Abstract
We describe the discovery and isolation of a paramyxovirus, feline morbillivirus (FmoPV), from domestic cat (Felis catus). FmoPV RNA was detected in 56 (12.3%) of 457 stray cats (53 urine, four rectal swabs, and one blood sample) by RT-PCR. Complete genome sequencing of three FmoPV strains showed genome sizes of 16,050 bases, the largest among morbilliviruses, because of unusually long 5′ trailer sequences of 400 nt. FmoPV possesses identical gene contents (3′-N-P/V/C-M-F-H-L-5′) and is phylogenetically clustered with other morbilliviruses. IgG against FmoPV N protein was positive in 49 sera (76.7%) of 56 RT-PCR–positive cats, but 78 (19.4%) of 401 RT-PCR–negative cats (P < 0.0001) by Western blot. FmoPV was isolated from CRFK feline kidney cells, causing cytopathic effects with cell rounding, detachment, lysis, and syncytia formation. FmoPV could also replicate in subsequent passages in primate Vero E6 cells. Infected cell lines exhibited finely granular and diffuse cytoplasmic fluorescence on immunostaining for FmoPV N protein. Electron microscopy showed enveloped virus with typical “herringbone” appearance of helical N in paramyxoviruses. Histological examination of necropsy tissues in two FmoPV-positive cats revealed interstitial inflammatory infiltrate and tubular degeneration/necrosis in kidneys, with decreased cauxin expression in degenerated tubular epithelial cells, compatible with tubulointerstitial nephritis (TIN). Immunohistochemical staining revealed FmoPV N protein-positive renal tubular cells and mononuclear cells in lymph nodes. A case-control study showed the presence of TIN in seven of 12 cats with FmoPV infection, but only two of 15 cats without FmoPV infection (P < 0.05), suggesting an association between FmoPV and TIN.
Paramyxoviruses are enveloped, negative-sense single-stranded RNA viruses that are divided into two subfamilies, Paramyxovirinae and Pneumovirinae. Viruses in the subfamily Paramyxovirinae have been associated with a number of emerging diseases in humans and various animals in the past two decades (1–9). There are currently five genera within the subfamily Paramyxovirinae, namely Respirovirus, Rubulavirus, Morbillivirus, Henipavirus, and Avulavirus, although some members of the subfamily remain unclassified. Among members of Paramyxovirinae, measles virus, mumps virus, and human parainfluenza viruses 1 through 4 are the most well known human paramyxoviruses, which cause outbreaks of respiratory to systemic infections (10–12). In addition, Hendra and Nipah viruses are also important zoonotic viruses that cause outbreaks in humans (13). Recently, we reported the discovery of three rubulaviruses, Tuhoko viruses 1, 2 and 3, from fruit bats in mainland China, and an unclassified paramyxovirus, Tailam virus, from Sikkim rats in Hong Kong (14, 15). Despite the presence of paramyxoviruses in a variety of animals, no paramyxoviruses have been naturally observed in cats, although there is controversial evidence that cats may be infected with parainfluenza 5 virus (16, 17).
Cats and dogs are the commonest domestic animals and pets worldwide. As a result of their close relatedness, interspecies jumping of viruses among these two kinds of animals is not uncommon. For coronaviruses, feline coronavirus and canine coronavirus are classified under the same species Alphacoronavirus 1, and feline coronavirus type II strains were generated by double homologous recombination between feline coronavirus type I strains and canine coronavirus (18). For parvoviruses, the fatal canine parvovirus that emerged in the 1970s also originated from a feline parvovirus, feline panleukopaenia virus (19, 20). As for herpesviruses, canid herpesvirus 1 and felid herpesvirus 1 are closely related and are classified under the genus Varicellovirus (21). Furthermore, for papillomaviruses, canine oral papillomavirus and feline papillomavirus are also closely related and are classified under the genus Lambdapapillomavirus (22). As dogs are well known hosts of a paramyxovirus, canine distemper virus, in the genus Morbillivirus (23), we hypothesized that there are previously unrecognized morbilliviruses in cats. To test this hypothesis, we carried out a molecular epidemiology study in stray cats in Hong Kong and diseased cats from mainland China for novel morbilliviruses, during which a virus was isolated and characterized. We also demonstrated that the virus is associated with tubulointerstitial nephritis (TIN) in cats. Based on the results of the present study, we propose a novel paramyxovirus in the genus Morbillivirus, named feline morbillivirus (FmoPV).
Results
Identification of a Paramyxovirus from Cats.
RT-PCR with consensus primers amplifying a 155-bp fragment in the L gene of morbilliviruses was positive in samples (53 urine, four rectal swabs, and one blood sample) from 56 (12.3%) of 457 stray cats captured from various locations in Hong Kong over a 2-y period (March 2009 to February 2011), with two stray cats positive on both the urine and rectal swabs. For the 16 diseased cats from mainland China, one (6.25%) was RT-PCR–positive in both its oral and rectal swabs. Real-time quantitative RT-PCR showed a median viral load of 3.9 × 103 (range, 0.037–1.4 × 106) copies/mL. Sequencing results suggested the presence of a previously undescribed paramyxovirus of the genus Morbillivirus, with fewer than 80% nt identities to known paramyxoviruses (Fig. S1). In the three cats for which RT-PCR was positive in two specimens, the sequences of the amplified fragments were identical in the two different specimens. We propose this paramyxovirus to be named FmoPV.
Genome Organization, Coding Potential, and mRNA Editing of FmoPV.
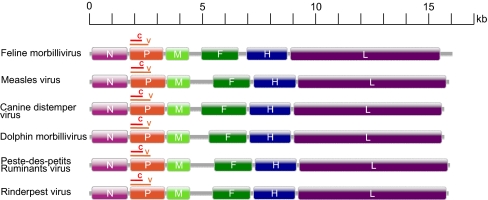
Complete genome sequences of three strains of FmoPV, from two urine (761U, 776U) and one rectal swab (M252A) samples, were determined. The genome size of FmoPV is 16,050 bases, and G+C contents are 35.1% to 35.3%, with FmoPV having the largest genome among all morbilliviruses with genome sequence available (Fig. 1). The genome of FmoPV conforms to the rule of six, as in other paramyxovirus genomes. It contains a 12-nt complementary 3′ leader and 5′ trailer sequence. The 3′ leader sequence is 55 nt. In contrast to other morbilliviruses, which have 5′ trailer sequences of only 40 or 41 nt, the genome of FmoPV has a trailer sequence of 400 nt, accounting for its bigger genome size. Such long trailer sequences of larger than 400 nt have been observed only in avian paramyxoviruses 3 (681–707 nt) and 5 (552 nt), and Tupaia paramyxovirus (590 nt; Fig. S2).
Fig. 1.
Genome organization of FmoPV and other morbilliviruses. The genes are shown as boxes that are drawn to scale. For P gene, the brown line above the orange box represents the region of V CDS and the red line represents the C CDS.
Similar to other morbilliviruses, the genome of FmoPV contains six genes (3′-N-P/V/C-M-F-H-L-5′; Fig. 1). Pairwise alignment of the predicted gene products among FmoPV and other paramyxoviruses showed the highest amino acid identities with members of the genus Morbillivirus, with the N, P/V/C(P), P/V/C(V), P/V/C(C), M, F, H, and L of FmoPV having 54.3% to 56.8%, 25.6% to 31.7%, 20.7% to 25.7%, 18.3% to 25.4%, 57.6% to 60.0%, 35.8% to 45.1%, 20.4% to 24.1%, and 55.2% to 57.3% amino acid identities to those of other morbilliviruses (Table S1). The lengths and characteristics of the major structural genes and intergenic regions are summarized in Table S2. Among all the structural genes, the N gene possessed the highest rate of nucleotide polymorphism, whereas the P gene possessed the highest rate of amino acid polymorphism (Fig. S3).
The conserved N-terminal motif MA(T/S)L in morbilliviruses was absent in the nucleoprotein (N) of FmoPV, which contained the sequence MSSL as a result of A-to-S (G-to-U at first codon position) substitution at the second amino acid (Fig. S4). Similar to the nuclear localization signal (NLS) of the N proteins in CdiPV, MeaPV, and RinPV but different from the classical NLS sequence, a leucine/isoleucine-rich motif at amino acid positions 70 to 77 is identified in the N protein of FmoPV (Fig. S4). Similar to the nuclear export signal of the N proteins in CdiPV and RinPV, a leucine-rich motif at amino acid positions 4 to 11 is also identified in the N protein of FmoPV (Fig. S4).
As in other morbilliviruses, the P/V/C gene of FmoPV contains two initiation codons, the first one for translation of P and V and the second for translation of C. Similar to most members of Paramyxovirinae, the P/V/C gene of FmoPV contains a UC-rich editing site that allows the addition of nontemplated G residues to mRNA products during P/V/C gene transcription, resulting in the production of different proteins with a common N-terminal region. In all three strains of FmoPV, this common N-terminal region consists of 226 aa.
To determine the exact location of P gene editing site and the number and frequency of G-residue insertions, a small cDNA fragment including the UC-rich region was amplified, cloned, and sequenced by using mRNA extracted from FmoPV infected CRFK cells. Among 23 independent clones sequenced, 13 contained the sequence TTAAAAGGGG (without G insertion, encoding P protein) and 10 contained the sequence TTAAAAGGGGG (one G inserted, encoding V protein). The sequence TTAnGn is conserved as in other paramyxovirus editing sites except for those of rubulaviruses. In contrast to other morbilliviruses in which the sequence is TTA5G3 (24), the TTAnGn sequence in FmoPV is TTA4G4.
Different from all other known morbilliviruses, the F protein of FmoPV has a single-basic proteolytic cleavage site, whereas the cleavage sites in other morbilliviruses are multibasic (25). Cellular trypsin-like protease cleaves the F protein into F1 and F2 before cell fusion occurs, which facilitates the isolation of these viruses in cell lines. Two heptad repeat sequences similar to those in F proteins of other paramyxoviruses were also identified in the F1 of FmoPV. The F protein of FmoPV also contains the 10 Cys residues that are highly conserved in other morbilliviruses and five potential N-glycosylation sites, most of which are located in the F2 peptide.
Phylogenetic Analyses.
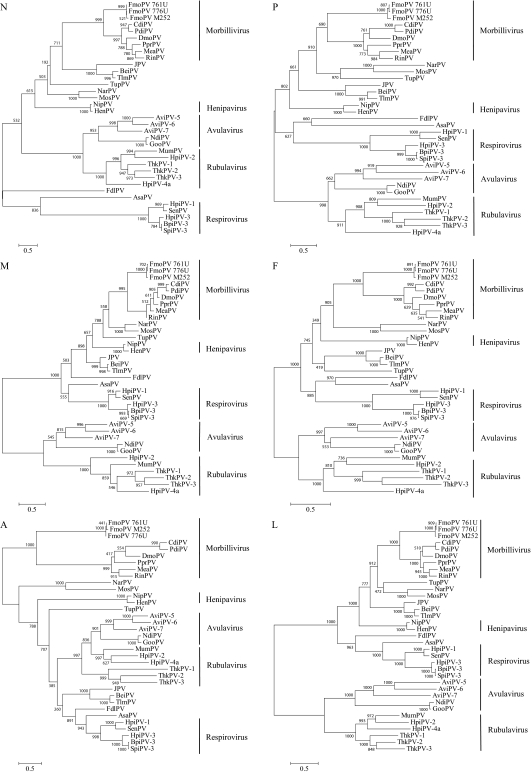
Phylogenetic trees constructed using the predicted amino acid sequences of N, P, M, F, H, and L genes of FmoPV and other members of Paramyxoviridae are shown in Fig. 2. In all six trees, the three viruses were clustered with morbilliviruses, with high bootstrap supports, forming a distinct subgroup (Fig. 2).
Fig. 2.
Phylogenetic analysis of N, P, M, F, A, and L amino acid sequences of FmoPV. The trees were constructed by maximum likelihood method with bootstrap values calculated from 1,000 trees and rooted on midpoint. The scale bars indicates the branch length that corresponds to 0.5 substitutions per site. Three strains from FmoPV were named as 761U, 776U, and M252A. Names and accession numbers of the other viruses are listed in Table S5.
Western Blot Analysis.
Among tested sera from the 56 cats that were RT-PCR–positive and 401 cats that were RT-PCR–negative for FmoPV, 49 (76.7%) and 78 (19.4%) tested positive for IgG against Escherichia coli expressed recombinant N protein of FmoPV by Western blot analysis, respectively (P < 0.0001; Fig. S5 and Table S3). Among tested sera from the 56 cats that were RT-PCR–positive for FmoPV, only five (8.9%) tested positive for IgM against N protein of FmoPV.
Viral Culture, Immunostaining, and Electron Microscopy (EM).
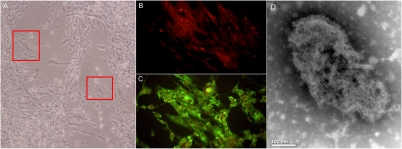
At the eighth passage, CRFK (feline kidney) cells inoculated with a urine sample (761U) positive for FmoPV showed cytopathic effects (CPEs) at day 14, in the form of cell rounding, followed by cell detachment from the monolayer and cell lysis. At the 16th passage, CPEs were evident at day 10, with syncytia formation (Fig. 3A). RT-PCR for FmoPV using the culture supernatants and cell lysates showed positive results in CRFK cells inoculated with urine sample 761U and VeroE6 (African green monkey kidney) cells inoculated with supernatant of CRFK cells positive for FmoPV. Specific apple green finely granular and diffuse cytoplasmic fluorescence was also observed by using serum from guinea pig immunized with recombinant N protein of FmoPV or corresponding serum of the infected cat (Fig. 3C). EM showed an enveloped virus with the typical “herringbone” appearance of the helical N in paramyxoviruses (Fig. 3D). Virions are highly variable in size, ranging approximately from 130 to 380 nm in diameter. No CPE and no viruses were detected by RT-PCR in NIH/3T3 (mouse embryo fibroblast), B95 (marmoset B-cell), and chicken embryo fibroblast cells inoculated with the samples.
Fig. 3.
(A) CPEs of FmoPV on CRFK cells. Red squares show the formation of giant cells. (B and C) Indirect immunofluorescent antigen detection in uninfected and infected CRFK cells using serum from guinea pig immunized with recombinant N protein of FmoPV, showing specific apple green cytoplasmic fluorescence in FmoPV-infected CRFK cells. (D) EM examination of infected CRFK cell culture supernatant showing enveloped virus with burst envelope and typical herringbone appearance of helical N in paramyxoviruses.
Histopathology and Immunohistochemistry.
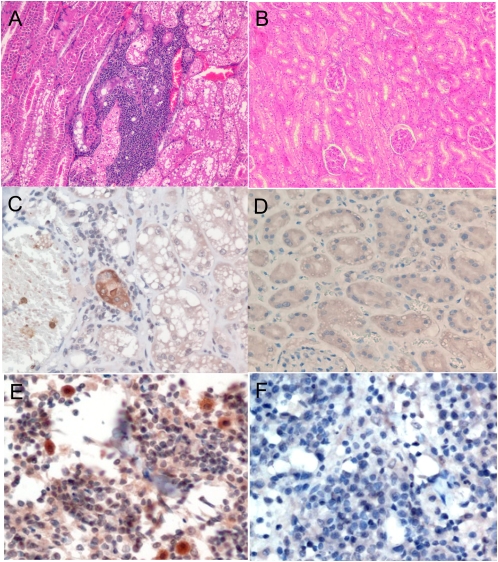
To identify possible diseases associated with FmoPV, necropsies were performed on two euthanized stray cats positive for FmoPV by RT-PCR. Histological examination of various organs revealed interstitial inflammatory infiltrate and renal tubular degeneration or necrosis in their kidneys (Fig. 4A). In addition, there was also marked decrease in cauxin expression in the degenerated tubular epithelial cells, compatible with TIN (Fig. S6). Immunohistochemical staining of their organs with guinea pig serum positive for anti-FmoPV N protein antibody revealed positive renal tubular cells in kidney sections (Fig. 4C) and positive mononuclear cells in lymph node sections (Fig. 4E). With the use of mouse anti-human myeloid/histiocyte antigen antiserum MAC387, the targets of FmoPV in lymph node sections were shown to be macrophages (Fig. S7).
Fig. 4.
(A and B) Histological section of kidneys stained by H&E from a stray cat with FmoPV detected in urine and a normal cat, showing aggregates of inflammatory cells in the interstitium and renal tubular degeneration in the infected cat. (C and D) Immunohistochemical staining of kidney sections of stray cat with FmoPV detected in urine using guinea pig serum positive for anti-FmoPV N protein antibody and preimmune guinea pig serum, showing positive renal tubular cells. (E and F) Immunohistochemical staining of lymph node sections of stray cat positive for FmoPV using guinea pig serum positive for anti-FmoPV N protein antibody and preimmune guinea pig serum, showing positive mononuclear cells.
Case-Control Study.
As the kidneys of the two stray cats showed histopathological features compatible with TIN, the kidneys, urine, and plasma were obtained from a total of 27 stray cats, including the two cats with necropsies performed, and were subjected to RT-PCR, histopathologic examination, and antibody detection by Western blot and immunofluorescence to examine for possible association between FmoPV infection (RT-PCR and/or antibody positive) and TIN. Among the 27 stray cats, TIN was observed in seven of 12 cats with evidence of FmoPV infection, but in only two of 15 cats without evidence of FmoPV infection (P < 0.05, Fisher exact test; Table S4).
Discussion
We report the discovery of a feline paramyxovirus, FmoPV, from stray cats in Hong Kong, which represents a documentation of paramyxoviruses found in the domestic cat (Felis catus). Although cats are known reservoirs of different viruses, the existence of paramyxoviruses in these common domestic animals was previously unknown. In this study, FmoPV was detected in the urine samples of 53 of 457 stray cats and in the rectal swab and blood samples of four and one of these cats, respectively. Western blot analysis revealed a seroprevalence of 27.8% among tested cats for IgG against recombinant N protein, and the presence of antibody is highly associated with the presence of virus. Analysis of the complete genomes of three FmoPV strains showed that they formed a distinct cluster among the morbilliviruses in all six phylogenetic trees constructed by using the N, P, M, F, H and L genes (Fig. 2). Immunohistochemistry also showed that, similar to other morbilliviruses such as measles virus, FmoPV infects mononuclear cells and parenchymal cells (Fig. 4). In the present study, the three strains of FmoPV exhibited high sequence similarity and identical genome organization, suggesting a single species of FmoPV and a high degree of species-specificity in FmoPV. Although no recombination was identified in the present FmoPV strains (Fig. S8), other viruses from cats, such as feline coronaviruses and feline papillomavirus, have been shown to be closely related to or recombine with their canine counterparts in dogs, suggesting that feline viruses may have the potential to cross species barrier in animals of similar living habitat (18, 26).
FmoPV is associated with TIN in cats. TIN involves primary injury to renal tubules and interstitium and is the most common cause of renal failure and one of the leading causes of deaths in cats. However, the cause of most cases of feline TIN remains unknown, and therefore treatment is mainly supportive and prevention is difficult. As FmoPV was mostly detected in urine samples, we hypothesized that FmoPV is associated with renal pathologic processes. In necropsy kidney tissues of two stray cats with positive FmoPV RT-PCR in their urine samples, we observed histopathological features compatible with TIN, as well as detection of N protein of FmoPV in the renal tubules by immunohistochemistry (Fig. 4). A case-control study showed that there is a positive association between FmoPV infection (RT-PCR and/or antibody positivity) and TIN in cats. Some recent studies suggested that feline TIN is mediated by an autoimmune mechanism because cats vaccinated with CRFK cell lysates developed antibodies to both CRFK and kidney cell lysates (27–29). Half these cats sensitized to CRFK lysates on multiple occasions developed TIN at 2 wk postsensitization. Sera from CRFK-inoculated cats were confirmed to recognize annexin A2 and α-enolase by Western blot. In humans, α-enolase antibodies are nephritogenic, and α-enolase and annexin A2 antibodies have been associated with autoimmune diseases. It is therefore possible that a feline nephrotropic virus, such as FmoPV, may trigger a self-sustained immunopathological process after this acute insult. Further study should be performed to ascertain the pathogenic role of this virus, and antiviral and vaccine development may be important in the prevention and treatment of TIN in cats. Notably, some morbilliviruses, such as Peste des Petits Ruminants virus, Rinderpest virus, and canine distemper virus, have also been found in kidney and/or urine (30–32). Further studies would delineate if these viruses are also associated with renal pathologic processes in these animals.
Although domestic cats have been associated with humans for almost 10,000 y, they usually pose little physical hazard to humans. However, as a result of cat bites or via other routes, cats can transmit a range of bacteria (e.g., Bartonella henselae), protozoa (e.g., Toxoplasma gondii), and, uncommonly, viruses (e.g., rabies virus), causing diseases in humans. Apart from the present paramyxovirus, viruses of at least 15 families have been found in cats, including our recent discovery of a picornavirus in cats (33). Moreover, the domestic cats have also been shown to be susceptible to infection by highly pathogenic avian influenza viruses H5N1 and H7N7 and SARS coronavirus, suggesting that they can be susceptible to viruses associated with serious infections (34–36). A previous survey in Hong Kong (http://www.aud.gov.hk/pdf_e/e54ch04.pdf) showed that one in every eight households was keeping pets, with 22.3% of the pet-keepers keeping cats. The number of locally licensed pet shops selling cats and dogs has increased from 77 in 2000 to 155 in 2009. This phenomenal increase is not unexpected, because the population is aging, the marriage rate is decreasing, the divorce rate is increasing, and the birth rate is decreasing. Many households are keeping pets as substitutes for children that do not require childcare. Besides sharing a bed with pets, these pet owners often kiss or are licked by pets. Such behavior may allow significant exposure to zoonotic agents carried by the pet or parasitizing arthropods (37). For FmoPV, serological studies for evidence of its possible spread in the human population could be performed in the context of broader epidemiological studies that target households with and without cats. Because of the possibility of cross-reactivity with other known and unknown morbilliviruses, testing a small number of owners and human contacts of sick cats may lead to unsubstantiated rumors and needless slaughter of healthy cats. On the other hand, host range restriction and pathogenicity may be determined by the difference in the NLS and nuclear export signal sequences and F protein cleavage sites between FmoPV and other morbilliviruses. For example, specific mutation in the NLS of PB2 polymerase of the influenza virus would significantly affect nuclear localization, replication, and host range of the virus (38, 39).
Materials and Methods
Sample Collection.
The Agriculture Fisheries and Conservation Department of Hong Kong provided samples collected from the 457 stray cats as part of a surveillance program. Nasal and rectal swabs, urine, and blood were collected by using procedures described previously (33). In addition, oral and rectal swabs from 16 diseased cats from mainland China were also collected. The use of samples from euthanized stray cats in this study was approved by the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong. Samples were collected immediately after euthanasia as routine policies for disposal of locally captured stray cats.
Necropsies of FmoPV-Infected Stray Cats.
Tissue samples were collected from the lungs; brain; heart; prescaspular, retropharyngeal, submandibular, and thoracic lymph nodes; spleen; liver; kidneys; urinary bladder; gall bladder; thymus; salivary gland; eyeball; nasal turbinate; intestine; pancreas; foot pads; testicles or ovary; and tonsil and adrenal gland. Half of each tissue sample was fixed in 10% neutral buffered formalin for histological processing and the other half was submerged in viral transport medium for RNA extraction and virus isolation.
RT-PCR of L Gene of Morbilliviruses and DNA Sequencing.
Viral RNA was extracted from nasal and rectal swabs, urine, and blood by using an EZ1 Virus Mini Kit (Qiagen) and from tissue samples by using a QIAamp Viral RNA Mini Kit (Qiagen). Morbillivirus detection was performed by amplifying a 155-bp fragment of L gene of morbilliviruses by using conserved primers (LPW12490, 5′-CAGAGACTTAATGAAATTTATGG-3′; and LPW12491, 5′-CCACCCATCGGGTACTT-3′) designed by multiple alignments of available L gene sequences of morbilliviruses. Reverse transcription, PCR, and sequencing were performed according to our previous publications (14, 15).
Real-Time Quantitative RT-PCR.
Real-time quantitative RT-PCR to detect L gene of FmoPV was performed on the 56 positive samples by using LightCyler FastStart DNA Master SYBR Green I Mix reagent kit (Roche), with primers LPW12490 and LPW12491. cDNA was amplified by LightCycler 2.0 (Roche) with 20-μL reaction mixtures containing FastStart DNA Master SYBR Green I Mix reagent kit (Roche), 2 μL of cDNA, 4 mmol/L MgCl2, and 0.5 mmol/L primers at 95 °C for 10 min, followed by 50 cycles at 95 °C for 10 s, 60 °C for 5 s, and 72 °C for 8 s. A plasmid containing the target sequence was used for generation of the standard curves.
Complete Genome Sequencing and Genome Analysis.
The complete genomes of FmoPV were amplified and sequenced by using RNA extracted directly from the specimens as templates with a strategy described in our previous publications (14, 15). Genome analysis was performed as described in our previous publications (14, 15, 33, 40, 41). Phylogenetic trees were constructed by maximum likelihood method by using PhyML 3.0 (42).
Analysis of P mRNA Editing.
To examine the number of G insertions at the P mRNA editing site, mRNA from original specimens was extracted by using the Oligotex mRNA Mini kit (Qiagen). First-strand cDNA synthesis was performed by a using SuperScript III kit (Invitrogen) with random hexamer primers. Primers (5′-TTCATCTCTTAGTTCCCAGGAA-3′ and 5′-TTTCAGACTCACCCTCGATATCT-3′) were used to amplify a 442-bp product of FmoPV covering the putative editing site. PCR, cloning and sequencing were performed as described in our previous publication (14).
Cloning and Purification of (His)6-Tagged Recombinant N from E. coli.
Primers (5′-ACGCGGATCCGATGTCTAGTCTA-3′ and 5′-CGGAATTCGGTTTTAGAAGGTCAGTA-3′) were used to amplify the N gene (519 aa) of FmoPV by RT-PCR. Cloning, expression and purification of (His)6-tagged recombinant N protein was performed as described in our previous publications (14, 41, 43).
Guinea Pig Sera.
Guinea pig antiserum against the N protein of FmoPV was produced by injecting 100 μg purified N protein of FmoPV, with an equal volume of complete Freund adjuvant (Sigma), s.c. to three guinea pigs. Incomplete Freund adjuvant (Sigma) was used in subsequent immunizations. Three inoculations at once every 2 wk per guinea pig were administered. Two weeks after the last immunization, 1 mL of blood was taken via the lateral saphenous vein of the guinea pigs to obtain the sera.
Western Blot Analysis.
Antibodies against the N protein of FmoPV were detected in plasma samples of the 56 cats that were RT-PCR–positive and 401 cats that were RT-PCR–negative for FmoPV by Western blot. Western blot analysis was performed as described in our previous publications (40, 41, 43), by using 1,000 ng purified (His)6-tagged recombinant N protein, 1:1,000 dilutions of cat plasma samples, 1:4,000 dilution of horseradish peroxidase-conjugated goat anti-cat IgG antibody, and 1:10,000 dilution of goat anti-cat IgM antibody (Bethyl).
Viral Culture.
Viral culture and EM were performed according to our previous publications (44, 45). Two hundred microliters of the three samples used for complete genome sequencing were subject to virus culture. After centrifugation, they were diluted fivefold with viral transport medium and filtered. Two hundred microliters of the filtrate was inoculated to 200 μL of MEM with Polybrene. Four hundred microliters of the mixture was added to 24-well tissue culture plates, with CRFK, B95, chicken embryo fibroblast, NIH/3T3, or Vero E6 cells, by adsorption inoculation. After 1 h of adsorption, excess inoculum was discarded, the wells were washed twice with PBS solution, and the mixture was replaced by 1 mL of serum-free MEM supplemented by 0.1 μg/mL of L-1-tosylamide-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma). Cultures were incubated at 37 °C with 5% CO2 and inspected daily by inverted microscopy for CPEs. After 2 to 3 wk of incubation, subculturing to fresh cell line was performed even if there were no CPEs, and culture lysates were collected for RT-PCR for FmoPV. Immunostaining and EM were performed on samples that were RT-PCR–positive for FmoPV.
Immunostaining.
CRFK and Vero E6 cells that were tested positive for FmoPV by RT-PCR were fixed in chilled acetone at −20 °C for 10 min. The fixed cells were incubated with 1:200 dilution of guinea pig antiserum against the N protein of FmoPV, followed by 1:50 diluted FITC-rabbit anti-guinea pig IgG (Invitrogen). Cells were then examined under a fluorescence microscope. Uninoculated cells were used as negative control.
Immunofluorescence Antibody Test.
CRFK cells infected with FmoPV were fixed in chilled acetone at −20 °C for 10 min. The fixed cells were incubated with fourfold dilutions of plasma from 1:10 to 1:10,240 from the 27 cats with necropsies, followed by 1:20 diluted FITC-goat anti-cat IgG (Sigma). Cells were then examined under a fluorescence microscope. Uninfected cells were used as negative control.
Histopathological Examination and Immunohistochemical Staining of FmoPV N Protein in Tissues and Cauxin Protein in Kidneys.
Fixed necropsy organs of the two stray cats were embedded in paraffin. Tissue sections of 5 μm were stained with H&E. Histopathological changes were observed by using a Nikon 80i microscope and imaging system. Expression of FmoPV N protein was examined by immunohistochemical staining. Tissue sections were deparaffinized and rehydrated, followed by blocking endogenous peroxidase with 3% (vol/vol) H2O2 for 20 min, and then with 10% (vol/vol) normal rabbit serum/PBS solution at room temperature for 1 h to minimize nonspecific staining. The sections were incubated at 4 °C overnight with 1:250 dilution of guinea pig anti-N protein antiserum, followed by incubation of 30 min at room temperature with 1:500 dilution of biotin-conjugated rabbit anti-guinea pig IgG, H and L chain (Abcam) (46). Streptavidin/peroxidase complex reagent (Vector Laboratories) was then added and incubated at room temperature for 30 min. Color development was performed by using 3,3′-diaminobenzidine and images were captured with a Nikon 80i imaging system and Spot-advance computer software. Double staining of lymph node was performed by using mouse anti-human myeloid/histiocyte antigen antiserum MAC387 (DakoCyomation) and labeled with Texas red-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) (47). Cauxin protein expression was detected according to a published protocol (48).
Supplementary Material
Acknowledgments
We thank Director Alan Chi-Kong Wong, Siu-Fai Leung, Thomas Hon-Chung Sit, and Howard Kai-Hay Wong [Agriculture Fisheries and Conservation Department (AFCD)]; Veterinary Officers of the AFCD Animal Management Centres for assistance and collection of samples; Dr. Chi-Kwan Yip, Dr. Cassius Chan, and Dr. King-Shun Lo for animal necropsy; Prof. Yamashita (Iwate University) for providing the anti-cat cauxin antibody; and Prof. K. C. Chan for the MAC387 antibody. We thank Mrs. Carol Yu, Prof. Richard Yu, Mr. Hui Hoy, and Mr. Hui Ming for support in genomic sequencing platform, and Ms. Eunice Lam for her donation for emerging infectious disease research. This work was supported in part by Research Grants Council Grant HKU 783611M; the Strategic Research Theme Fund and University Development Fund of the University of Hong Kong; the Providence Foundation Limited in memory of the late Dr. Lui Hac Minh; and the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the Hong Kong Special Administrative Region Department of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see Table S5.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119972109/-/DCSupplemental.
References
- 1.Barrett T. Morbillivirus infections, with special emphasis on morbilliviruses of carnivores. Vet Microbiol. 1999;69:3–13. doi: 10.1016/s0378-1135(99)00080-2. [DOI] [PubMed] [Google Scholar]
- 2.Chua KB, et al. Nipah virus: A recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 3.Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J Gen Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- 4.Moreno-López J, Correa-Girón P, Martinez A, Ericsson A. Characterization of a paramyxovirus isolated from the brain of a piglet in Mexico. Arch Virol. 1986;91:221–231. doi: 10.1007/BF01314282. [DOI] [PubMed] [Google Scholar]
- 5.Osterhaus AD, et al. Morbillivirus infections of aquatic mammals: newly identified members of the genus. Vet Microbiol. 1995;44:219–227. doi: 10.1016/0378-1135(95)00015-3. [DOI] [PubMed] [Google Scholar]
- 6.Philbey AW, et al. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg Infect Dis. 1998;4:269–271. doi: 10.3201/eid0402.980214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidona CA, Kurz HW, Gelderblom HR, Darai G. Isolation and molecular characterization of a novel cytopathogenic paramyxovirus from tree shrews. Virology. 1999;258:425–434. doi: 10.1006/viro.1999.9693. [DOI] [PubMed] [Google Scholar]
- 8.Stone BM, et al. Fatal cetacean morbillivirus infection in an Australian offshore bottlenose dolphin (Tursiops truncatus) Aust Vet J. 2011;89:452–457. doi: 10.1111/j.1751-0813.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 9.Young PL, et al. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg Infect Dis. 1996;2:239–240. doi: 10.3201/eid0203.960315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau SK, et al. Human parainfluenza virus 4 outbreak and the role of diagnostic tests. J Clin Microbiol. 2005;43:4515–4521. doi: 10.1128/JCM.43.9.4515-4521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SK, et al. Clinical and molecular epidemiology of human parainfluenza virus 4 infections in Hong Kong: Subtype 4B as common as subtype 4A. J Clin Microbiol. 2009;47:1549–1552. doi: 10.1128/JCM.00047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virtue ER, Marsh GA, Wang LF. Paramyxoviruses infecting humans: The old, the new and the unknown. Future Microbiol. 2009;4:537–554. doi: 10.2217/fmb.09.26. [DOI] [PubMed] [Google Scholar]
- 13.Eaton BT, Broder CC, Middleton D, Wang LF. Hendra and Nipah viruses: Different and dangerous. Nat Rev Microbiol. 2006;4:23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau SK, et al. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology. 2010;404:106–116. doi: 10.1016/j.virol.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo PC, et al. Complete genome sequence of a novel paramyxovirus, Tailam virus, discovered in Sikkim rats. J Virol. 2011;85:13473–13474. doi: 10.1128/JVI.06356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bart M, Guscetti F, Zurbriggen A, Pospischil A, Schiller I. Feline infectious pneumonia: A short literature review and a retrospective immunohistological study on the involvement of Chlamydia spp. and distemper virus. Vet J. 2000;159:220–230. doi: 10.1053/tvjl.1999.0451. [DOI] [PubMed] [Google Scholar]
- 17.Chatziandreou N, et al. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5) J Gen Virol. 2004;85:3007–3016. doi: 10.1099/vir.0.80200-0. [DOI] [PubMed] [Google Scholar]
- 18.Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegl G, et al. Characteristics and taxonomy of Parvoviridae. Intervirology. 1985;23:61–73. doi: 10.1159/000149587. [DOI] [PubMed] [Google Scholar]
- 20.Truyen U. Evolution of canine parvovirus—a need for new vaccines? Vet Microbiol. 2006;117:9–13. doi: 10.1016/j.vetmic.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 21.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Family Herpesviridae. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. pp. 111–122. [Google Scholar]
- 22.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Family Papillomaviridae. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. pp. 235–248. [Google Scholar]
- 23.Griffin DE. In: Measles Virus. Fields Virology. Knipe DM, et al., editors. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1551–1586. [Google Scholar]
- 24.Chard LS, Bailey DS, Dash P, Banyard AC, Barrett T. Full genome sequences of two virulent strains of peste-des-petits ruminants virus, the Côte d'Ivoire 1989 and Nigeria 1976 strains. Virus Res. 2008;136:192–197. doi: 10.1016/j.virusres.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 25.Visser IK, et al. Fusion protein gene nucleotide sequence similarities, shared antigenic sites and phylogenetic analysis suggest that phocid distemper virus type 2 and canine distemper virus belong to the same virus entity. J Gen Virol. 1993;74:1989–1994. doi: 10.1099/0022-1317-74-9-1989. [DOI] [PubMed] [Google Scholar]
- 26.Terai M, Burk RD. Felis domesticus papillomavirus, isolated from a skin lesion, is related to canine oral papillomavirus and contains a 1.3 kb non-coding region between the E2 and L2 open reading frames. J Gen Virol. 2002;83:2303–2307. doi: 10.1099/0022-1317-83-9-2303. [DOI] [PubMed] [Google Scholar]
- 27.Whittemore JC, Hawley JR, Jensen WA, Lappin MR. Antibodies against Crandell Rees feline kidney (CRFK) cell line antigens, alpha-enolase, and annexin A2 in vaccinated and CRFK hyperinoculated cats. J Vet Intern Med. 2010;24:306–313. doi: 10.1111/j.1939-1676.2010.0476.x. [DOI] [PubMed] [Google Scholar]
- 28.Lappin MR, et al. Investigation of the induction of antibodies against Crandell-Rees feline kidney cell lysates and feline renal cell lysates after parenteral administration of vaccines against feline viral rhinotracheitis, calicivirus, and panleukopenia in cats. Am J Vet Res. 2005;66:506–511. doi: 10.2460/ajvr.2005.66.506. [DOI] [PubMed] [Google Scholar]
- 29.Lappin MR, Basaraba RJ, Jensen WA. Interstitial nephritis in cats inoculated with Crandell Rees feline kidney cell lysates. J Feline Med Surg. 2006;8:353–356. doi: 10.1016/j.jfms.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kul O, Kabakci N, Atmaca HT, Ozkul A. Natural peste des petits ruminants virus infection: Novel pathologic findings resembling other morbillivirus infections. Vet Pathol. 2007;44:479–486. doi: 10.1354/vp.44-4-479. [DOI] [PubMed] [Google Scholar]
- 31.Liess B, Plowright W. Studies on the pathogenesis of rinderpest in experimental cattle. I. Correlation of clinical signs, viraemia and virus excretion by various routes. J Hyg (Lond) 1964;62:81–100. doi: 10.1017/s0022172400039796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito TB, et al. Detection of canine distemper virus by reverse transcriptase-polymerase chain reaction in the urine of dogs with clinical signs of distemper encephalitis. Res Vet Sci. 2006;80:116–119. doi: 10.1016/j.rvsc.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Lau SK, et al. Identification of a novel feline picornavirus from the domestic cat. J Virol. 2012;86:395–405. doi: 10.1128/JVI.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marschall J, Hartmann K. Avian influenza A H5N1 infections in cats. J Feline Med Surg. 2008;10:359–365. doi: 10.1016/j.jfms.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martina BE, et al. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Riel D, Rimmelzwaan GF, van Amerongen G, Osterhaus AD, Kuiken T. Highly pathogenic avian influenza virus H7N7 isolated from a fatal human case causes respiratory disease in cats but does not spread systemically. Am J Pathol. 2010;177:2185–2190. doi: 10.2353/ajpath.2010.100401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chomel BB, Sun B. Zoonoses in the bedroom. Emerg Infect Dis. 2011;17:167–172. doi: 10.3201/eid1702.101070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resa-Infante P, et al. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS ONE. 2008;3:e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarendeau F, et al. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 2008;4:e1000136. doi: 10.1371/journal.ppat.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau SK, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo PC, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 43.Woo PC, et al. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li IW, et al. Differential susceptibility of different cell lines to swine-origin influenza A H1N1, seasonal human influenza A H1N1, and avian influenza A H5N1 viruses. J Clin Virol. 2009;46:325–330. doi: 10.1016/j.jcv.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Peiris JS, et al. SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan KH, et al. Wild type and mutant 2009 pandemic influenza A (H1N1) viruses cause more severe disease and higher mortality in pregnant BALB/c mice. PLoS ONE. 2010;5:e13757. doi: 10.1371/journal.pone.0013757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susta L, Torres-Velez F, Zhang J, Brown C. An in situ hybridization and immunohistochemical study of cytauxzoonosis in domestic cats. Vet Pathol. 2009;46:1197–1204. doi: 10.1354/vp.08-VP-0132-B-FL. [DOI] [PubMed] [Google Scholar]
- 48.Miyazaki M, et al. Tubulointerstitial nephritis causes decreased renal expression and urinary excretion of cauxin, a major urinary protein of the domestic cat. Res Vet Sci. 2007;82:76–79. doi: 10.1016/j.rvsc.2006.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.