci USA. 2004;101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodani S, Lodato MA, Durrant MC, Picart F, Willey JM. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol Microbiol. 2005;58:1368–1380. doi: 10.1111/j.1365-2958.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- 31.Goto Y, et al. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010;8:e1000339. doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci USA. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology. 2001;147:643–651. doi: 10.1099/00221287-147-3-643. [DOI] [PubMed] [Google Scholar]
- 34.Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: A novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 35.McClerren AL, et al. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci USA. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caetano T, Krawczyk JM, Mosker E, Süssmuth RD, Mendo S. Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem Biol. 2011;18:90–100. doi: 10.1016/j.chembiol.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Cooper LE, McClerren AL, Chary A, van der Donk WA. Structure-activity relationship studies of the two-component lantibiotic haloduracin. Chem Biol. 2008;15:1035–1045. doi: 10.1016/j.chembiol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majchrzykiewicz JA, et al. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother. 2010;54:1498–1505. doi: 10.1128/AAC.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Meer JR, et al. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenkarev ZO, et al. Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry. 2010;49:6462–6472. doi: 10.1021/bi100871b. [DOI] [PubMed] [Google Scholar]
- 41.Challis GL. Genome mining for novel natural product discovery. J Med Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 42.Velásquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol. 2011;15:11–21. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kersten RD, et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dischinger J, Josten M, Szekat C, Sahl HG, Bierbaum G. Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13. PLoS One. 2009;4:e6788. doi: 10.1371/journal.pone.0006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Fewer DP, Sivonen K. Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria. PLoS One. 2011;6:e22384. doi: 10.1371/journal.pone.0022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabuki T, Uenishi H, Seto Y, Yoshioka T, Nakajima H. A unique lantibiotic, thermophilin 1277, containing a disulfide bridge and two thioether bridges. J Appl Microbiol. 2009;106:853–862. doi: 10.1111/j.1365-2672.2008.04059.x. [DOI] [PubMed] [Google Scholar]
- 48.Li B, Cooper LE, van der Donk WA. In vitro studies of lantibiotic biosynthesis. Methods Enzymol. 2009;458:533–558. doi: 10.1016/S0076-6879(09)04821-6. [DOI] [PubMed] [Google Scholar]
- 49.Velásquez JE, Zhang X, van der Donk WA. Biosynthesis of the antimicrobial peptide epilancin 15X and its unusual N-terminal lactate moiety. Chem Biol. 2011;18:857–867. doi: 10.1016/j.chembiol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.