Abstract
CRM197 is an enzymatically inactive and nontoxic form of diphtheria toxin that contains a single amino acid substitution (G52E). Being naturally nontoxic, CRM197 is an ideal carrier protein for conjugate vaccines against encapsulated bacteria and is currently used to vaccinate children globally against Haemophilus influenzae, pneumococcus, and meningococcus. To understand the molecular basis for lack of toxicity in CRM197, we determined the crystal structures of the full-length nucleotide-free CRM197 and of CRM197 in complex with the NAD hydrolysis product nicotinamide (NCA), both at 2.0-Å resolution. The structures show for the first time that the overall fold of CRM197 and DT are nearly identical and that the striking functional difference between the two proteins can be explained by a flexible active-site loop that covers the NAD binding pocket. We present the molecular basis for the increased flexibility of the active-site loop in CRM197 as unveiled by molecular dynamics simulations. These structural insights, combined with surface plasmon resonance, NAD hydrolysis, and differential scanning fluorimetry data, contribute to a comprehensive characterization of the vaccine carrier protein, CRM197.
Diphtheria is a contagious respiratory disease that was a major cause of death among children around the world until mass vaccination was introduced in the 1920s. Although diphtheria has now been virtually eliminated in the industrialized world, rare outbreaks still occur worldwide (1, 2). Corynebacterium diphtheriae was shown to be the causative agent of diphtheria by Loeffler in 1885 (3), and Roux and Yersin showed that an extracellular toxin, diphtheria toxin (DT) secreted by C. diphtheriae is responsible for toxicity (4). The formaldehyde-treated detoxified form of DT, diphtheria toxoid, has been successfully used for mass vaccination and is still widely used as a component of combination vaccines (5, 6).
A major contribution to the understanding of the mode of action of DT was the discovery of mutated forms in the early 1970s (7). Several phages encoding mutants of DT, named cross-reactive materials (CRMs), were isolated following nitrosoguanidine-based mutagenesis of the phage containing the gene encoding DT. Being naturally nontoxic, CRMs were immediately recognized as having great potential for vaccine development. The most important CRM identified was CRM197, an enzymatically inactive and nontoxic form of DT that contains a single amino acid substitution from Glycine to Glutamate in position 52 (8). Subsequently CRM197 was found to be an ideal carrier for conjugate vaccines against encapsulated bacteria. Here, the carrier protein is covalently linked to poorly immunogenic and T-cell-independent capsular polysaccharides, thus creating T-cell-dependent conjugate antigens that are highly immunogenic in infants (9–11).
Vaccines containing CRM197 as a carrier protein have been successfully used to immunize hundreds of millions of children. Such vaccines currently include Menveo®, a recently approved tetravalent conjugate vaccine against serogroups A-C-W135-Y of Neisseria meningitidis, Menjugate® and Meningitec® (against serotype C of N. meningitidis), Vaxem-Hib® and HibTITER® (against Haemophilus influenzae type B, Hib), and the multivalent pneumococcal conjugate Prevnar™ (12).
The widespread use of diphtheria toxoid and CRM197 has prompted many investigations of DT and related proteins. Diphtheria toxin is an ADP-ribosylating enzyme that is secreted as a proenzyme of 535 residues and is processed by trypsin-like proteases with release of two fragments (A and B). Fragment A uses NAD as a substrate, catalyzing the cleavage of the N-glycosidic bond between the nicotinamide ring and the N-ribose and mediating the covalent transfer of the ADP-ribose (ADPRT activity) to the modified Histidine 715 (diphthamide) of the elongation factor EF-2. This posttranslational diphthamide modification inactivates EF-2, halting protein synthesis and resulting in cell death. Extensive structural studies elucidated the molecular architecture of DT (13–19). The A fragment of DT (also named C domain) carries the catalytic active site and is the only fragment of the toxin required for the final step of intoxication, while the B fragment carries the R and T domains, which mediate binding to receptors on the host cell surface and promote the pH-dependent transfer of fragment A to the cytoplasm, respectively. An Arginine-rich disulfide-linked loop connects fragment A to fragment B (or domain C to domains TR), and this interchain disulfide bond is the only covalent link between the two fragments after proteolytic cleavage of the chain at position 186.
While much progress has been made on the molecular characterization of DT over the last two decades, an understanding of the molecular basis for the lack of toxicity of CRM197 has so far been elusive. Here we present the crystal structures of full-length nucleotide-free (NF)-CRM197, and of CRM197 in complex with the NAD hydrolysis product nicotinamide (NCA). In addition, the differences between CRM197 and DT were elucidated using surface plasmon resonance (SPR), a NAD-glycohydrolase (NADase) activity assay, molecular dynamics (MD) simulations, and differential scanning fluorimetry (DSF).
Results
Overall Structure of Nucleotide-Free-CRM197.
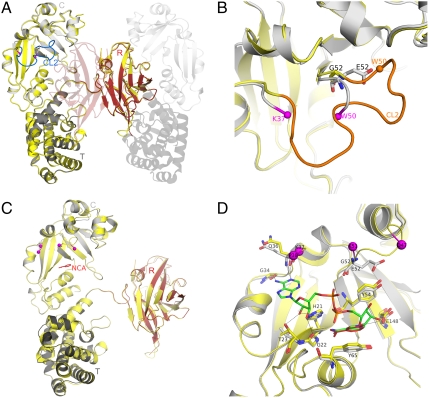
Crystals of a noncovalent dimer of CRM197 were obtained at pH 9 after freezing the protein in PBS buffer as previously reported for crystal forms III and IV of DT (20). The first crystal structure of DT was also solved as a noncovalent dimer that was later shown to be a nontoxic form. This arose as an artifact of freezing the toxin in sodium phosphate (21, 22). The structure of NF-CRM197 was solved at 2-Å resolution by molecular replacement using as input model the coordinates of NF-DT (PDB ID code 1SGK) (18). The asymmetric unit contains one molecule of CRM197, which interacts with a second (symmetry-related) molecule by “domain swapping” (Fig. 1A and Fig. S1), as observed previously in dimeric DT (23). Thus, the monomer of CRM197 has an “open” conformation, where an individual monomer is noncovalently associated to a second chain by swapping of their R domains (Fig. S1B). Nonetheless, the swapping of the R domains makes the assembled CT domains from one chain and the R domain from the second chain virtually identical to the monomeric “closed” DT reported previously (13) (Fig. S1C).
Fig. 1.
NF- and NCA-CRM197. (A) CRM197 catalytic (C) (residues 1–187), transmembrane (T) (residues 201–384), and receptor (R) (residues 387–535) domains are colored as light gray, dark gray, and red, respectively, and labeled. Nucleotide-free DT (PDB ID code 1SGK) is shown as yellow cartoon after superposition onto nucleotide-free CRM197. The active-site loop (CL2) of DT is shown in blue. The symmetry-related, domain-swapped molecule that forms the dimer of CRM197 is depicted using transparent cartoons. (B) Zoom into the superimposed catalytic domain only, showing the active-site loop (CL2) from DT in orange, the position of E52 of CRM197 and G52 of DT as sticks with carbon atoms colored in gray and yellow, respectively. The Cα atoms of the last visible residues of the disordered active-site loop of CRM197 (K37 and W50) are shown as spheres and colored in magenta. Nitrogen and oxygen atoms are colored in blue and red, respectively. (C) NAD-bound DT (PDB ID code 1TOX) is depicted as yellow cartoon after superposition onto NCA-CRM197 that is colored by domains as in Fig. 1A. NCA bound to CRM197 is shown as red sticks and labeled, and the last visible residues of the active-site loop of CRM197 and DT complexed with NAD are shown as spheres and colored in magenta. (D) The NAD-binding site in CRM197 (gray) compared to DT (yellow). Residues known to be involved both in NAD-binding (G22, T23, G34, Q36, Y54, Y65) or in catalysis (H21, E148) are shown as sticks for DT (carbon atoms in yellow) and CRM197 (carbon atoms in gray). E52 and G52 in CRM197 and DT are shown as sticks, while last visible residues of the active-site loop in both NCA-CRM197 (K37, K51) and NAD-DT (P38, D47) are shown as spheres and colored magenta. The NAD bound to DT is shown as sticks with carbon atoms colored in green, and nitrogen, oxygen, and phosphorous atoms, colored in blue, red, and orange, respectively. The NCA bound to CRM197 is shown as red sticks and labeled.
The overall fold of NF-CRM197 is nearly equivalent to the fold of a monomer from dimeric NF-DT, with an overall rmsd of 0.5 Å for 498 equivalent Cα atoms (Fig. 1A). Therefore, no major conformational changes affecting the overall fold were observed when comparing the apo-structure of CRM197 to DT.
Active-Site Loop of Nucleotide-Free-CRM197 and position of Glutamate 52.
Electron-density maps were of excellent overall quality, readily allowing model building of CRM197. One region of major interest partly lacking electron density is the active-site loop (CL2) of the C domain (Fig. 1B). In DT, this loop is positioned over the NAD-binding pocket and is thought to be important for access of NAD to the binding cleft and for EF-2 recognition (16). In NF-CRM197, there is no electron density for the CL2 loop residues 38–49, while clear density resumes for the backbone atoms of the flanking residues K37 and W50, suggesting that dislocation of the active-site loop takes place upon the substitution G52E. In summary, although NF-CRM197 has an overall fold virtually identical to NF-DT, one major difference is observed in the displacement of the active-site loop that covers the NAD-binding pocket of DT.
Structure of Nicotinamide-CRM197.
Despite previous reports that CRM197 has a significantly lower affinity for NAD when compared with DT (24–28), comparisons of NF-DT and NAD-DT with NF-CRM197 show that, except for the disordered active-site loop, the NAD-binding pocket is precisely maintained in CRM197. This was confirmed by superimposing the A fragment of CRM197 on the corresponding residues of NF-DT (1SGK) (rmsd of 0.4 Å for 171 aligned Cα atoms) or on those of NAD-DT (1TOX) (rmsd 0.5 Å). Therefore, the apo-structure presented herein suggested that CRM197 might also be expected to bind NAD. To assess this possibility, an excess of NAD (5 mM) was used to perform soaking experiments using crystals of NF-CRM197. After soaking for approximately 5 min, crystals were transferred to a cryo-protectant solution and flash cooled in liquid nitrogen at -180°C (see Methods).
The structure of CRM197 from crystals soaked with NAD was solved at 2.0-Å resolution by molecular replacement using as input model the coordinates of NF-DT (PDB ID code 1SGK). The asymmetric unit contains two molecules of CRM197 in the open conformation, interacting by “domain swapping” as described above for NF-CRM197 and NF-DT. The overall structure is highly similar to NF- and NAD-DT, and to NF-CRM197, with rmsds of 0.5 Å in all cases, comparing 501 and 494 identical Cα atoms, respectively (Fig. 1C). Also in this new crystal form, a lack of electron density for the active-site loop residues 38–50 again suggested a high degree of flexibility in this region of CRM197.
Molecular Details of the Nicotinamide-CRM197 Interaction.
After the first cycles of restrained refinement, clear peaks of 2Fo - Fc electron-density at a contour level of 1σ and of Fo - Fc at 3σ became visible in the active-site region of CRM197 (Fig. S2). Superposition of DT complexed with NAD (PDB ID code 1TOX) was used as a guide to locate and model NAD into this additional electron density and to verify if the NAD from soakings was indeed incorporated into the crystals of CRM197. Modeling and refinement resulted in well-resolved electron density for NCA, while electron density for the remaining ADP-ribose was diffuse and thus difficult to interpret and model (Fig. S2). Group occupancy refinement in PHENIX (29) was used after modeling ADP-ribose or whole NAD in order to test if partial occupancy could explain the ambiguous densities. However, this approach did not result in improved maps. Therefore, only NCA was included and refined in the final model, hereafter termed NCA-CRM197. The remaining unexplained blobs of electron density are likely a consequence of the presence of a mixture of substrate and hydrolyzed products in the catalytic pocket of CRM197.
Comparison of NCA-CRM197 and NAD-DT (PDB ID code 1TOX) reveals the NCA binding mode to be identical (Fig. 1D and Fig. S2). Also, the position and relative orientations of protein residues making contacts with NCA or directly involved in the ADPRT activity are conserved among DT and CRM197 as shown from their superposition (Fig. 1D). Y54 and Y65 line the catalytic cleft of DT, forming a hydrophobic groove that accommodates NCA. Y65 is essential for NAD binding (30), and as in NAD-DT, it forms an aromatic stacking interaction with the ring of NCA and Y54 (Fig. 1D). E148 is a catalytic residue, involved in the cleavage of the N-glycosidic bond that releases nicotinamide and ADP-ribose. The side chain of residue H21 forms hydrogen bonds to NAD and to the backbone of DT and was previously postulated to be involved in maintaining the structural integrity of the NAD-binding pocket (18). Other residues known to be directly or indirectly involved in NAD binding (G22, T23, G34, Q36) (16) are topologically equivalent between CRM197 and DT (Fig. 1D). In summary, the G52E mutation does not directly affect the architecture of the NAD-binding pocket.
NAD Binding to CRM197 and DT.
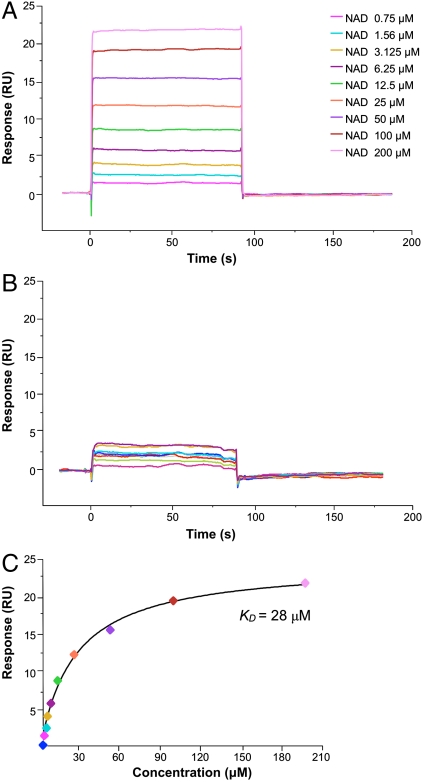
To verify and compare binding of NAD to CRM197 and DT in solution, we measured the affinity of both proteins for NAD by SPR (Fig. 2). Binding of NAD was observed both for immobilized DT (Fig. 2A) and immobilized CRM197 (Fig. 2B). However, a significant dose response to increasing NAD concentration was observed only for DT, while the binding of NAD to CRM197 was almost negligible. A steady-state binding analysis of the NAD-DT interaction yielded an equilibrium dissociation constant (KD) of 28 ± 3 μM, which matches the previously reported values (Fig. 2C) (31). In contrast, the lower response of immobilized CRM197 to increasing concentrations of NAD resulted in data scattered within a low resonance unit interval, which did not allow determination of a binding affinity. Thus, although the structures reveal that CRM197 possesses an intact active site, the SPR data show that its ability to effectively bind NAD in solution is significantly reduced. These observations are in agreement with our initial structure-based hypothesis that the dislocation of the active-site loop of CRM197, while not altering the overall architecture of the active site, has a direct influence on the ability of CRM197 to bind its substrate.
Fig. 2.
CRM197 has residual affinity for NAD. Increase of resonance units upon binding of NAD to immobilized DT and CRM197 is shown in A and B, respectively. The plateau line represents the steady-state equilibrium phase of interactions between the proteins and NAD, whereas the decrease in resonance from the plateau represents the dissociation phase. (C) Steady-state analysis of NAD binding to DT.
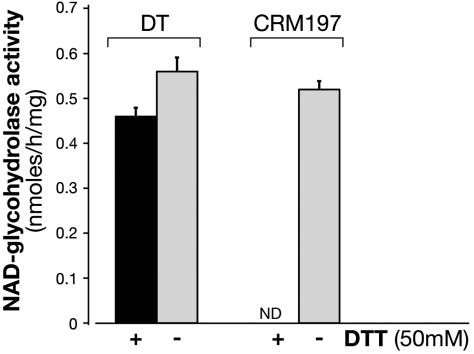
To support the structurally informed hypothesis of residual NAD binding and cleavage by CRM197, we also measured the ability of CRM197 to hydrolyse NAD in solution and compared it to DT. A radioactive NADase activity assay was performed in the presence or absence of the reducing agent dithiothreitol (DTT). While DT shows comparable, high levels of NADase activity both in the presence or absence of DTT (Fig. 3), CRM197 is inactive in the presence, and partially active (approximately 80%) in the absence of the reducing agent. Considering that crystals of the complex NCA-CRM197 were obtained in nonreducing conditions, this result is in support of a further destabilizing effect by the reducing agent. The expected effect that DTT has on CRM197, or DT, is to reduce the disulfide bond that holds together fragment A (C domain) and B (T/R domain). However, no release of fragment A can take place upon addition of DTT alone; for this to happen, trypsin would also be required (24). On the contrary, either in presence or absence of reducing agent, DT is equally able to cleave NAD.
Fig. 3.
CRM197 in nonreducing conditions has NAD-glycohydrolase activity. The NAD-glycohydrolase activity of DT (20 μg) and CRM197 (20 μg) was carried out in 50 mM potassium phosphate, pH 7.5, 0.1 mM [carbonyl-14C]NAD (0.05 μCi) in a total volume of 0.15 ml. The activity was measured in the presence (black bars) or in the absence (gray bars) of 50 mM DTT. Samples (50 μL) after incubation at 37 C for 18 h were applied to 1-mL column of Dowex AG 1-X2 and 14C-nicotinamide eluted with 5 mL of H2O for liquid scintillation counting. NAD-glycohydrolase is expressed as nmol/h/mg and the incorporated radioactivity was measured in a Packard Top counter. ND, not detectable. Data are the means SD of values from two independent experiments.
Together, the SPR and NADase data confirm the structural observation that CRM197 has an intact NAD-binding pocket, equivalent to that present in DT. CRM197 is still able to bind NAD, although with significantly less affinity than DT and only in the absence of a further destabilizing agent (i.e., reducing agents that likely detach fragment A from fragment B without fully releasing it) it is also able to hydrolyze NAD.
Molecular Dynamics of the Active-Site Loop and Overall Stability of CRM197.
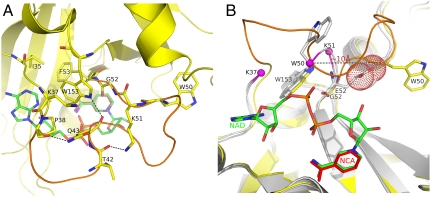
In order to understand the direct influence of the G52E substitution on the conformation of the active-site loop of CRM197, we performed molecular dynamics simulations with a time step of 2 fs. As initial coordinates for DT we used NF-DT (PDB ID Code 1SGK), while for CRM197 we modified the NF-DT coordinates by mutating the residue G in position 52 to E as observed in our crystal structures. Residues 38–52 of the active-site loop in the structure of CRM197 have rmsd values greater than 90% of the values for wild-type DT, supporting the notion that this loop becomes more flexible as a direct consequence of the G52E mutation. The enhanced flexibility of the loop is associated with the loss of intraloop hydrogen bonds that help to stabilize the native loop conformation in NF-DT. Hydrogen bonds between the backbone carbonyl of P38 and the side chain NH of Q43 and between the backbone carbonyl of T42 and the Nε of K51 were observed with frequency greater than 50% in the NF-DT simulation and less than 15% in CRM197 (Fig. 4A). The loop destabilization is likely induced by the direct impact of the G52E mutation on two neighboring tryptophan residues (W50 and W153), which surround residue 52 (Fig. 4B). In the NF-DT structure, W153 interacts with the active-site loop via an H-bond between its side chain NεH and the backbone carbonyl of Q43. Also, W153 is involved in hydrophobic interactions with G52 and with the side chains of K37, I35, P38, Q43, and F53 (Fig. 4A). The analysis of the MD trajectories demonstrates that the frequency with which the bond between W153 and Q43 is formed is 40% in NF-DT but only 1.6% in NF-CRM197.
Fig. 4.
Active-site loop conformation of CRM197 sampled by MD. (A) The active-site loop of NF-DT (PDB ID code 1SGK) is shown in orange. Residues of the loop involved in hydrogen bonds (P38, T42, K51, W153) are shown in sticks with carbon atoms colored in yellow, and the H-bond shown with dashes. Residues of the active site (I35, K37, F53) contributing to a hydrophobic environment around W153 are also shown as sticks. As reference, the NAD from the complex NAD-DT (PDB ID code 1TOX) is superimposed on NF-DT and shown as sticks in the background with carbon atoms colored in green. (B) Reoriented view of the active site. NF-CRM197 and NCA-CRM197 are shown in gray, after superposition on NF-DT (yellow). W50, G52, E52, and W153 from NF-DT, and from NCA-CRM197 and NF-CRM197 are shown in sticks with carbon atoms colored in yellow and gray for DT and CRM197, respectively. OE1 and OE2 atoms of E52 of CRM197 are shown as dots.
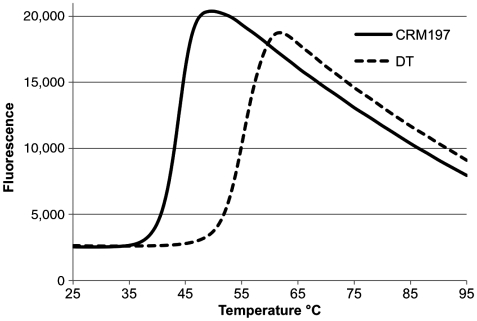
To evaluate if the disorder of the active-site loop might have any direct influence on the overall stability of CRM197, we measured the thermal stability of CRM197 and DT by DSF, observing a lower melting temperature (-10°C) for CRM197 (Fig. 5). This, together with the SPR and MD data above, suggests that the G52E mutation contributes not only to the conformational flexibility of the active-site loop and the reduced affinity for NAD, but it also has an influence on the overall stability of CRM197.
Fig. 5.
CRM197 is less thermostable than DT. Thermal denaturation was induced heating CRM197 and DT from 25 °C to 95 °C. CRM197 and DT displayed Tm values of 44.3 °C and 55.2 °C respectively and 10.9 °C ΔTm , indicating a greater stability of DT in comparison to CRM197.
Discussion
An explanation for the lack of toxicity in CRM197 has eluded scientists since it was discovered that the difference from DT is the single amino acid substitution G52E (8). G52 was already known as a noncatalytic residue, and the crystal structures of DT showed that residue G52 is not directly involved in either NAD binding or EF-2 recognition. However, several biochemical experiments supported the notion of conformational differences between CRM197 and DT. For example, CRM197 and DT were shown to possess different susceptibilities to proteolysis (32); monoclonal antibodies against CRM197 showed different binding properties when compared with binding to DT (33); CRM197 was also shown to be less stable in the cytoplasm than DT (34); and ATP was shown to bind to DT inhibiting binding to the receptors (28) but not to CRM197 (35). However, more recent studies also showed that CRM197 overexpression in the cytosol can be toxic to the cell and may be able to weakly inhibit protein synthesis (36–38).
The crystal structures of CRM197 presented here show that the only difference with DT is in the conformation of the active-site loop in front of the NAD-binding site. While this loop was previously observed well ordered in the structure NF-DT (18) and disordered in the complex NAD-DT (16), CRM197 shows an intrinsic flexibility of this region regardless of the presence or absence of substrate. In addition to an identical overall fold, CRM197 and DT also have an identical organization and architecture of the NAD-binding site, and accordingly the co-crystal structure NCA-CRM197 shows that CRM197 is still able to hydrolyze NAD and release NCA, which binds to CRM197 in an equivalent way to the complex NAD-DT.
One consequence of the flexible active-site loop of CRM197 is a reduced ability to bind NAD. Partially in agreement with previous studies, SPR shows that CRM197 has much lower affinity for NAD than DT. However, confirming the structural observations of a somewhat catalytically competent CRM197, NAD hydrolysis by CRM197 in solution could be detected, but only in nonreducing condition (same as the co-crystal structure conditions). It is likely that an already destabilized CRM197 becomes completely inactive when DTT, reducing the interfragment disulfide bond between fragments A and B, introduces further instability in the domain organization of CRM197. Despite the instability caused by the G52E mutation and the ensuing active-site loop flexibility, in the absence of DTT CRM197 is identical to the catalytically competent conformation of DT and therefore able to hydrolyze NAD. As an additional verification, we were unable to either obtain crystals of CRM197, or to measure any NADase activity, in the presence of the reducing agent DTT.
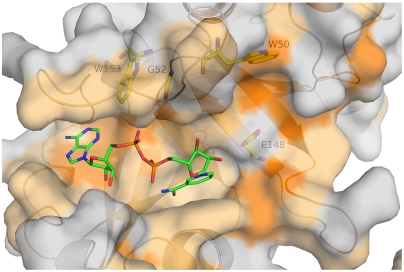
Molecular dynamics simulations reveal the molecular basis for the active-site loop disorder, which originates from the disruption of hydrogen-bond interactions between W153 and the backbone atoms of Q43 of the active-site loop. Notably, this translates into appreciably less frequent H bonds in CRM197 between residues of the active-site loop. A second tryptophan located nearby, W50, is partially exposed to the solvent, and its indole side chain contributes to form a hydrophobic pocket that accommodates the catalytic residue E148 (Fig. 6). Mutagenesis of W50 and W153 to alanine has dramatic effects both on the ADPRT and NADase activity of DT (39). The W50A mutant shows a decrease of ADPRT activity by almost 105-fold and it completely abolishes the NADase activity, while the W153A substitution shows an approximately 40-fold and an approximately 10-fold decrease of ADPRT and NADase activity, respectively. Instead, when the tryptophans are mutated to phenylalanine, these mutants (W50F and W153F) show only minimal reduction in both activities, confirming that aromatic stacking interactions play a significant role in substrate binding.
Fig. 6.
Hydrophobicity of the NAD-binding pocket of DT. NF-DT (1SGK) is shown as gray surface and hydrophobic residues are colored using a scale from light to bright orange. The superimposed NAD molecule from the structure NAD-DT (PDB ID code 1TOX) is shown as sticks with carbon, oxygen, nitrogen, and phosphorous atoms colored in green, red, blue, and orange, respectively. Residues W153, G52, W50, and E148 are shown as sticks under the transparent surface, with carbon, oxygen, and nitrogen atoms colored in yellow, red, and blue, respectively.
Our structures show that in addition to introducing steric clashes with the backbone of W50 (Fig. 4B), because G52 is packed between the hydrophobic W50 and W153 (Fig. 4A and Fig. 6) the polar side chain of E52 is displaced toward the solvent. As a direct consequence, although this entire region is characterized by high flexibility, we can observe the dislodgment of the indole side chain of W50 in NF-CRM197, which is almost 10 Å distal to the same residue in DT (Fig. 4B).
An additional effect of the flexibility of the active-site loop is overall instability of CRM197, as observed by DSF. This is in agreement with the different properties of CRM197 and DT, such as: different interaction with the toxin-receptors (24); less stability in the cytosol (34); more susceptibility to proteases (32); and almost complete lack of toxicity. However, because our data show that there are no major conformational changes in the overall structure of the proteins but rather increased flexibility and instability, it is not surprising that when highly expressed in the reducing environment of the cytosol, CRM197 has residual toxicity (36–38).
Our results outline the molecular events that take place upon mutation of Glycine 52 to Glutamate to produce CRM197, providing experimental evidence for both localized and overall destabilization. The combination of structural, dynamics, and functional studies described in this work promote a better understanding of the differences between CRM197 and DT that make CRM197 an ideal carrier protein for use in vaccine development.
Methods
Protein Production and Crystallization.
CRM197 was obtained from Manufacturing of Novartis Vaccines and Diagnostics. Purified DT was purchased from Sigma-Aldrich Corporation. Purification and crystallization methods are outlined in SI Methods.
Data Collection and Structure Determination.
Data were collected at beamline 5.0.3 of the ALS and both the structure of nucleotide-free and nicotinamide-bound CRM197 were solved at 2.0 Å by molecular replacement in PHASER (40). Data collection and refinement statistics are shown in Table S1. Figures were generated with the program PYMOL (http://www.pymol.org). Details of data collection and structure determination are described in SI Methods.
Surface Plasmon Resonance (SPR), Biacore.
All SPR experiments were performed using a Biacore T200 (GE Healthcare) equilibrated at 25 °C. Monomeric forms of DT and of CRM197 were immobilized on the sensor surface and NAD (Sigma) was injected in running buffer. Full details of the experimental setup, titration series, and data analysis methods are presented in SI Methods.
NAD-Glycohydrolase Activity Assay.
An enzymatic assay based on nicotinamide release was employed to measure the NAD-glycohydrolase activity of monomeric CRM197 and DT (41). Full details are outlined in SI Methods.
Molecular Dynamics Simulation.
For the DT simulation we used the apo crystal structure from PDB ID 1GSK. In order to set up the mutant simulation, the residue G52 was substituted for E in 1SGK. The time step for integration was 2 fs, and the coordinates of all atoms were saved every 5 ps. The full protocol employed can be found in SI Methods.
Thermofluor Assay/Differential Scanning Fluorimetry.
Full details of the experimental setup are presented in SI Methods.
Supplementary Material
Acknowledgments.
We thank Daniele Veggi for technical assistance, Silvana Savino, Scott Lesley, and Martin Seidel for continued support, and Catherine Mallia for editorial assistance. The work in this paper is based on experiments conducted at beamline 5.0.3 of the Advanced Light Source (ALS). The ALS is supported by the director, Office of Science, Office of Basic Energy Sciences, Material Sciences Division of the US Department of Energy under contract DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
Footnotes
Conflict of interest statement: The sponsor is a full-time employee of Novartis Vaccines
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201964109/-/DCSupplemental.
References
- 1.Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. The pathology of diphtheria. J Infect Dis. 2000;181:S116–S120. doi: 10.1086/315551. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Diphtheria epidemic—new independent states of the former Soviet Union, 1990–1994. MMWR Morb Mortal Wkly Rep. 1995;44:177–181. [PubMed] [Google Scholar]
- 3.Pappenheimer AM, Gill DM. Diphtheria. Science. 1973;182:353–358. doi: 10.1126/science.182.4110.353. [DOI] [PubMed] [Google Scholar]
- 4.Roux E, Yersin A. Contribution a l’etude de la diphthérie [Contribution to the study of diphtheria] Ann Inst Pasteur (Paris) 1888;2:629–661. in French. [Google Scholar]
- 5.FitzGerald JG. Diphtheria toxoid as an immunizing agent. Can Med Assoc J. 1927;17:524–529. [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Diphtheria vaccine. Weekly Epidemiological Record. 2006;81:24–32. [Google Scholar]
- 7.Uchida T, Gill DM, Pappenheimer AM. Mutation in the structural gene for diphtheria toxin carried by temperate phage. Nature New Biol. 1971;233:8–11. doi: 10.1038/newbio233008a0. [DOI] [PubMed] [Google Scholar]
- 8.Giannini G, Rappuoli R, Ratti G. The amino-acid sequence of two non-toxic mutants of diphtheria toxin: CRM45 and CRM197. Nucleic Acids Res. 1984;12:4063–4069. doi: 10.1093/nar/12.10.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol. 2010;28:107–130. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- 10.Pollard AJ, Perrett KP, Beverley PC. Maintaining protection against invasive bacteria with protein-polysaccharide conjugate vaccines. Nat Rev Immunol. 2009;9:213–220. doi: 10.1038/nri2494. [DOI] [PubMed] [Google Scholar]
- 11.Ada G, Isaacs D. Carbohydrate-protein conjugate vaccines. Clin Microbiol Infect. 2003;9:79–85. doi: 10.1046/j.1469-0691.2003.00530.x. [DOI] [PubMed] [Google Scholar]
- 12.Shinefield HR. Overview of the development and current use of CRM(197) conjugate vaccines for pediatric use. Vaccine. 2010;28:4335–4339. doi: 10.1016/j.vaccine.2010.04.072. [DOI] [PubMed] [Google Scholar]
- 13.Choe S, et al. The crystal structure of diphtheria toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 14.Bennett MJ, Eisenberg D. Refined structure of monomeric diphtheria toxin at 2.3 A resolution. Protein Sci. 1994;3:1464–1475. doi: 10.1002/pro.5560030912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett MJ, Choe S, Eisenberg D. Refined structure of dimeric diphtheria toxin at 2.0 A resolution. Protein Sci. 1994;3:1444–1463. doi: 10.1002/pro.5560030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 17.Louie GV, Yang W, Bowman ME, Choe S. Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor. Mol Cell. 1997;1:67–78. doi: 10.1016/s1097-2765(00)80008-8. [DOI] [PubMed] [Google Scholar]
- 18.Bell CE, Eisenberg D. Crystal structure of nucleotide-free diphtheria toxin. Biochemistry. 1997;36:481–488. doi: 10.1021/bi962214s. [DOI] [PubMed] [Google Scholar]
- 19.Weiss MS, Blanke SR, Collier RJ, Eisenberg D. Structure of the isolated catalytic domain of diphtheria toxin. Biochemistry. 1995;34:773–781. doi: 10.1021/bi00003a010. [DOI] [PubMed] [Google Scholar]
- 20.Fujii G, Choe SH, Bennett MJ, Eisenberg D. Crystallization of diphtheria toxin. J Mol Biol. 1991;222:861–864. doi: 10.1016/0022-2836(91)90577-s. [DOI] [PubMed] [Google Scholar]
- 21.Carroll SF, Barbieri JT, Collier RJ. Dimeric form of diphtheria toxin: Purification and characterization. Biochemistry. 1986;25:2425–2430. doi: 10.1021/bi00357a019. [DOI] [PubMed] [Google Scholar]
- 22.Collier RJ. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 23.Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchida T, Pappenheimer AM, Harper AA. Reconstitution of diphtheria toxin from two nontoxic cross-reacting mutant proteins. Science. 1972;175:901–903. doi: 10.1126/science.175.4024.901. [DOI] [PubMed] [Google Scholar]
- 25.Uchida T, Pappenheimer AM, Greany R. Diphtheria toxin and related proteins. I. Isolation and properties of mutant proteins serologically related to diphtheria toxin. J Biol Chem. 1973;248:3838–3844. [PubMed] [Google Scholar]
- 26.Uchida T, Pappenheimer A, Jr, Harper A. Diphtheria toxin and related proteins. II. Kinetic studies on intoxication of HeLa cells by diphtheria toxin and related proteins. J Biol Chem. 1973;248:3845–3850. [PubMed] [Google Scholar]
- 27.Michel A, Dirkx J. Fluorescence studies of nucleotides binding to diphtheria toxin and its fragment A. Biochim Biophys Acta. 1974;365:15–27. doi: 10.1016/0005-2795(74)90246-3. [DOI] [PubMed] [Google Scholar]
- 28.Lory S, Carroll SF, Bernard PD, Collier RJ. Ligand interactions of diphtheria toxin. I. Binding and hydrolysis of NAD. J Biol Chem. 1980;255:12011–12015. [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Blanke SR, Huang K, Collier RJ. Active-site mutations of diphtheria toxin: Role of tyrosine-65 in NAD binding and ADP-ribosylation. Biochemistry. 1994;33:15494–15500. doi: 10.1021/bi00255a031. [DOI] [PubMed] [Google Scholar]
- 31.Collins CM, Barbieri JT, Collier RJ. Interaction of diphtheria toxin with adenylyl-(3′,5′)-uridine 3′-monophosphate. I. Equilibrium and kinetic measurements. J Biol Chem. 1984;259:15154–15158. [PubMed] [Google Scholar]
- 32.Hu VW, Holmes RK. Single mutation in the A domain of diphtheria toxin results in a protein with altered membrane insertion behavior. Biochim Biophys Acta. 1987;902:24–30. doi: 10.1016/0005-2736(87)90132-5. [DOI] [PubMed] [Google Scholar]
- 33.Bigio M, et al. Conformational changes in diphtheria toxoids. Analysis with monoclonal antibodies. FEBS Lett. 1987;218:271–276. doi: 10.1016/0014-5793(87)81060-8. [DOI] [PubMed] [Google Scholar]
- 34.Yamaizumi M, Uchida T, Takamatsu K, Okada Y. Intracellular stability of diphtheria toxin fragment A in the presence and absence of anti-fragment A antibody. Proc Natl Acad Sci USA. 1982;79:461–465. doi: 10.1073/pnas.79.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mekada E, Uchida T. Binding properties of diphtheria toxin to cells are altered by mutation in the fragment A domain. J Biol Chem. 1985;260:12148–12153. [PubMed] [Google Scholar]
- 36.Qiao J, Ghani K, Caruso M. Diphtheria toxin mutant CRM197 is an inhibitor of protein synthesis that induces cellular toxicity. Toxicon. 2008;51:473–477. doi: 10.1016/j.toxicon.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Kimura Y, Saito M, Kimata Y, Kohno K. Transgenic mice expressing a fully nontoxic diphtheria toxin mutant, not CRM197 mutant, acquire immune tolerance against diphtheria toxin. J Biochem. 2007;142:105–112. doi: 10.1093/jb/mvm115. [DOI] [PubMed] [Google Scholar]
- 38.Kageyama T, et al. Diphtheria toxin mutant CRM197 possesses weak EF2-ADP-ribosyl activity that potentiates its anti-tumorigenic activity. J Biochem. 2007;142:95–104. doi: 10.1093/jb/mvm116. [DOI] [PubMed] [Google Scholar]
- 39.Wilson BA, Blanke SR, Reich KA, Collier RJ. Active-site mutations of diphtheria toxin. Tryptophan 50 is a major determinant of NAD affinity. J Biol Chem. 1994;269:23296–23301. [PubMed] [Google Scholar]
- 40.McCoy A, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moss J, Manganiello VC, Vaughan M. Hydrolysis of nicotinamide adenine dinucleotide by choleragen and its A protomer: possible role in the activation of adenylate cyclase. Proc Natl Acad Sci USA. 1976;73:4424–4427. doi: 10.1073/pnas.73.12.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.