Abstract
The internal ribosome entry site (IRES) in the hepatitis C virus (HCV) RNA genome is essential for the initiation of viral protein synthesis. IRES domains adopt well-defined folds that are potential targets for antiviral translation inhibitors. We have determined the three-dimensional structure of the IRES subdomain IIa in complex with a benzimidazole translation inhibitor at 2.2 Å resolution. Comparison to the structure of the unbound RNA in conjunction with studies of inhibitor binding to the target in solution demonstrate that the RNA undergoes a dramatic ligand-induced conformational adaptation to form a deep pocket that resembles the substrate binding sites in riboswitches. The presence of a well-defined ligand-binding pocket within the highly conserved IRES subdomain IIa holds promise for the development of unique anti-HCV drugs with a high barrier to resistance.
Keywords: crystallography, hepatitis C virus inhibitor, RNA structure
Infection with hepatitis C virus (HCV), which affects over 170 million individuals worldwide, is a leading cause of liver failure and hepatocellular carcinoma (1). Until earlier this year, when two protease inhibitors were approved as the first direct antiviral drugs for the treatment of HCV infection (2), the standard anti-HCV therapy consisted of an immunostimulatory regimen of pegylated interferon-α and the nucleoside analog ribavirin, which suffered from low efficacy as well as serious side effects (3). The prevalence of preexisting drug-resistance mutations in HCV quasispecies due to the low fidelity of the viral RNA-dependent RNA polymerase (NS5B) creates an urgent need for combination therapy with unique antiviral agents directed at distinct HCV targets (4).
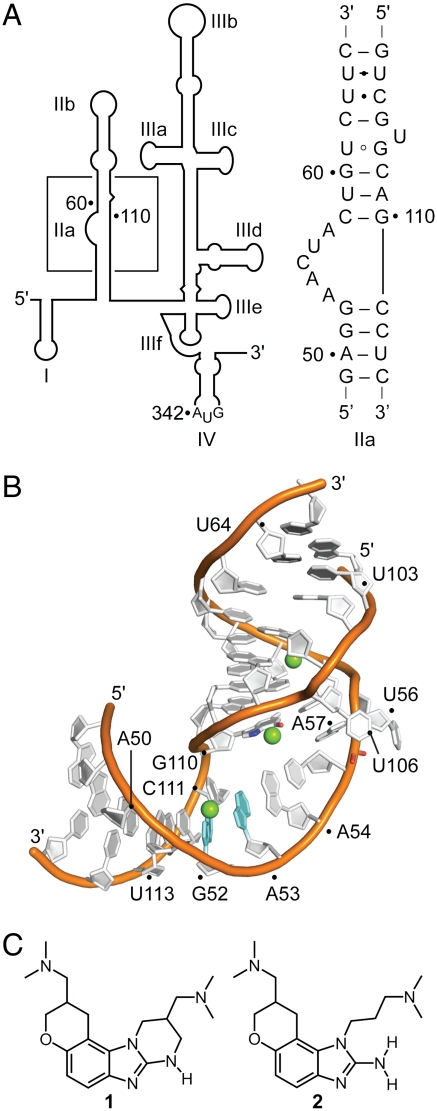
Among the potential targets for HCV inhibitors, the 5′ untranslated region (UTR) of the viral RNA genome stands out for its high sequence conservation within virus clinical isolates (5), which exceeds the conservation of the HCV protein reading frames. The HCV 5′ UTR harbors an internal ribosome entry site (IRES) which recruits host cell 40S ribosomal subunits and ultimately initiates translation of virus proteins via a 5′ cap-independent mechanism (6, 7). The function of the IRES relies on a structured RNA element, which contains several independently folding domains (Fig. 1A) (8, 9). The three-dimensional structure of the subdomain IIa target was previously determined in our laboratory revealing an overall bent architecture around an RNA internal loop (Fig. 1B) (10), in agreement with NMR analyses of the full domain II (11) and cryoelectron microscopy studies of IRES-40S complexes (12, 13). The L-shaped conformation of subdomain IIa directs the apical hairpin loop IIb toward the ribosomal E site in proximity of the active site. Ribosomal association of domain II induces a conformational change in the 40S head and closes the mRNA binding cleft. Both, the correct positioning of the viral mRNA initiation codon as well as the joining of the ribosomal subunits to form functional 80S units depend critically on the L-shaped architecture of the domain II (7, 12–14).
Fig. 1.
The HCV IRES RNA target. (A) Secondary structure of the 5′ UTR (nucleotides 1–341 of HCV genotype 1b). The boxed region indicates the subdomain IIa whose sequence is shown. (B) Crystal structure of the subdomain IIa (10). Mg2+ ions are shown as green spheres. (C) Benzimidazole translation inhibitors of the HCV IRES (15, 17).
Results and Discussion
Recognition of the Benzimidazole Inhibitor in the Ligand-Binding Pocket.
The subdomain IIa is the target for benzimidazole inhibitors (Fig. 1C, compounds 1, 2) that reduce viral RNA levels in the HCV replicon at low micromolar concentration (15–17). We previously used FRET methods to demonstrate that binding of the benzimidazole inhibitors induces widening of the interhelical angle in the bend of IRES subdomain IIa, which may facilitate undocking of subdomain IIb from the ribosome and thereby likely inhibits IRES-driven translation in HCV-infected cells (16). The ligand-induced conformational change in the IIa RNA is supported by a structural model of a subdomain IIa RNA in complex with a related benzimidazole, which was derived from NMR data (18). The NMR model gave insight into global conformational changes that occur upon ligand association but could not elucidate details of the binding interaction. We have now used X-ray crystallography to determine the high resolution structure of the subdomain IIa RNA target in complex with the benzimidazole HCV translation inhibitor 2.
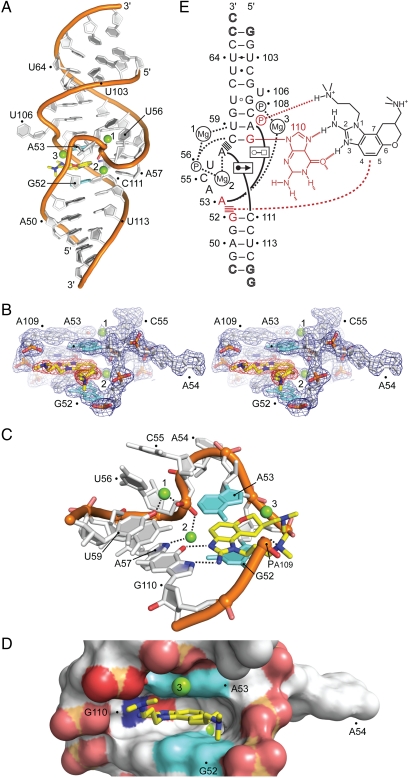
An oligonucleotide corresponding to the subdomain IIa was cocrystallized with a racemic mixture of 2 (SI Appendix, Table S1) and the structure was determined by X-ray diffraction at 2.2 Å resolution (Fig. 2; SI Appendix, Fig. S1 and Tables S2, S3). In agreement with previous findings of a ligand-induced straightening of the subdomain IIa (16, 18), the RNA in the complex adopts an overall linear architecture with the helices that are flanking the internal loop coaxially stacked (Fig. 2A). The RNA internal loop refolds from its curved conformation in the free RNA (Fig. 1B) to form a tightly fitting cavity that deeply encapsulates the ligand (Fig. 2 C and D) and is not participating in crystal packing contacts (SI Appendix, Fig. S2). The benzimidazole 2 docks at the RNA target via hydrogen bonding to the guanine Hoogsteen edge in the C58-G110 base pair as well as stacking interactions between A53 and the G52-C111 pair which form, respectively, the roof and floor of the binding pocket (Fig. 2 C–E; SI Appendix, Table S3). An additional intramolecular hydrogen bond occurs between the protonated dimethylamino-propyl side chain of the ligand and the phosphate group of A109. The hydrogen bonds to G110, which are shielded from competition with hydration inside the binding pocket, in conjunction with the hydrophobic stacking interactions explain the ability of the polar benzimidazole ligand to bind at the RNA target even in high salt under the crystallization conditions (2 M ammonium sulfate). Analysis of atom thermal factors in the crystal structure supports a picture of the ligand-binding site as the exceptionally rigid centerpiece of the complex (SI Appendix, Fig. S3). Whereas the resolution of the electron density map at 2.2 Å did not allow to unambiguously assign the stereochemistry of the bound ligand (Fig. 2B; SI Appendix, Fig. S1), slightly better refinement statistics were obtained for the (R)-2 enantiomer. However, conformational analysis of 2 suggested that the structures of the enantiomers are very similar and binding of either form to the subdomain IIa target would be compatible through the interactions seen in the crystal structure. An NMR-based study of the subdomain IIa in complex with a related benzimidazole compound is in agreement with respect to the selection of the bound enantiomer (18), however, except for the ligand intercalation between G52 and A53, the NMR model is lacking key features of the binding pocket observed in the high resolution crystal structure. In the NMR model, the ligand-binding site is relatively open, exposing the ligand to solvent on both groove sides of the flanking RNA helix, and the hydrogen bonding interactions with the Hoogsteen edge of G110 are absent.
Fig. 2.
Crystal structure of the subdomain IIa RNA inhibitor complex. (A) Overall view of the complex. The benzimidazole ligand (2) is in yellow. Mg2+ ions are shown as green spheres. (B) Stereoview of the 2Fo-Fc electron density contoured at 1σ around the ligand-binding site. (C) Detail view of the ligand-binding site. The bases of G52 and A53, which form the intercalation site for the benzimidazole scaffold, are shown in cyan. The purine of G110 provides the docking edge for the amino-imidazole group. Hydrogen bond interactions are indicated by dashed lines. (D) Surface representation, highlighting the deep ligand-binding pocket. (E) Schematic of the interactions in the ligand-binding site (SI Appendix, Table S3). Hydrogen bonds are shown as dashed lines. Formation of non-Watson–Crick base pairs is indicated with solid lines and symbols according to Leontis et al. (45). Stacked lines (≡) indicate stacking of bases and intercalation of the ligand. Residues interacting with the benzimidazole are highlighted in red.
The crystal structure explains conclusively structure-activity relationships for related benzimidazole derivatives (SI Appendix, Fig. S4). Compounds that lack the amino substituent at the 2-position or the N,N-dimethylamino-propyl chain off the N1, both of which participate in key hydrogen bonding interactions with the RNA, do not bind to the subdomain IIa target. Modifications at the 6-position improve binding with basic substituents showing the best activities whereas larger nonpolar groups having little beneficial effect. The tight fit of the ligand into the substrate binding site does not accommodate even small substituents at the 4- and 5-position of the benzimidazole and derivatives with such modifications are indeed inactive.
RNA Architecture of the Ligand-Binding Pocket.
The intricate architecture of the ligand-binding pocket is organized by a metal spine at the back of the cavity, consisting of the phosphate of U56, which is rotated into the RNA helix major groove, and two magnesium ions anchored between the prochiral phosphate oxygen atoms and the bases of A57 and U59 (Fig. 2 C and E). The extreme contortion of the RNA backbone that directs the U56 phosphate into the helix interior is further stapled in place by hydrogen bonds between the C55 phosphate and 2′ hydroxyl groups of the flanking residues A54 and U56. The elaborate network of backbone and metal interactions forces the base of A54 to project away from the RNA whereas residues C55 and U56 tightly pack in the RNA helix major groove as well as against the backbone of C58 and U59. A third magnesium ion closes part of the solvent exposed mouth of the ligand site by bridging the RNA strands with interactions at the base of A53 and the C108 phosphate. The prominent role that magnesium ions play in the stabilization of the subdomain IIa (10) is thus maintained for the RNA-ligand complex. Both, the free RNA target as well as the complex contain three magnesium ions as intrinsic structural components, which, along with the RNA, undergo adaptive reorganization upon binding of the benzimidazole ligand.
Roof and floor of the binding pocket are stabilized by base triples which form through cross-bracing interactions along the RNA helix with A53 docking at the Hoogsteen edge of A109 and A57 interacting with the sugar edge of C111 (Fig. 2E; SI Appendix, Table S3). Additional hydrogen bonds that stabilize the distorted internal loop in the benzimidazole complex include interactions of the U56 phosphate with the C58 base and of the C55 2′ hydroxyl with the A53 base (SI Appendix, Table S3).
The Subdomain IIa RNA-Ligand Complex Resembles a Conserved Riboswitch.
The formation of a deep binding pocket in the subdomain IIa through adaptive ligand recognition bears resemblance to the substrate complexation mechanism in aptamers and riboswitches (19–21). In ribosomal binding sites for antibiotics, the largest and most diverse class of natural RNA targets for small molecules, adaptive ligand recognition is not observed apart from localized conformational changes affecting single nucleotides (22–24). It is intriguing to speculate that the ligand-induced conformational change in the HCV IRES subdomain IIa might have a biological function that involves a cellular trigger, perhaps, such as a protein interacting with the IRES RNA. We propose that the amino-imidazole scaffold in the viral translation inhibitor 2 is a structural mimetic of an arginine side chain, which, in the context of an IRES- or ribosome-binding protein, could induce the conformational switch of the IIa subdomain from an L-shaped to an extended structure. The interaction motif of the guanidine-like moiety in the benzimidazole with the Hoogsteen edge of G110 is analogous to that seen for the arginine side chain in numerous peptide and protein complexes of both RNA and DNA (SI Appendix, Fig. S5) (25–28). Interestingly, two sulfate ions are bound at the RNA minor groove of the subdomain IIa complex, one of them directly interacting with the N2 amino group of the highly conserved G51, in proximity of the benzimidazole ligand pocket. Such guanine-bound sulfate ions might be indicative of a putative protein binding surface (29), perhaps providing further support for our hypothesis of the subdomain IIa containing a protein binding site. Previously, ribosomal protein S5 (rpS5) and heterogeneous ribonucleoprotein D (hnRNP D) have been identified by UV-crosslinking and immunoprecipitation as direct binding partners of the HCV domain II RNA (30, 31).
It has been noted that the curved topology of the IRES domain II, which directs the apical loop of subdomain IIb at the ribosomal E site, would prevent the progression of the ribosome from initiation to elongation (12, 13). In the transition to productive translation, domain II has to be moved out of the E site to make room for deacylated tRNA. Perhaps a conformational change in the subdomain IIa as observed in the crystal structure reported here but triggered by adaptive recognition of a protein is involved in the release of the ribosome from the IRES-bound complex. Consistent with this hypothesis, we have recently discovered modular diamino-piperidine inhibitors of the HCV replicon that affect HCV translation by binding and arresting subdomain IIa in the L-shaped state of the free RNA and thereby interfering with translation initiation (32).
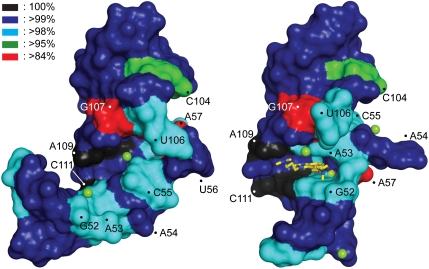
The notion that the subdomain IIa plays a key functional role in HCV IRES-driven translation beyond providing a purely architectural RNA motif is supported by the extraordinary sequence conservation in this region of the viral genome (Fig. 3; SI Appendix, Fig. S6 and Table S4). With the exception of three residues (A57, C104, G107), the sequence of subdomain IIa is conserved > 98% within 1,600 HCV clinical isolates and across all genotypes (33). Residues around the ligand-binding site show > 99% conservation, including A109 and C111, which are 100% conserved. Whereas in the unliganded conformation the base of the bulged-out residue U56 does not participate in stabilizing the subdomain IIa architecture (10) (Fig. 1B) and mutations at this position do not affect formation of the L-shaped fold (16), this nucleotide is > 99% conserved. The high degree of conservation at U56 becomes explainable in the context of the ligand-bound conformation of subdomain IIa in which this residue is involved in tight packing interactions (see above) that can only be achieved with a uridine base at this position.
Fig. 3.
Binding site conservation in the HCV IRES target. The sequence conservation in clinical isolates of HCV has been mapped on the surface of the RNA structure. (Left) The unliganded RNA, (Right) the complex with the viral translation inhibitor 2. The degree of conservation is indicated by color coding. See also SI Appendix, Table S3, Fig. S6).
Although the sequences of domain II in closely related HCV-like IRES elements diverge, the internal loop secondary structure, which provides the basis for a bent architecture, is a conserved feature (9). IRES elements in other pestiviruses as well as picornaviruses derived from simian, avian, and porcine hosts contain domains that are recognizably similar to the HCV IRES domain II (34, 35). It is tempting to speculate that the role of subdomain IIa as a protein ligand-triggered release switch for translation initiation is a conserved feature of these viral IRES elements.
Ligand-Induced Conformational Switching.
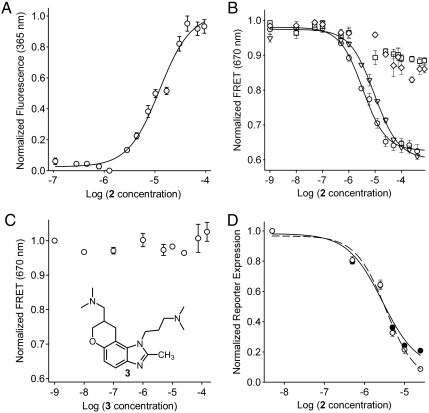
To investigate if conformational switching between the free and ligand-bound states of subdomain IIa occurs in solution we studied the interaction of the benzimidazole 2 with fluorescently labeled RNA, probing both the local environment of the binding pocket as well as the overall conformation of the L-shaped fold. We had previously identified the conformation of the A54 residue as a sensitive probe of the RNA folding state (10). Replacement of A54 by the fluorescent nucleobase analog 2-aminopurine (2AP) was used to monitor metal ion binding as well as RNA folding. Comparison of the subdomain IIa crystal structures shows that the A54 base is packed inside the fold in the free RNA while it is rotated out in the ligand-bound state of the target (Fig. 1B, Fig. 2C). Titration of 2AP-54-labeled subdomain IIa with benzimidazole 2 led to a dose-dependent increase of 2AP fluorescence, suggesting that the fluorescent base analog is displaced from the RNA interior upon ligand-triggered switching of the target conformation (Fig. 4A). This observation confirmed that ligand binding in solution induces a transition between conformational states of the A54 residue consistent with the crystal structures of free and complexed subdomain IIa.
Fig. 4.
Binding and biological activity of benzimidazole 2 at the HCV IRES target. (A) Fluorescence signal for a titration of 2AP54-labeled IIa RNA with compound 2 in the presence of 10 mM Mg2+. Error bars represent ± 1 SD calculated from three independent titrations. Fitting of a dose-response curve resulted in an EC50 value of 12 ± 1 μM. (B) FRET signal for titrations of Cy3/Cy5-labeled IIa RNA with benzimidazole 2 in the presence of 2 mM Mg2+. Symbols represent WT RNA (○), A57U mutant (∇), and two double mutants, C58G/G110C (□) and C58U/G110A (◊). Fitting of dose-response curves resulted in EC50 values for ligand binding of 3.4 ± 0.3 μM (WT) and 9.3 ± 1.1 μM (A57U). The double mutants did not bind the inhibitor (EC50 > 100 μM). (C) FRET signal for a titration of Cy3/Cy5-labeled IIa WT RNA with the analog compound 3 that has the 2-amino functionality replaced by a methyl group. The analog does not bind the RNA target. (D) Inhibition of HCV translation in human Huh-7.5 cells. Titrations of compound 2 were performed against BM4-5 FEO subgenomic replicon (•) and JFH1 full-length virus (○). Fitting of dose-response curves resulted in EC50 values for translation inhibition of 2.8 ± 0.4 μM (replicon) and 3.4 ± 0.5 μM (virus). In all graphs error bars represent ± 1 SD calculated from triplicate experiments, except for D where four physical replicates of triplicate experiments were used.
To investigate ligand-triggered changes in the overall conformation of the L-shaped fold we used a previously established FRET assay (16) in which subdomain IIa RNA, terminally labeled with a cyanine dye pair, was titrated with the benzimidazole 2. The assay monitors the interhelical angle between the stems flanking the internal loop in subdomain IIa via measurement of the distance between the stem termini. Addition of the benzimidazole 2 resulted in dose-dependent quenching of FRET with an EC50 value of 3.4 ± 0.3 μM (Fig. 4B), suggesting that ligand binding induced a conformational change, which led to widening of the interhelical angle of the L-shaped RNA fold. Specific high-affinity binding of the ligand required the presence of magnesium ions, which are key structural elements of both the ligand-bound as well as the ligand-free form of the IIa RNA (10, 16). When the target RNA was titrated with benzimidazole 2 in the absence of magnesium, a weak induction of FRET was observed albeit at a much higher concentration of the ligand (EC50 = 117 ± 8 μM) (SI Appendix, Fig. S7), which suggests unspecific folding perhaps through electrostatic interactions with the cationic compound 2.
Because the crystal structure showed that binding of the ligand 2 involves key hydrogen bonds to G110 (Fig. 2E), we tested two double mutants in which this anchor residue was exchanged to a cytosine or adenine whereas the Watson–Crick pairing was retained. The mutated RNAs carrying the base pair reversal (in C58G/G110C) or exchange (C58U/G110A) were both proficient for folding into the L-shaped architecture of the wild type subdomain IIa (SI Appendix, Fig. S8). Neither double mutant displayed ligand-triggered FRET quenching upon addition of the benzimidazole 2 up to a concentration of 100 μM (Fig. 4B) demonstrating the essentiality of the G110 residue for ligand binding. For further confirmation of the docking mode of 2 observed in the crystal the methyl analog 3 was tested in which the 2-amino functionality of 2 was replaced by a methyl group (Fig. 4C). As expected, removal of the hydrogen bond donating amino group ablated binding to the IIa target. The A57U mutant, which was previously identified as conferring some resistance to the related benzimidazole translation inhibitor 1 (Fig. 1C) in the HCV replicon (16), affected binding of 2 as well, showing a roughly threefold lower affinity (EC50 = 9.3 ± 1.1 μM) (Fig. 4B). This weaker binding might be an indirect effect due to destabilization of the ligand-binding pocket by disruption of the A57⋯G52-C111 triple because the base at position 57 does not directly contact the benzimidazole in the complex.
Both, the titration of 2AP54-labeled RNA as well as the FRET studies confirm that binding of the viral translation inhibitor 2 in solution induces a transition between conformational states of the subdomain IIa, which are locally as well as globally consistent with the crystal structures. Addition of compound 2 to cells infected with either subgenomic HCV replicon or full-length virus led to dose-dependent inhibition of viral translation (Fig. 4D), demonstrating the functional consequences of the conformational induction at the subdomain IIa target. This observation is in agreement with our earlier findings for the structurally related translation inhibitor 1 (16).
Implications for Development of Anti-HCV Therapies.
The findings reported here of a deep solvent-excluding inhibitor binding pocket in the highly conserved subdomain IIa of the HCV IRES add a unique dimension to the repertoire of targets for anti-HCV therapy. The architecture of the well-defined benzimidazole binding site will be a valuable starting point for the structure-based design of HCV inhibitors, supported by the notion of viral translation as an attractive therapeutic target (36, 37). The extremely high conservation of the subdomain IIa RNA in HCV clinical isolates suggests that mutations around the benzimidazole binding pocket will be difficult to reconcile with IRES function. Inhibitors directed at this target will potentially benefit from selection of low-fitness resistance mutants with reduced replication rates and reduced frequency of occurrence in the treatment-naïve population.
Materials and Methods
Compound Synthesis.
The benzimidazole HCV translation inhibitor 2 was synthesized as a mixture of enantiomers according to a previously published procedure (17). Synthesis of the methyl analog 3, which served as an inactive control compound, proceeded over two steps from the published intermediate 4 (17) as outlined in SI Appendix. The identity and purity of the compounds were confirmed by mass spectrometry as well as 1H and 13C NMR.
RNA Preparation.
RNA for crystallization and fluorescently labeled RNA (2AP and cyanine dyes) was purchased from Integrated DNA Technologies as chemically synthesized and HPLC-purified oligonucleotides. See SI Appendix, Table S1 for a list of oligonucleotides. Stock solutions were prepared by dissolving lyophilized oligonucleotides in 10 mM sodium cacodylate buffer, pH 6.5. RNA was annealed from stoichiometric amounts of strands in the presence of 5 mM MgCl2 by heating to 75 °C for 5 min followed by slow cooling to room temperature.
Crystallization and Data Collection.
Subdomain IIa RNA was cocrystallized with benzimidazole 2 at 16 °C by hanging drop vapor diffusion after mixing 1 μL of 0.33 mM RNA and 2 mM of 2 with an equal volume of precipitating solution containing 10 mM magnesium sulfate, 50 mM sodium cacodylate, pH 6.5, and 2 M ammonium sulfate. Cube-shaped crystals grew over 3 mo of equilibration against 700 μL of well solution containing precipitating solution. Crystals were flash-cooled in liquid nitrogen before data collection. Diffraction data were collected in a nitrogen stream at 110 K on a Rigaku rotating anode X-ray generator (λ = 1.54 Å) equipped with a MAR345 imaging plate detector system. Datasets collected were processed, integrated, and scaled with the HKL2000 package (38).
Structure Solution and Refinement.
The three-dimensional structure of the subdomain IIa complex was solved by molecular replacement with the program Phaser (39) using search models derived from the structure of the free RNA target (10) and refined by the program Refmac (40) both within the Collaborative Computational Project 4 (CCP4) package (41). Subsequent iterative rounds of manual building and refinement, alternating between Refmac and manual rebuilding in Coot (42), were based on the obtained 2Fo-Fc and Fo-Fc maps. Positions of compound 2, metal and sulfate ions were initially assigned based on electron density as well as geometry of coordinating ligands. Final refinement was carried out in PHENIX (43) with individual isotropic atomic displacement parameters and water picking (SI Appendix, Table S1). The omit map for compound 2 (SI Appendix, Fig. S1B) was obtained by restrained refinement based on the experimental phases with compound 2 removed from the structure.
2AP Fluorescence Experiments.
Fluorescence measurements on the 2AP-labeled subdomain IIa RNA were performed as described previously (10, 16) on a thermostated RF-5301PC spectrofluorometer (Shimadzu) at 25 °C. The 2AP-labeled RNA was excited at 310 nm and emission was read at 365 nm.
FRET Experiments.
FRET experiments were performed as described previously (16) on a Spectra Max Gemini monochromator plate reader (Molecular Devices) at 25 °C. RNA was at 100 nM concentration in 10 mM Hepes buffer (pH 7.0). Emission filters were set at 550 and 665 nm. Cy3 label was excited at 520 nm and transferred fluorescence was read as Cy5 emission at 670 nm. Data were analyzed and FRET calculated as described previously (16).
HCV Replicon Assay.
BM4-5 FEO RNA (genotype 1b HCV replicon) was generated from the corresponding DNA plasmid using T7 RNA polymerase as previously described (44). The impact of compound 2 on HCV subgenomic replicon replication was assessed, using the method previously described (16), in cells stably expressing the BM4-5 FEO replicon in 96-well plates (10,000 cells/well). Cells were incubated with the compound for 48 h and the results expressed as the mean (± SEM) of the relative light units for each condition.
HCV Full-Length Virus Assay.
The Jc1-Luc2A plasmid was constructed from the J6/JFH(p7-Rluc2A) plasmid (a gift from Charles Rice, The Rockefeller University, New York, NY) using restriction digestion and ligation of the photinus pyralis luciferase gene amplified from the BM4-5 FEO replicon. Following verification of the Jc1-Luc2A sequence it was used to generate in vitro transcribed RNA. Ten micrograms of Jc1-Luc2A RNA were used to transfect Huh-7.5.1 cells via electroporation. Following electroporation cells were cultured in 10 cm dishes at 37 °C and 5% CO2 until cell culture supernatant was harvested at day 3 to 4. Cell culture supernatants were clarified by centrifugation at 3,000 × g for 10 min and then concentrated. The infectivity of supernatants was determined by counting infected foci formed using serial dilutions of supernatant. The impact of compound 2 on full-length HCV was tested in 96-well plates. Ten thousand Huh-7.5.1 cells were seeded into wells and incubated for 4 h to allow for attachment. Jc1-Luc2A virus was added at a multiplicity of infection of 0.01 along with the compound. Luciferase activity was determined after 72 h of incubation (OneGlo, Promega) using a microplate luminometer (Turner Biosystems).
Supplementary Material
Acknowledgments.
We thank J. Parsons for help with the fluorescence assays during the early phase of the project and N. Nguyen for help during X-ray data collection. Supported in part by the National Institutes of Health Grants R01 AI72012 (to T.H.) and K08 AI069989 (to D.L.W.), as well as the San Diego State University Research Foundation (B.M.B. and M.A.P.). Support of the University of California, San Diego NMR facility by the National Science Foundation is acknowledged (Chemistry Research Instrumentation and Facilities Grant CHE-0741968).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors of the IIa RNA-ligand complex have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, www.rcsb.org (PDB ID code 3TZR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118699109/-/DCSupplemental.
References
- 1.Jang JY, Chung RT. Chronic hepatitis C. Gut Liver. 2011;5:117–132. doi: 10.5009/gnl.2011.5.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enserink M. Infectious diseases. First specific drugs raise hopes for hepatitis C. Science. 2011;332:159–160. doi: 10.1126/science.332.6026.159. [DOI] [PubMed] [Google Scholar]
- 3.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 4.Robinson M, Tian Y, Delaney WEt, Greenstein AE. Preexisting drug-resistance mutations reveal unique barriers to resistance for distinct antivirals. Proc Natl Acad Sci USA. 2011;108:10290–10295. doi: 10.1073/pnas.1101515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis DR, Seth PP. Therapeutic targeting of HCV internal ribosomal entry site RNA. Antiviral Chem Chemother. 2011;21:117–128. doi: 10.3851/IMP1693. [DOI] [PubMed] [Google Scholar]
- 6.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 8.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 9.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew Chem Int Ed Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 11.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 12.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 13.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA. 2011;17:1258–1273. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth PP, et al. SAR by MS: Discovery of a new class of RNA-binding small molecules for the hepatitis C virus: Internal ribosome entry site IIA subdomain. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 16.Parsons J, et al. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol. 2009;5:823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker MA, Satkiewicz E, Hermann T, Bergdahl BM. An efficient new route to dihydropyranobenzimidazole inhibitors of HCV replication. Molecules. 2011;16:281–290. doi: 10.3390/molecules16010281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen RB, et al. Inhibitor-induced structural change in the HCV IRES domain IIa RNA. Proc Natl Acad Sci USA. 2010;107:7263–7268. doi: 10.1073/pnas.0911896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 20.Serganov A. Determination of riboswitch structures: Light at the end of the tunnel? RNA Biol. 2010;7:98–103. doi: 10.4161/rna.7.1.10756. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Lau MW, Ferre-D’Amare AR. Ribozymes and riboswitches: Modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermann T. Drugs targeting the ribosome. Curr Opin Struct Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 24.Yonath A. Antibiotics targeting ribosomes: Resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 25.Puglisi JD, Chen L, Frankel AD, Williamson JR. Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss MA, Narayana N. RNA recognition by arginine-rich peptide motifs. Biopolymers. 1998;48:167–180. doi: 10.1002/(SICI)1097-0282(1998)48:2<167::AID-BIP6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman MM, et al. AANT: The amino acid-nucleotide interaction database. Nucleic Acids Res. 2004;32:D174–D181. doi: 10.1093/nar/gkh128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo J, Westhof E. Classification of pseudo pairs between nucleotide bases and amino acids by analysis of nucleotide-protein complexes. Nucleic Acids Res. 2011;39:8628–8637. doi: 10.1093/nar/gkr452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masquida B, Sauter C, Westhof E. A sulfate pocket formed by three GoU pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA. 1999;5:1384–1395. doi: 10.1017/s1355838299991173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukushi S, et al. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 31.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol. 2008;82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carnevali M, Parsons J, Wyles DL, Hermann T. A modular approach to synthetic RNA binders of the hepatitis C virus internal ribosome entry site. ChemBioChem. 2010;11:1364–1367. doi: 10.1002/cbic.201000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuiken C, Hraber P, Thurmond J, Yusim K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008;36:D512–D516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belsham GJ. Divergent picornavirus IRES elements. Virus Res. 2009;139:183–192. doi: 10.1016/j.virusres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C R Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Gallego J, Varani G. The hepatitis C virus internal ribosome-entry site: A new target for antiviral research. Biochem Soc Trans. 2002;30:140–145. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman B, Liu Q. Hepatitis C viral protein translation: Mechanisms and implications in developing antivirals. Liver Int. 2011;31:1449–1467. doi: 10.1111/j.1478-3231.2011.02543.x. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Wyles DL, Kaihara KA, Vaida F, Schooley RT. Synergy of small molecular inhibitors of hepatitis C virus replication directed at multiple viral targets. J Virol. 2007;81:3005–3008. doi: 10.1128/JVI.02083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leontis NB, Stombaugh J, Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.