Abstract
Signaling pathways depend on regulatory protein-protein interactions; controlling these interactions in cells has important applications for reengineering biological functions. As many regulatory proteins are modular, considerable progress in engineering signaling circuits has been made by recombining commonly occurring domains. Our ability to predictably engineer cellular functions, however, is constrained by complex crosstalk observed in naturally occurring domains. Here we demonstrate a strategy for improving and simplifying protein network engineering: using computational design to create orthogonal (non-crossreacting) protein-protein interfaces. We validated the design of the interface between a key signaling protein, the GTPase Cdc42, and its activator, Intersectin, biochemically and by solving the crystal structure of the engineered complex. The designed GTPase (orthoCdc42) is activated exclusively by its engineered cognate partner (orthoIntersectin), but maintains the ability to interface with other GTPase signaling circuit components in vitro. In mammalian cells, orthoCdc42 activity can be regulated by orthoIntersectin, but not wild-type Intersectin, showing that the designed interaction can trigger complex processes. Computational design of protein interfaces thus promises to provide specific components that facilitate the predictable engineering of cellular functions.
Keywords: computational modeling and design, signal transduction, synthetic biology
Most approaches to engineering cellular systems with new functions have taken advantage of the relative ease with which DNA elements can be used to control gene expression (1–3). In contrast, few studies have attempted to directly engineer protein-protein interaction networks. Recent pioneering examples include engineered control of input/output relationships in protein circuits (4, 5), protein-based logic gates (6), and control of protein activity in biological processes by light (7–9). Essentially all of these approaches create fusions of existing modular protein elements to yield diverse functions (10).
Nonetheless, our ability to create new functions by domain recombination is constrained by the toolkit of domains that are naturally available. Reuse of the same or closely related domains can yield undesired or unanticipated crosstalk, complicating the ability to predictably modify function within the context of a complex cellular protein interaction network. A potential solution to this problem would be to modify protein-protein interfaces directly by tuning interaction affinity and specificity as well as by creating orthogonal protein pairs (11). In its simplest form, an orthogonal pair consists of two engineered proteins that specifically interact with each other, but avoid significant crosstalk with their native wild-type counterpart proteins (Fig. 1A). Such orthogonal interactions are useful for achieving predictable biological control in a variety of contexts. For example, orthogonal interactions could be used to insulate a desired functional pathway from another competing process. Orthogonal protein pairs could also allow more precise control if they can be specifically triggered by a small molecule to rapidly activate their function. One approach to engineering orthogonal systems is to borrow molecular components from a different organism. However, components from other organisms might not properly interface with existing cellular machinery and require further engineering to control multicomponent cellular pathways. Instead, it may be advantageous to “rewire” existing protein interactions to create orthogonal pairs that can be externally controlled. Such protein network engineering strategies are not only useful to reengineer cells to perform new functions, but also to delineate the existing functional interaction networks.
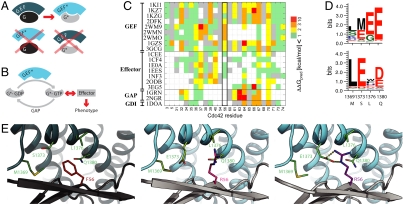
Fig. 1.
Strategy for computational design of an orthogonal signaling interaction. (A) Schematic representation of design requirements for orthogonality: the interface between the GTPase Cdc42 (G) and ITSN (GEF) is modified to generate a pair G*/GEF* with new specificity. (B) Simplified schematic representation of the core GTPase signaling circuit to define the design requirements for a functional G*/GEF* pair that interfaces correctly with other cellular components (GAP and effector proteins that are required for phenotypic output). (C) Computational alanine scanning. Shown are the estimated effects on binding energy of replacing each residue in the Cdc42/ITSN interface (PDB code 1KI1) with alanine in the context of 19 co-complex structures of Cdc42 with partner proteins (white indicates residues not in the interface in the respective structure). Altering position F56 of Cdc42 mainly affects interaction with GEFs. (D) Comparison of fixed backbone (top) and flexible backbone (bottom) computational design predictions for four residues in ITSN (wild-type residues are indicated on the x axis) in the vicinity of position 56 of Cdc42 for a F56R mutation. (E) Model of designed orthoCdc42/orthoITSN interface from fixed (middle) and flexible (right) backbone modeling compared to the wild-type complex (left). Gray: Cdc42; Teal: ITSN; shown in sticks are the five designed interface residues. Small backbone changes modeled by backrub motions (0.53 Å Cα rmsd) allowed the sidechains of R56 and E1373 to adopt conformations that can form hydrogen bonds (dashed lines).
Computational design has been successfully applied to many protein engineering applications (11, 12), including design of proteins with new or altered protein-protein interactions (11, 13, 14). A clear next challenge is to design protein interfaces to create orthogonal proteins that can perform and control complex biological functions in the context of cells and organisms. Here we describe such a proof-of-concept application of computational protein design, which generated an engineered pair of interacting proteins that is orthogonal to the wild-type proteins. The orthogonal interaction can be specifically triggered by a small molecule, and can interface with existing cellular components to control complex biological responses both in an in vitro reconstituted system and in mammalian cells.
Results
The GTPase Model System and Design Principles.
As our model system we chose the interactions of Rho-type GTPases that function as binary switches in signal transduction networks controlling key biological functions such as establishment of cell polarity and cell motility via regulation of the actin cytoskeleton (15). GTPases control signaling by cycling between the GDP-bound, inactive state, and the GTP-bound, active state that can bind to downstream effector proteins and propagate signaling information (Fig. 1B). The GTPase switch (16) is modulated by accessory proteins: GTPase Activating Proteins (GAPs) accelerate the hydrolysis of GTP to GDP (inhibiting signal transduction), GTPase Exchange Factors (GEFs) accelerate the exchange of GDP and GTP (promoting signal transduction), and GDP Dissociation Inhibitors (GDIs) modulate the distribution of cytosolic and membrane-bound pools of GTPase. Therefore, the core GTPase signaling circuit comprises interactions of the GTPase with several different binding partners (GAP, GEF, GDI, and effector). Because the main regulatory process activating the GTPase switch involves regulation of the interaction between the GTPase and GEFs (17), we chose one such interaction, between the GTPase Cdc42 and a Cdc42-specific GEF Intersectin (ITSN), as our target.
We sought to design a functionally orthogonal interaction between a mutant Cdc42 (orthoCdc42) and a mutant ITSN (orthoITSN) that is buffered from the native pair (Cdc42WT and ITSNWT). This process should create two functionally interacting cognate pairs (Cdc42WT/ITSNWT and orthoCdc42/orthoITSN) and two noncognate pairs with no observable interaction specificity (Cdc42WT/orthoITSN and orthoCdc42/ITSNWT) (Fig. 1A). To allow the newly created cognate pair to perform its biological function, we constrained the design to minimally perturb residues implicated in the recognition interfaces of GDP, GTP, GAPs, effectors, and GDIs by Cdc42. Cellular activation of Cdc42 leads to considerable changes in cell morphology via induction of actin polymerization through the effector WASP. We thus expected that activation of orthoCdc42 by orthoITSN should be able to trigger WASP binding in vitro and morphology changes in cells.
Computational Design Strategy.
We first sought to identify residues in Cdc42 that affect the binding interface between Cdc42 and ITSN, but do not affect interactions with other known Cdc42 binding partners. We performed a computational alanine scan (18) on 19 co-complex structures of Cdc42 with its binding partners (nine GEFs, two GAPs, seven effectors, and one GDI), to estimate the contribution of each interface residue to binding each partner (Fig. 1C, SI Appendix, Fig. S1A). The alanine scan identified position 56 as the main candidate that affects GEF binding without perturbing the interactions with other binding partners. Additionally the F56 sidechain is spatially separated from the nucleotide binding and catalytic sites of Cdc42, suggesting that changes at F56 should not affect the affinity of Cdc42 for GDP or GTP.
Next we wanted to identify appropriate mutations around F56 in the Cdc42/ITSN interface that would lead to a functional interaction between orthoCdc42 and orthoITSN without introducing crosstalk between the noncognate pairs. An initial application of our previously developed computational second-site suppressor protocol (19) to the structure of Cdc42 and ITSN (PDB ID: 1KI1) (20) suggested that almost all substitutions of F56 would be destabilizing to the interaction (SI Appendix, Fig. S1B). However, none of the predicted compensatory changes of neighboring residues in ITSN was specific to the identity of the mutated amino acid modeled at position 56. Failure to correctly predict precise details of sidechain-sidechain interactions is a known problem with computational design approaches that leave the protein backbone fixed, such as the original second-site suppressor protocol (11, 19, 21, 22). Thus, we applied a recently developed flexible backbone design method, RosettaBackrub (23), to predict residue changes on ITSN that would compensate for mutations at Cdc42 F56. Although the flexible backbone design method has been benchmarked on existing data (23, 24), this application represents the first forward engineering test of the method’s efficacy (see Methods). Using a flexible backbone ensemble created with RosettaBackrub, we allowed all possible mutations (except cysteine) at the four sites in ITSN adjacent to Cdc42 position 56 (1,369; 1,373; 1,376; 1,380) and searched for specific interactions with a variant residue at the 56 site of Cdc42. The predicted sequence logo in Fig. 1D shows that the mutation of position 1,373 in ITSN from serine to glutamate was distinctly enriched when F56 in Cdc42 was mutated to arginine [the residue with the largest predicted difference between destabilization in the noncognate and stabilization in the cognate pair (SI Appendix, Fig. S1B)]. This interaction replaces the hydrophobic interactions (F56-L1376) observed in the original pair with a defined polar interaction (R56-E1373) in the designed pair (Fig. 1E). Importantly, a computational model of the complex of Cdc42 (F56R) and ITSN (S1373E) now showed specific hydrogen bonds formed between these two engineered sidechains that were not observed with fixed backbone simulations performed under identical conditions (Fig. 1E). Changing only one of these interacting residues in either Cdc42 (F56R) or ITSN (S1373E) is predicted to significantly destabilize the interactions between noncognate pairs. In the following, the specific Cdc42 (F56R) and ITSN (S1373E) variants are named orthoCdc42 and orthoITSN, respectively. (For designed variants other than the orthoCdc42/orthoITSN pair, see SI Appendix, Results, Table S1).
In Vitro Nucleotide Exchange Activity and Binding Affinity.
We first determined the ability of the orthoITSN DH-PH domains to catalyze nucleotide exchange in orthoCdc42 by following the dissociation (Fig. 2A) and association (Fig. 2B) of fluorescently labeled nucleotide analogs. orthoITSN specifically catalyzed exchange in orthoCdc42 but not in Cdc42WT. Similarly, exchange in orthoCdc42 was only catalyzed by orthoITSN but not by ITSNWT. These results demonstrate that only one substitution in each protein is sufficient to engineer an orthoCdc42/orthoITSN pair that is indeed functionally orthogonal in vitro. This result is remarkable, given that such dramatic switches in protein-protein interaction specificity often require many changes (19, 21, 25). In our case, other modeled substitutions in ITSN, such as M1369L, did not change the exchange activity in orthoCdc42/orthoITSN (SI Appendix, Table S1). Moreover, the ITSN Q1380E mutation (a prominent prediction of the fixed backbone protocol, Fig. 1D) was not active towards orthoCdc42 in combination with S1373E (SI Appendix, Results, Fig. S2), further confirming the importance of the specific R56-E1373 interaction.
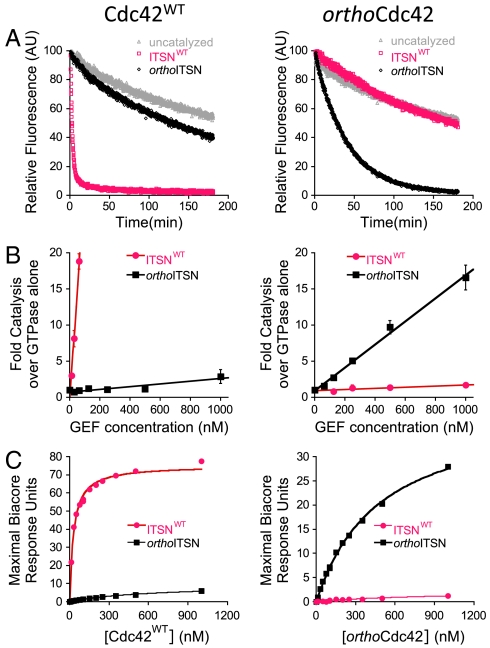
Fig. 2.
The designed interaction is orthogonal in vitro. In (A)–(C), Cdc42WT is shown on the left and orthoCdc42 on the right. Pink: data for ITSNWT; black: data for orthoITSN. (A) Catalysis of nucleotide exchange by ITSNWT and orthoITSN, monitored by dissociation of fluorescent mant-GDP from Cdc42WT and orthoCdc42. Gray: intrinsic exchange in Cdc42 in the absence of any ITSN. (B) Catalysis of nucleotide exchange from initial rates of mant-GDP association at varying GEF concentrations. Data represent averages and standard deviations from at least three experiments. (C). Binding affinity monitored by Surface Plasmon Resonance equilibrium analysis (SI Appendix, Fig. S4).
While the designed mutations essentially eliminated cross-reactivity with the wild-type partners in noncognate complexes (Fig. 2B), orthoITSN was a weaker nucleotide exchange catalyst for orthoCdc42 compared to ITSNWT for Cdc42WT. To explain this weaker activity, we analyzed both the stability of the engineered variants and their binding affinity. Neither mutation significantly destabilized the engineered proteins, as indicated by similar apparent melting temperatures monitored using circular dichroism (SI Appendix, Fig. S3). However, the weaker functional interactions were consistent with direct binding affinity measurements of cognate and noncognate Cdc42 and ITSN complexes determined by surface plasmon resonance (Fig. 2C, SI Appendix, Fig. S4). The interaction between Cdc42WT and ITSNWT had a KD of 29 ± 2 nM, similar to that determined in a previous study (33 nM) (26). The KD of orthoCdc42 and orthoITSN was 478 ± 22 nM, approximately 16-fold weaker. Importantly, essentially no binding was observed under our conditions between the noncognate Cdc42WT/orthoITSN or orthoCdc42/ITSNWT, directly demonstrating the physical origin of the orthogonal relationship between cognate pairs.
Structural Basis of the Designed Specificity.
To assess the accuracy of the design model, we determined the crystal structure of the complex between orthoCdc42 and the DH-PH domains of orthoITSN (Fig. 3, SI Appendix, Results, Fig. S5, Table S2). The structure confirms the engineered salt bridge interaction between the sidechains of R56 in orthoCdc42 and E1373 in orthoITSN (Fig. 3B, SI Appendix, Fig. S5A). However, there are notable downstream rearrangements of sidechains extending up to about 10 Å from the designed site, where sidechains of N39 and Y40 in orthoCdc42 essentially switch positions (Fig. 3C, SI Appendix, Fig. S5B), concomitant with backbone changes in the interface.
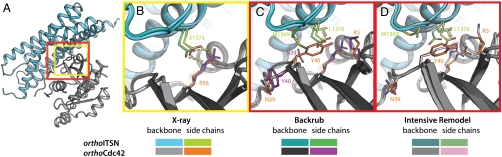
Fig. 3.
The crystal structure of the orthoCdc42/orthoITSN complex confirms the designed interaction, but also highlights requirements for advanced flexible-backbone remodeling protocols. (A) Overview of the structure of the designed complex between orthoCdc42 (gray) and the orthoITSN DH domain (teal). Boxes highlight the location of the designed site near the center of the protein-protein interface (yellow) as well as the area of backbone and side-chain rearrangements (red), magnified in (B–D). Sidechain and backbone colors are as indicated in the figure. (B) Comparison of the R56-E1373 interaction in the backrub flexible-backbone computational model (as in Fig. 1E, right) and in the crystal structure of the designed orthoCdc42/orthoITSN complex. Dashed lines represent hydrogen bonds. (C, D) Comparison of the network of residues surrounding the designed site that were rearranged to accommodate the mutations, as predicted by the backrub model (C) and the intensive remodeling protocol (D, details in SI Appendix, Results) vs. their observed position in the crystal structure of the designed complex. The remodeling protocol (D) was able to capture both sidechain and backbone conformational changes in the crystal structure of orthoCdc42/orthoITSN that were missed by the initial backrub predictions (C).
While the RosettaBackrub prediction successfully captured the defined interaction between the two designed residues by allowing small backbone adjustment and brought the backbone conformation slightly closer to that of the designed structure (SI Appendix, Fig. S6B), it had not captured the larger conformational change accompanying the sidechain rearrangements around Y40. Such conformational changes are a possible reason for the reduced biochemical activity in our case, and are also likely to occur more generally in response to designed mutations in interfaces. We thus tested whether a new remodeling protocol (SI Appendix, Fig. S6, Methods, Results) that switches between diversifying conformations and intensifying sampling, while iterating between energy functions using soft and hard repulsive forces, could model such interface changes. Intensive sampling around the designed interface site indeed yielded a conformation (the lowest energy structure in one of six resulting clusters) that was very close (0.56 Å Cα rmsd in the region of interest) to the solved crystal structure of the design and recapitulated the experimentally observed switch in the sidechains positions of N39 and Y40 (Fig. 3D).
Interactions with Other GTPase Binding Partners.
The substitution in orthoCdc42 was designed to minimize effects on other known binding partners of the GTPase (Fig. 1C). One of the most important interactions in the Cdc42 activation cycle is the binding of GTP-bound Cdc42 to the effector protein WASP, which allows for activation of the Arp2/3 complex, and induces actin polymerization. A second key interaction is with GAPs that accelerates the hydrolysis of GTP bound to GTPases. Consistent with the design strategy, orthoCdc42 binds to a fragment of N-WASP (residues 201–321) (although with an approximately fourfold weaker KD than Cdc42WT, SI Appendix, Fig. S7A), and p50RhoGAP can enhance nucleotide hydrolysis in orthoCdc42 (SI Appendix, Fig. S7B). Full-length orthoCdc42 (containing a prenylated C-terminal CAAX motif) can also bind the Guanine Dissociation Inhibitor RhoGDI (SI Appendix, Fig. S7C). In addition to the interaction with ITSN, Cdc42 has intrinsic specificity for other exchange factors, which is preserved in orthoCdc42 (SI Appendix, Results, Table S3). Taken together, these results suggest that orthoCdc42 can still interact with core components of the GTPase signaling circuit, and that the designed substitutions in orthoCdc42 and orthoITSN have not introduced new and undesirable crosstalk with other known GTPases and GTPase signaling circuit components (SI Appendix, Table S3).
In Vitro Reconstitution of a Partial Signaling Pathway.
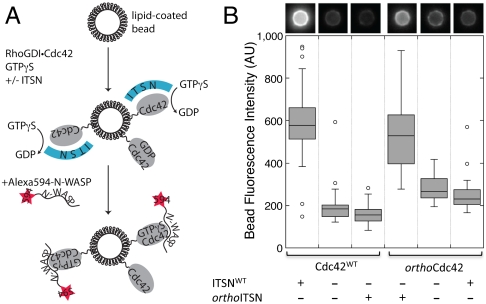
The biochemical analysis above suggests that the engineered substitutions of orthoCdc42 and orthoITSN have generated a new protein pair that does not interact with the wild-type proteins, but where orthoCdc42 maintains binary interactions with other Cdc42 regulation factors. To test the function of the designed pair in the context of a larger Cdc42 pathway, we used an in vitro assay with purified components to monitor N-WASP recruitment to lipid-coated beads (27) (Fig. 4A). This assay mimics activation of membrane-bound Cdc42 by GEF-catalyzed nucleotide exchange and subsequent interaction of GTP-bound Cdc42 with the effector N-WASP. As designed, the localization of fluorescently labeled N-WASP (residues 137–502) to the surface of lipid-coated beads increased only in the presence of the Cdc42WT/ITSNWT or the orthoCdc42/orthoITSN cognate pairs, but not with the noncognate pairs (Fig. 4B). Kolmogorov-Smirnov testing of the bead fluorescence intensity distributions indicated that these differences were significant (p < 1.5e - 6 for each condition, three independent experiments with at least 20 individual beads counted per experiment). Consistent with the previously noted weaker affinity of the designed pair, the required concentration of orthoITSN was higher (2.5 μM) than ITSNWT (1 μM) in each respective condition.
Fig. 4.
The designed orthoCdc42/orthoITSN interaction mediates specific GTPase activation and effector binding in an in vitro reconstituted system. Alexa 594 labeled N-WASP (residues 137–502) translocation to a lipid-coated glass bead is specifically increased in the presence of a cognate interaction between Cdc42 and ITSN. (A) Schematic illustrating the assay and the order of addition of the components. (B) The total fluorescence intensity of individual beads relative to the background was measured, and the distributions of the fluorescence intensities from multiple beads (n > 23 for each condition) are shown in box plot representation. Boxes enclose the first and third quartile of the distribution and display a line at the median; whiskers extend outward no more than 1.5 times the size of the box and data points outside this range are drawn individually. A representative bead image is shown above each condition.
Pathway Activity with Designed Components in Mammalian Cells.
We next tested whether the designed orthoCdc42/orthoITSN pair, despite its lower exchange activity and weakened affinity compared to the wild-type complex, still functions in endogenous signaling networks of GTPases and GEFs in mammalian cells. We coupled the designed protein-protein interaction with a small molecule-based inducible localization system similar to that described in (28). Using this method, the cell-permeable small molecule Rapamycin can be added to recruit FK506 binding protein (FKBP)-linked ITSN to the plasma membrane by inducing Rapamycin-mediated binding of FKBP to FK506-rapamycin-binding (FRB) protein, which is localized to the membrane using the membrane-targeting domain from the Lyn protein (Fig. 5A). Activated Cdc42 is known to induce the formation of filopodia in NIH 3T3 mouse fibroblast cells (29), as well as lamellopodia by activating the GTPase Rac through interaction with the IRSp53 protein (30). Thus, increasing the local ITSN concentration near the membrane should lead to nucleotide exchange and activation of membrane-localized inactive Cdc42, which in turn actives Cdc42 signaling to induce cell morphological changes. In this way, because Cdc42 activation should be triggered by Rapamycin-dependent ITSN recruitment, any change in cellular phenotype can be observed in the same background before and after the addition of the small molecule.
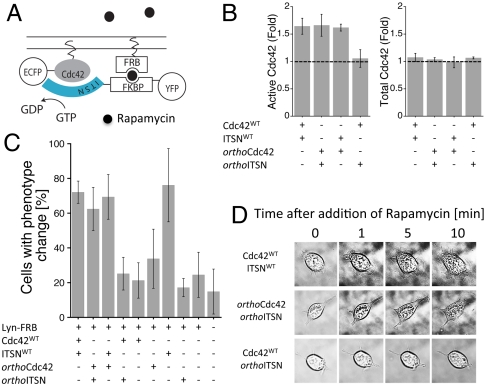
Fig. 5.
The orthoCdc42/orthoITSN pair is functional in mammalian cells. (A) Schematic representation of the cell-based assay using a Rapamycin-based recruitment system (FRB, FKPB) to colocalize fluorescently tagged GTPase and GEF constructs at the membrane. (B) Fold increase in active Cdc42 (comparing samples with and without addition of Rapamycin for 60 s) from lysed NIH 3T3 cells measured with a G-LISA assay (left). The total Cdc42 loaded in the G-LISA assay was determined by an ELISA assay, and is also shown in fold change, again comparing samples with and without Ramamycin addition (right). All samples had Lyn-FRB transfected. Error bars represent the standard deviation of three experiments. (C) Percentage of NIH 3T3 cells that showed morphological changes (filopodia/lamellipodia) after addition of Rapamycin, determined by live cell microscopy. All samples had Lyn-FRB transfected. Error bars represent the standard deviation of three experiments. The total numbers of counted cells for each condition, from left to right, are: 103, 111, 133, 120, 57, 62, 55, 50, 71, and 84. (D) Representative images of cell morphological changes upon Rapamycin addition.
We first determined whether orthoITSN could activate orthoCdc42 in cells by measuring the levels of activated and total Cdc42 before and after the addition of Rapamycin (see Methods). orthoITSN indeed activated orthoCdc42, but not Cdc42WT, as expected (Fig. 5B). Overall, the activation of orthoCdc42 by orthoITSN was similar to the activation of Cdc42WT by ITSNWT, and the active Cdc42 was at the highest level in the first 60–90 s after the addition of Rapamycin (SI Appendix, Fig. S8A). Finally, to determine whether activation of orthoCdc42 by orthoITSN could result in morphological changes (filopodia and/or lamellopodia) in NIH 3T3 cells, we counted cells that showed induced morphological changes after the addition of Rapamycin (Fig. 5C) using fluorescence microscopy of living cells (Fig. 5D). Consistent with the Cdc42 activation assay (Fig. 5B), increased filopodia/lamellipodia were observed in cells transfected with either the orthoCdc42 and orthoITSN designed pair or the wild-type pair, but not with the noncognate Cdc42WT/orthoITSN pair. Similarly, transfection of orthoITSN in the presence of the Rapamycin recruitment system but in the absence of orthoCdc42 resulted in considerably less phenotypic change.
We note that these assays (as also apparent in Fig. 5B) cannot determine orthogonality with respect to the other noncognate pair orthoCdc42/ITSNWT, as Rapamycin-induced localization of ITSNWT most likely leads to activation of endogenous Cdc42WT. Consistent with this idea, transfection with the other noncognate pair orthoCdc42/ITSNWT had levels of morphological change similar to the cognate pairs. Furthermore, transfection of ITSNWT alone (but including the membrane-recruiting construct Lyn-FRB) shows an equivalent level of morphological change. Taken together, including additional results monitoring morphological changes by impedance (SI Appendix, Results, Fig. S8B), the cellular assays indicate that the designed orthoCdc42/orthoITSN interaction functions within cells to trigger production of filopodia/lamellipodia.
Discussion
In this work, we used advanced computational protein design methods to reengineer a signaling circuit by direct modification of an interaction interface; this approach stands in contrast to previous work that either engineered expression control at the gene level or recombined existing modular protein domains. We show that the designed proteins function orthogonally in vitro and trigger responses in cells. Therefore, the engineered interacting orthogonal pair still interfaces with existing cellular machinery to direct changes in cell morphology, a complex phenotypic outcome.
Engineering orthogonality of specific interactions, while at the same time maintaining correct interfaces with existing machinery, is challenging in multiple respects. The orthogonality of the designed interaction is remarkable, given that it was achieved with only one residue change on either partner, but it comes at the price of reduced affinity. Detailed structural analysis of designed proteins is critical for evaluating inaccuracies in the design model. The defined interaction of the designed R-E pair in a central interface location, on which our predictions were based, was correctly captured in the model. However, deviations further away from the designed site illustrate the difficulty of predicting energetics and conformations of interacting residues, in particular polar networks in protein interfaces. It is not unlikely that the different conformations of the polar interaction network (SI Appendix, Fig. S5B) are approximately isoenergetic and that small changes in the surroundings, including long-range effects, can cause population shifts resulting in coordinated conformational changes. It may be difficult to predict these changes computationally in part because the relative free energy differences may be small. In this context, it is remarkable that a new intensive backbone remodeling protocol is capable of sampling conformations close to the observed structure (Fig. 3D). Currently, the Rosetta energy does not distinguish between these models, and structural clustering is necessary to reveal the diversity of the sampled conformations (SI Appendix, Fig. S6).
It is difficult to find sites in multifunctional proteins such as GTPases that can be engineered without pleiotropic consequences on many interactions or detrimental effects on function altogether. In fact, position 56, identified here by computational design as the major engineerable site (Fig. 1), may be one of a few sites that can be mutated in Cdc42 without dramatically affecting multiple partner interactions. F56 of Cdc42 has previously been implicated as a residue that defines the specificity of Cdc42 for various GEFs including ITSN (20, 31, 32). In contrast to previous studies that switched between existing interaction preferences, however, our design has created a different specificity. This finding prompts the question of whether the F56R and S1373E substitutions are present in any other existing GTPase-GEF interactions. Of the 23 Rho subfamily GTPases in the human genome, none have arginine at the position equivalent to F56 (33). In the 66 characterized human GTPase exchange factor sequences, only five have glutamate at the position equivalent to S1373. All five have either been shown to not catalyze exchange in Cdc42, or are members of the Lbc subfamily that in general does not catalyze exchange in Cdc42 (34, 35). These results suggest that the substitutions designed by computational methods are unique.
Almost every protein is involved in a number of interactions with different binding partners. The ability to design new specificities into target interfaces without affecting other interactions is useful both for the biological interrogation of protein interactions and for the design of circuits that could produce new biological behaviors. This study indicates that computational methods can become an essential tool for the design of new protein interfaces. Improving computational design methodologies, including approaches to more accurately model structural and sequence plasticity in interfaces (11), will allow protein engineers and synthetic biologists to create new interactions of increasing complexity and specificity.
Methods
Computational Protein Interface Design.
The crystal structure of Cdc42WT/ITSNWT (PDB ID: 1KI1) (20) was used as starting conformation for structure-based computational protein design. Computational alanine scanning was performed as described (18). For fixed backbone design, we used the computational second-site suppressor protocol as described (19) (SI Appendix, Fig. S1B). These simulations aimed to identify substitutions in one protein that are significantly destabilizing to the complex formed with the wild-type partner protein but can be compensated for by complementary changes in the partner. Flexible-backbone protein design used RosettaBackrub (23, 36) and the sequence tolerance protocol developed in (23, 24). One hundred low-scoring backrub structures were generated from the starting structure of Cdc42WT/ITSNWT, and used as a backbone ensemble in design simulations to determine sequence tolerated at the Cdc42/ITSN interface. In the design step, the amino acid identity at Cdc42 position 56 was fixed but the residue was allowed to change its rotameric conformation, and the four neighboring residues (M1369, S1373, L1376, Q1380) in ITSN were allowed to change to any other residues (designed) except cysteine. The intensive flexible-backbone design and remodeling strategy (SI Appendix, Results, Fig. S6) begins with modeling the F56R and S1373E mutations, followed by backbone diversification using RosettaBackrub (36) and kinematic closure (KIC) methods (37), and final intensified sampling and refinement using KIC. Soft and hard repulsive forces are iterated similar to a recently described protocol for protein folding (38). Simulation details and all Rosetta command lines are given in SI Appendix, Methods.
Protein Biochemistry.
All in vitro assays except the N-WASP translocation experiments used soluble forms of the GTPases (residues 1–179 in Cdc42) lacking the C-terminal prenylation sites. All exchange factor sequences were derived from human or mouse cDNA and encoded both the DH and PH domains (SI Appendix, Table S4). Proteins for in vitro experiments were expressed and purified from Escherichia coli, and nucleotide dissociation and association assays were performed as detailed in SI Appendix, Methods. Cdc42—ITSN binding affinities were determined by surface plasmon resonance (SPR) experiments similar to those described in Smith, et al. (26), and the N-WASP translocation assay was performed as described by Co, et al. (27). (For more details on protein in vitro assays see SI Appendix, Methods).
Crystallography.
Crystals were grown at room temperature as hanging drops above a well of 100 mM Tris pH 7.5, 25% PEG 3350, 150 mM ammonium sulfate, and 1 mM DTT. Crystals were harvested using a solution of 20% glycerol and 17% PEG 3350 as a cryoprotectant. Details on data collection, analysis and structure determination are given in SI Appendix, Methods. The PDB model was deposited as: 3QBV.
Cell-Based Assays.
The Cdc42 G-LISA Kit (Cytoskeleton) was used to detect active GTP-bound Cdc42 in NIH 3T3 cells, and an ELISA assay was used to measure the total Cdc42 loaded (SI Appendix, Methods). For live cell fluorescence microscopy, NIH 3T3 cells were cultured in 8-well Lab-Tek II Chambered Coverglass wells. After serum starvation, pictures were taken on a Nikon Eclipse Ti Microscope with a 60X or 100X objective at 37 °C (SI Appendix, Methods).
Supplementary Material
Acknowledgments.
We thank Orion Weiner, Anselm Levskaya, Ben Rhau, and Alex Watters for helpful suggestions, Colin Smith and Shane O’Connor for help with design simulations, Kris Kuchenbecker and Peter Hwang for help with SPR, Farid Ahmad for help with crystallography, and James Onuffer, Benjamin Rhau, and Jason Park for help with cell-based assays. T.K. is supported by awards from the National Science Foundation (MCB-CAREER 0744541, EF-0849400), the Sandler Foundation, and the UC Lab Research Program. W.A.L., J.T. and T.K. were supported by the US National Institutes of Health Roadmap Initiative (PN2 EY016546, W.A.L., principal investigator). W.A.L. was supported by awards from the National Science Foundation (EEC-0540879), and the National Institute of Health (RO1 GM062583, P50 GM081879). J.S.F is a QB3@UCSF Fellow. A.S. is an EMBO long-term fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3QBV).
See Commentary on page 5140.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114487109/-/DCSupplemental.
References
- 1.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 4.Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature. 2007;447:596–600. doi: 10.1038/nature05851. [DOI] [PubMed] [Google Scholar]
- 5.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 6.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 7.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung DW, Otomo C, Chory J, Rosen MK. Genetically encoded photoswitching of actin assembly through the Cdc42-WASP-Arp2/3 complex pathway. Proc Natl Acad Sci USA. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 12.Pokala N, Handel TM. Review: protein design-where we were, where we are, where we’re going. J Struct Biol. 2001;134:269–281. doi: 10.1006/jsbi.2001.4349. [DOI] [PubMed] [Google Scholar]
- 13.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 16.Cherfils J, Zeghouf M. Chronicles of the GTPase switch. Nat Chem Biol. 2011;7:493–495. doi: 10.1038/nchembio.608. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 18.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortemme T, et al. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 20.Snyder JT, et al. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- 21.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Sammond DW, Eletr ZM, Purbeck C, Kuhlman B. Computational design of second-site suppressor mutations at protein-protein interfaces. Proteins. 2010;78:1055–1065. doi: 10.1002/prot.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CA, Kortemme T. Predicting the tolerated sequences for proteins and protein interfaces using RosettaBackrub flexible backbone design. PLoS One. 2011;6:e20451. doi: 10.1371/journal.pone.0020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CA, Kortemme T. Structure-based prediction of the peptide sequence space recognized by natural and synthetic PDZ domains. J Mol Biol. 2010;402:460–474. doi: 10.1016/j.jmb.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 25.Levin KB, et al. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009;16:1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- 26.Smith WJ, et al. A Cdc42 mutant specifically activated by intersectin. Biochemistry. 2005;44:13282–13290. doi: 10.1021/bi050591b. [DOI] [PubMed] [Google Scholar]
- 27.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krugmann S, et al. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 30.Ladwein M, Rottner K. On the Rho'd: the regulation of membrane protrusions by Rho-GTPases. FEBS Lett. 2008;582:2066–2074. doi: 10.1016/j.febslet.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Xing J, Streuli M, Leto TL, Zheng Y. Trp(56) of rac1 specifies interaction with a subset of guanine nucleotide exchange factors. J Biol Chem. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- 32.Karnoub AE, et al. Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nat Struct Biol. 2001;8:1037–1041. doi: 10.1038/nsb719. [DOI] [PubMed] [Google Scholar]
- 33.Colicelli J. Human RAS superfamily proteins and related GTPases. Science Signaling Signal Transduction Knowledge Environment. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 35.Glaven JA, Whitehead IP, Nomanbhoy T, Kay R, Cerione RA. Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J Biol Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 36.Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol. 2008;380:742–756. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khatib F, et al. Algorithm discovery by protein folding game players. Proc Natl Acad Sci USA. 2011;108:18949–18953. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.