Abstract
The circadian clock governs a large array of physiological functions through the transcriptional control of a significant fraction of the genome. Disruption of the clock leads to metabolic disorders, including obesity and diabetes. As food is a potent zeitgeber (ZT) for peripheral clocks, metabolites are implicated as cellular transducers of circadian time for tissues such as the liver. From a comprehensive dataset of over 500 metabolites identified by mass spectrometry, we reveal the coordinate clock-controlled oscillation of many metabolites, including those within the amino acid and carbohydrate metabolic pathways as well as the lipid, nucleotide, and xenobiotic metabolic pathways. Using computational modeling, we present evidence of synergistic nodes between the circadian transcriptome and specific metabolic pathways. Validation of these nodes reveals that diverse metabolic pathways, including the uracil salvage pathway, oscillate in a circadian fashion and in a CLOCK-dependent manner. This integrated map illustrates the coherence within the circadian metabolome, transcriptome, and proteome and how these are connected through specific nodes that operate in concert to achieve metabolic homeostasis.
Keywords: metabolism, diurnal, CircadiOmics, hepatic
Circadian rhythms exist within a wide range of biological processes and control numerous aspects of physiology, including the sleep/wake cycle, eating, hormone and neurotransmitter secretion, and even cognitive function (1–4). Integral to the modern lifestyle is the ability to eat, sleep, work, exercise, and socialize around the clock and yet these allowances may serve as a preamble to obesity and other metabolic disorders. Recent studies reveal that a distorted circadian cycle can lead to aberrations in metabolism, producing symptoms such as obesity, insulin resistance, and others consistent with the metabolic syndrome (5–9). Whereas studies focused on night-shift and rotating shift workers emphasize the link between circadian rhythmicity and metabolism, rodent models of circadian arrhythmia also support this link (10–13). As an essential modulator of metabolism in vivo, the liver provides a cardinal domain in which to study interactions between the clock and metabolism as much of the liver transcriptome and proteome oscillates in expression or activity. These circadian characteristics of the liver are dependent largely on the zeitgeber (ZT), food (14–21).
Precision within the circadian clock depends on nuclear transcriptional and translational feedback loops, but recent evidence that circadian, nontranscriptional, and nontranslational cytosolic rhythms crosstalk with nuclear rhythms to maintain circadian timing provides support for the idea that metabolites may affect the transcriptional–translational canonical clock system and vice versa (22, 23). Some biochemical oscillations have been shown to cycle in a circadian fashion, such as calcium, 3′–5′-cyclic adenosine monophosphate (cAMP) and nicotinamide (NAD+) (24–27). These oscillations feed into the clock system and can modulate circadian dynamics in vivo. Several blood metabolites have also been observed to oscillate, the oscillation of which depends on an intact endogenous clock (28). A comprehensive diurnal liver metabolome has never been deciphered, however. As an organ involved in glycogen storage, the production of bile acids, and the storage and provider of numerous amino acids and vitamins for the rest of the body, the circadian oscillation of metabolic function within the liver is seminal to understanding circadian physiology.
To determine whether liver metabolites oscillate in a diurnal fashion and to determine the extent to which their presence is Clock controlled, liver samples from wild-type and Clock-deficient (Clock−/−) mice were prepared for GC/MS and LC/MS/MS analysis. We present a computational platform that encompasses this metabolite data, validated data concerning oscillating metabolic enzymes, and data covering interactions between oscillating metabolites from different metabolic pathways. To convey this information, we created a database entitled “CircadiOmics,” which forms a consolidated model of how the daily metabolome, transcriptome, and proteome might work together to achieve metabolic homeostasis. Validation of these nodes experimentally reveals the remarkable interconnectedness of the liver metabolome and the hepatic circadian clock system.
Results
Liver Metabolome Undergoes Daily Oscillations.
To determine whether liver metabolites oscillate in a 24-h fashion in vivo, livers from WT and Clock−/− mice were analyzed by liquid and gas chromatography and tandem mass spectrometry (n = 5 per time point per genotype). Animals were maintained in a 12-h light/dark (LD) cycle for the duration of the experiment and tissues were collected every 6 h throughout the day. For each sample, metabolic analyses were performed using ultrahigh performance liquid chromatography/tandem mass spectrometry (UHPLC/MS/MS) and GC/MS. MS/MS data were searched against a standard library (Methods) (29). (For a full list of metabolites, see Dataset S1 and http://circadiomics.igb.uci.edu/). Spectra for 538 metabolites were obtained. Metabolites that showed time or genotype main effects were grouped on the basis of their major pathway (Fig. 1 and Fig. S1). A majority of amino acid and xenobiotic metabolites peaked at night (ZT15–ZT21), whereas peak abundance of nucleotide, carbohydrate, and lipid metabolites occurred at ZT9 (Fig. 2). Of the 538 total metabolites measured, 172 showed a time main effect (P < 0.05, ANOVA) and 132 showed a genotype main effect (P < 0.05, ANOVA) (SI Methods). Of 309 named metabolites (detectable compounds of known identity), 100 showed a time main effect and 73 showed a genotype main effect. A nonparametric algorithm, JTK_CYCLE, was used to detect metabolites that displayed 24-h rhythmicity (Datasets S2, S3, and S4) (30). By JTK_CYCLE, 60 metabolites were detected as oscillating in a diurnal fashion in WT livers (P < 0.05). (In Clock-deficient livers, 21 metabolites were detected as oscillatory when P < 0.05, but only 7 were oscillatory when P ≤ 0.01.) Of those 7, 6 also oscillated in WT livers (Fig. S2).
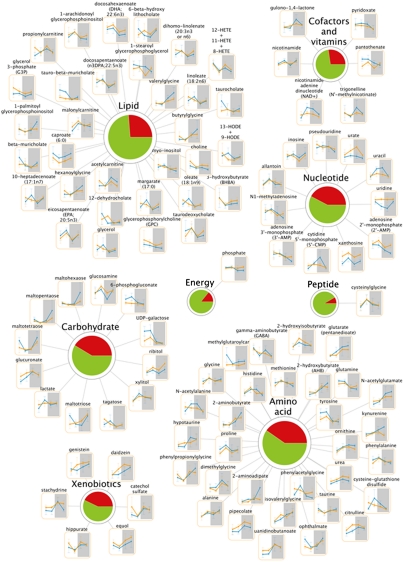
Fig. 1.
Metabolic pathways of the liver containing diurnally regulated metabolites. Major metabolic pathways are represented in the liver by numerous metabolites that change in abundance throughout the 24-h cycle (orange lines, Clock−/− metabolites; blue lines, WT metabolites). Metabolites that vary in abundance over time are shown. Pie charts depict the percentage of metabolites that changed over time (red) vs. those that did not (green). (N = 5 per genotype per time point.)
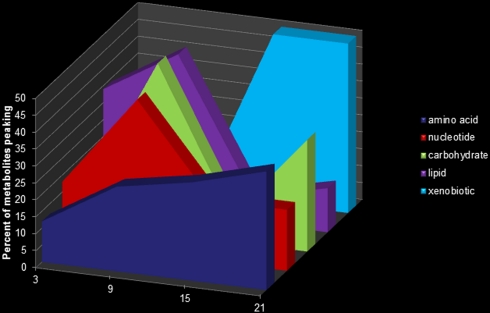
Fig. 2.
Metabolic pathway metabolites show disparate temporal peaks. Nucleotide and carbohydrate metabolites generally peaked at ZT9, whereas amino acid and xenobiotic metabolism-related biochemicals peaked at night (ZT15–ZT21).
Clock-Driven Metabolome.
It has been reported that Clock-deficient mice are rhythmic in the brain due to the presence of the CLOCK paralog, NPAS2 (31). However, peripheral clocks are arrhythmic in the absence of systemic cues. In vivo, the patterns of liver gene expression are altered in a gene-specific manner (31–33). Comparisons between metabolite abundance in the livers of WT and Clock-deficient mice revealed multiple deviations at all zeitgeber times compared (Fig. S3).
Metabolic pathways were broken down into general subpathways to determine whether some specificity is attained within the diurnal metabolome in vivo. As many metabolites are involved in multiple different metabolic pathways, subpathways convey the pathways to which a given metabolite is most closely associated, not exclusively associated. Variations in the percentage of subpathway metabolites were noted within each metabolic pathway. For example, within lysine metabolism all six metabolites showed either a time or genotype main effect, whereas only one of five metabolites in the tryptophan metabolic pathway demonstrated a time or genotype main effect (Fig. 3 and Fig. S2). All three metabolites associated with nicotinate metabolism demonstrated a time main effect (NAD+, P < 0.001; NAM, P = 0.008; and N′-methylnicotinate, P = 0.03). NAD+ levels also demonstrated a genotype:time interaction. NAD+ levels were depressed in Clock−/− mice at ZT9 (Clock−/−/WT = 0.71, P = 0.038). These findings are important because rhythmicity of NAD+ and its contribution to clock maintenance has previously been demonstrated (26, 27), and NAD+ occupies a central position connecting most circadian metabolic pathways.
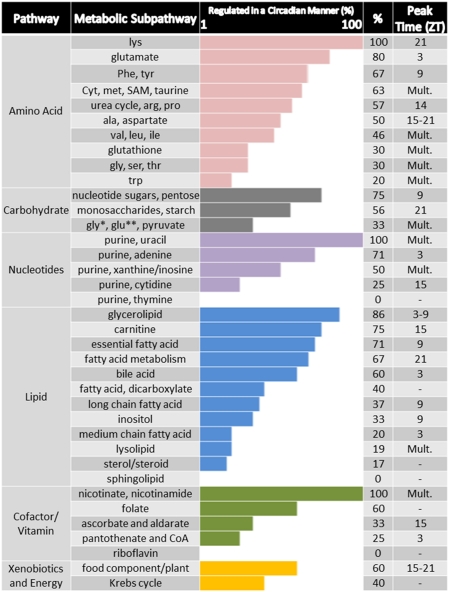
Fig. 3.
Rhythmicity in metabolic subpathways. Metabolic pathway representation by subpathways and the percent of metabolites altered within each relative to genotype or time (only subpathways with ≥3 metabolites are graphed). The observed peak time (ZT hour) of metabolites within specific subpathways is shown on the right. ZT hour is denoted if all or the majority of metabolites identified within the category peak at one ZT. “Mult.” stands for “multiple” and refers to metabolite peaks within the subpathway that were distributed across multiple zeitgeber times. Dashes indicate that no metabolite within the subfamily showed altered abundance over time. Such subpathways include metabolites that were only altered in the Clock−/− livers, and were not changed over time or oscillatory in expression patterns (where % greater than 0 is indicated) (*, glycolysis; **, gluconeogenesis).
Some subpathways showed cohesiveness among metabolites in terms of peak time expression. Specifically, the long chain fatty acids that changed over time [margarate (17:0), 10-heptadecenoate (17:1n7), and oleate (18:1n9)] all peaked at ZT9. All five of the fatty acids identified that are predominantly acquired from the diet or are essential [linoleate (18:2n6), dihomolinolenate (20:3n3 or n6), eicosapentaenoate (EPA; 20:5n3), docosapentaenoate (n3 DPA; 22:5n3), and docosahexaenoate (DHA; 22:6n3)] also changed cohesively over time. Of these, 18:2n6 and 20:5n3 demonstrated 24-h rhythmicity in WT but not Clock-deficient livers (Datasets S1, S2, S3, and S4). Furthermore, all four of the food component/plant subpathway metabolites (daidzein, genistein, equol, and stachydrine) peaked during the night, with three of the four peaking at ZT21. Bile acids also showed a preference for peak time expression, which occurred at ZT3 for five of the six bile acids having a time main effect.
Prediction of Diurnal Phenotypes by Metabolome Analysis.
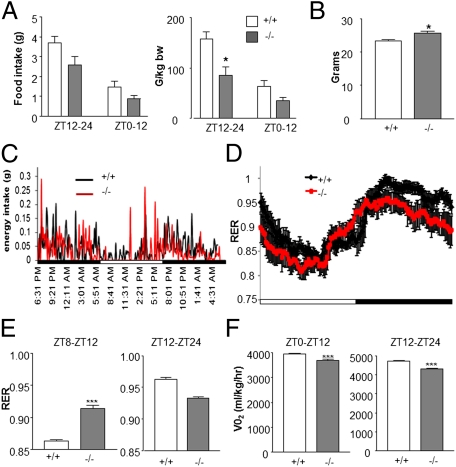
Several food-derived biochemicals were altered in Clock-deficient livers and bile acids generally failed to oscillate (Fig. S4). As food functions as a strong zeitgeber for the liver (18, 19, 34), these data suggested that Clock−/− mice may have a yet unappreciated metabolic phenotype. Metabolic attributes of Clock-deficient animals have been less studied compared with the Clock (c/c) mutant animal (but see ref. 35), which is arrhythmic in constant darkness, obese, and shows attributes consistent with the metabolic syndrome (11, 13). WT and Clock−/− mice were analyzed by indirect calorimetry for CO2 emission, O2 consumption, and energy intake. The respiratory exchange ratio (RER) and heat were also calculated (Fig. 4 and Fig. S4). Whereas Clock−/− animals showed a slight elevation in body weight (WT (+/+) = 23.62 g ± 0.4475 g; Clock−/− (−/−) = 26.07 g ± 0.5789 g; P = 0.0152, Mann-Whitney u test), the 24-h energy intake was unchanged (Fig. 4 A, Left and B). However, when normalized to body weight, Clock−/− mice appear to consume less food during the night (Fig. 4A, Right) (ZT12–ZT24, WT (+/+) = 3.658 g; Clock−/− (−/−) = 2.394 g, P < 0.05, Bonferroni posttest). Interestingly, Clock−/− animals woke several hours before lights off and began consuming food (Fig. 4C, red; arrow ∼ZT8). This premature activity is in keeping with previously reported data showing an early onset of nightly bout activity (32). Energy intake during the period ZT8–ZT12 in Clock−/− animals corresponded with an increase in RER, indicating that Clock−/− mice burned more carbohydrates during this late-phase resting period (Fig. 4D). However, RER was generally lower during the night in Clock−/− animals (Fig. 4E), (main effect of genotype, P < 0.001; main effect of time, P < 0.001; two-way ANOVA, day and night values an average of two 24-h periods) and oxygen consumption was also dampened throughout the daily cycle, revealing decreased energy demands (Fig. 4F). Interestingly, metabolic analysis revealed that, whereas hepatic food-derived biochemicals increased in WT animals at ZT15 and ZT21, these metabolites did not oscillate in Clock−/− mice and generally showed increased concentrations at ZT9. Thus, the liver metabolome provided valuable information regarding the metabolic and physiological functions of the clock in vivo.
Fig. 4.
Energy intake in clock knockout mice precedes the dark cycle. (A) Energy intake in 8-wk-old Clock-deficient (−/−) mice and WT littermate controls (+/+) (error ± SEM, *P < 0.05, Bonferroni posttest compared with WT). (B) Body weight in grams (g) of WT (+/+) and Clock-deficient (−/−) animals (error ± SEM, *P < 0.05, Mann-Whitney test compared with WT ZT12–24). (C) Average energy intake patterns in WT (+/+) and Clock-deficient (−/−) animals. (D and E) Respiratory exchange ratios (VCO2/VO2) of WT (+/+) and Clock-deficient (−/−) animals across a 24-h period (***P < 0.001, genotype; ***P < 0.001, time; two-way ANOVA) as well as during the hours of ZT8–12 and ZT12–24 (error ± SEM, ***P < 0.001 compared with WT). (F) Oxygen consumption in WT and Clock-deficient (−/−) animals during the resting (ZT0–ZT12) and active (ZT12–ZT24) periods. N = 7–8 animals per genotype (error ± SEM, ***P < 0.001, unpaired t test compared with WT).
Constructing a Map of the Diurnal Metabolome.
To better understand how the diurnal metabolome might function on the backdrop of already known oscillating and metabolic events in the liver, we developed a computational platform called CircadiOmics to integrate and visualize circadian rhythm and omic data from multiple sources. Specifically, CircadiOmics integrates data encompassing the following: (i) metabolic pathways and reactions from Kyoto Encyclopedia of Genes and Genomes (KEGG) (36–38), (ii) oscillating gene expression patterns from previously reported high-resolution DNA microarrays (39, 40), (iii) protein–protein interactions from multiple protein–protein interaction databases (41–43), (iv) binding data from hepatic Bmal1 ChIP-seq experiments (44), (v) putative transcription factor–enzyme regulatory relationships from MotifMap (45, 46), and (vi) the metabolomic profile from wild-type and Clock−/− liver samples. Whereas each of these datasets comes with its own noise and limitations, it is through their integration that different lines of evidence can be combined to achieve a more global and robust view of 24-h rhythms. This integrated platform enabled us to derive a unique comprehensive map of the in vivo daily metabolome in liver cells (SI Methods and Figs. S5–S8). The 3 nodes in the map are composed of the following: (i) named metabolites, (ii) enzymes directly related to the named metabolites, and (iii) transcription factors that may regulate the enzymes, adding up to over 2,700 nodes. The four edges in the map correspond to the following: (i) enzyme–metabolite edges extracted from KEGG, (ii) metabolite–metabolite edges drawn between any pair of metabolites that participate in a common reaction, (iii) protein–protein interaction edges from protein–protein interaction databases, and (iv) transcription factor–enzyme edges extracted from MotifMap or ChIP-seq data. Over 9,500 edges are depicted. The mapping process is summarized as a flowchart (Fig. S6). To manage the complexity of this map, we have provided metabolite centric views of the subnetworks that surround each named metabolite (Figs. S5 and S7). These views are freely accessible through the CircadiOmics Website (http://circadiomics.igb.uci.edu). The Website allows users to search for any of the named metabolites and view experimental metabolite levels and statistics for WT vs. Clock−/− livers, as well as the subnetwork associated with each metabolite. The subnetwork includes all of the enzymes that catalyze a reaction in which the metabolite participates, other metabolites that coreact with the metabolite of interest, transcription factors with putative binding sites in the promoter regions (up to 8 kbp upstream) of the genes encoding the enzymes, and protein–protein interactors of these enzymes. The corresponding edges are displayed together with the Bmal1 ChIP-seq mouse liver binding data edges. The gene node graphics show the circadian mouse liver gene expression, and the metabolite node graphics show the metabolite levels in WT and Clock−/− livers. The network and its subnetworks can be explored and rearranged interactively, providing a comprehensive system-level view of the circadian molecular pathways.
Connections Between Metabolic Nodes and Transcription Nodes.
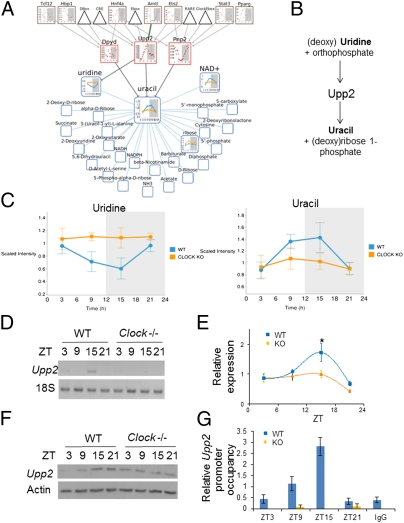
The validity of our created metabolite nodes was tested experimentally. Fig. 5 provides an example of a node that typifies the prominent interfaces found in this study between the diurnal liver metabolome and transcriptome. Within the uracil-containing pyrimidine metabolic pathway, uridine oscillated in WT livers, whereas a remarkable absence in oscillation was observed in Clock−/− mice (Fig. 5 A and C). Uracil showed an oscillation in WT mice that was antiphase to uridine and like uridine, it did not oscillate in Clock−/− livers (Fig. 5C). Uridine levels were elevated in the mutant livers (ZT9 Clock−/− /WT = 1.550, q = 0.054, P < 0.001; ZT15 Clock−/− /WT, P < 0.001). Values of uracil showed time and genotype main effects (P = 0.012 and P < 0.001, respectively, two-way ANOVA), and uridine values showed time and genotype main effects (P = 0.008 and P = 0.001, respectively, two-way ANOVA) as well as a genotype:time interaction. Within the metabolic node, a number of regulatory enzymes, including uridine phosphorylase 2 (UPP2), were implicated (Fig. 5A). UPP2 is essential for pyrimidine salvage and catalyzes the cleavage of uridine to uracil and ribose-1-phosphate (Fig. 5B) (47, 48). To assess the expression of Upp2 in vivo, mRNA was analyzed in WT and Clock−/− livers. Upp2 was found to oscillate in a diurnal fashion (n = 5–8 livers per zeitgeber time, per genotype) with a peak at ZT15. Upp2 oscillation was considerably reduced in Clock−/− mice (Fig. 5E) (main effect of genotype, P = 0.020, two-way ANOVA), correlating with the increased uridine but depressed uracil levels. UPP2 protein also showed an oscillation throughout the diurnal cycle (Fig. 5F). Analysis of the Upp2 promoter by MotifMap showed numerous E boxes upstream of the transcriptional start site. Chromatin immunoprecipitation analysis revealed that CLOCK binds to this region of the Upp2 promoter in a diurnal fashion, with a peak at ZT15 (Fig. 5G). To verify that CLOCK binding followed the expected profile of binding on other target sites in the same livers, CLOCK binding to the Dbp promoter was analyzed in the same WT and Clock−/− livers. As expected, CLOCK binding to upstream Dbp regulatory elements showed the expected circadian profile, with a peak at ZT9 (Fig. S9) and Dbp and Per2 expression profiles matched those previously reported (32). Whereas Upp2 appears to be directly regulated by the circadian clock machinery, the elevated levels of uridine at ZT21 may reflect further circadian regulation of this metabolite, as UPP2 protein remains high at this time point. Elevated levels of ribose-1-phosphate can drive the reaction in the opposite direction; therefore, one possibility is that the reaction begins to reverse between ZT15 and ZT21 (49). Another possibility is that the rapid increase in uridine levels at ZT21 reflects the rate of uridine taken up by the liver from the portal drainage (50).
Fig. 5.
Cohesiveness between computational networks and in vivo uracil metabolism. (A) The uracil network predicts an interaction with uridine phosphorylase 2 (UPP2). (B) UPP2 participates in the reversible reaction whereby uridine and orthophosphate are converted to uracil and ribose 1-phosphate. (C) Uridine and uracil oscillate in an antiphase pattern in WT livers but are nonoscillatory in Clock−/− livers. (D and E) Semiquantitative PCR of Upp2 and quantification of Upp2 mRNA in WT (+/+) and Clock−/− (−/−) livers (error ± SEM, *P < 0.05, genotype main effect; ***P < 0.001, time main effect; two-way ANOVA; *, P < 0.05 Bonferroni posttest). (F) UPP2 and Actin protein expression in WT and Clock−/− livers (each band represents five pooled livers) (G) Diurnal binding of CLOCK to the Upp2 promoter. Immunoprecipitation of CLOCK from liver homogenates and quantification of CLOCK-bound target DNA by qPCR normalized to Upp2 input DNA. (For binding of CLOCK to the Dbp promoter in the same livers, see Fig. S9. N = 3–10 livers per data point.).
Some metabolite nodes, such as the urea node, contained several metabolites having a time main effect. Urea, ornithine, and citrulline all showed diurnal variation (http://circadiomics.igb.uci.edu). Exploration of other nodes revealed that some metabolites were not rhythmic in WT livers yet were altered in Clock−/− livers. The citrate and arachidate (C20:0) nodes fit this description (Figs. S7A and S8A). Within these networks, citrate synthase and several acyl-CoA-thioesterases (ACOTs) were identified, the expressions of which were found to be severely abnormal in Clock−/− livers (Figs. S7 and S8).
Circadian Clock Connects the Hepatic Metabolome and Transcriptome.
Circadian physiology reveals that strong connections between the circadian clock and cellular metabolism exist (1, 51–56), but the extent to which these occur and the nature of these interactions have been largely unappreciated. This study reveals a comprehensive and integrative map of the diurnal metabolome in the liver and provides a detailed depiction of the dynamic interfaces between the metabolome, proteome, and transcriptome. In addition to revealing scores of oscillating metabolites in the liver, this study demonstrates that the liver metabolome is dependent in part on local effects of the circadian clock transcriptional machinery, as exemplified by CLOCK binding to the Upp2 promoter. As evidenced by metabolites that oscillate in WT and Clock−/− mouse livers but whose normalized expression differs, however, it is likely that some metabolic pathways depend more on zeitgebers (such as food or light) or an interaction of zeitgeber and CLOCK-driven mechanisms. For example, NAD+ has been shown to increase in the liver during fasting but decrease after refeeding (57). Consistent with this finding, our data show levels of NAD+ rising in the liver of WT mice during the late phase of the resting period but failing to do so in Clock−/− mice. Whereas this reduction of NAD+ may reflect the diurnal consumption of food in Clock−/− mice, our work has previously demonstrated the role of CLOCK in the regulation of the NAD+ salvage pathway (27).
Interestingly, the aberrant pattern of food-related biochemicals and bile acids in the Clock−/− livers strongly suggests that the suprachiasmatic nucleus and hypothalamic cues associated with rhythmic food intake are disrupted in these animals. The molecular events responsible for this advanced daily food consumption may be related to the causes underlying human diseases such as night eating syndrome (NES) where individuals wake during the night to consume food (58–60). The molecular events responsible for this diurnal eating pattern are not yet clear.
Discussion
The growing evidence that circadian disruption impinges on metabolic homeostasis has created the need for a deeper understanding of the interplay between circadian and metabolic processes. From the context of the liver's role in energy homeostasis and drug metabolism, the integration of the circadian transcriptome and metabolome should provide valuable insights into circadian physiology and disease treatment. For example, uridine homeostasis is important for spermatogenesis, vascular resistance, and neuronal function, and uridine levels are critically important in preventing the toxicity normally associated with 5-fluorouracil-based drug regimens (49). Furthermore, our study shows the urea cycle to be controlled in a 24-h manner. Because the deamination of amino acids produces levels of ammonia that could be life threatening, its elimination via conversion to urea is essential for survival. Proteomic analysis reveals that members of the urea cycle oscillate in the liver (14, 28), and urea has been reported to oscillate in the blood of several species (28, 61–65). As blood flow into the liver is a circadian event, it is possible that some metabolites represent physiological oscillations that are independent of hepatocyte production; however, these still reflect the changing diurnal landscape of hepatic metabolism.
Node validation studies reveal a striking number of enzymes implicated in deregulation of metabolites in the Clock−/− livers. Enzymes contributing to pyrimidine salvage (UPP2), lipid metabolism (ACOTs), and the Krebs cycle (citrate synthase, CS) pathways are deregulated in Clock−/− livers. Importantly, aberrant regulation of these enzymes was predicted by metabolite nodes, expressly demonstrating the interconnectedness of the hepatic circadian clock with the hepatic metabolome and transcriptome as well as the utility of the CircadiOmics dataset. Whereas this interconnectedness is clearly demonstrated in the nodes investigated thus far, the presence of a few rhythmic metabolites in the Clock−/− livers underscores the complexity of circadian rhythmicity at the systems level. For example, catechol sulfate abundance is rhythmic in Clock−/− livers. Catechol is found naturally in fruits and vegetables; however, its sulfation occurs in the intestine. As energy intake is primarily (although not exclusively) centrally controlled, substances such as catechol sulfate are either a reflection that some aspects of both central rhythmicity (energy intake) and peripheral rhythmicity (i.e., sulfation) are maintained or that the lack of precision centrally can be compensated by an intact peripheral pathway.
In summary, a diurnal metabolome exists in the liver, where it is controlled by zeitgebers and the transcriptional machinery of the circadian clock itself. CircadiOmics promises to advance our understanding of the global networks that exist within the liver and allow a deeper comprehension of the many and inseparable links between metabolism and circadian rhythmicity.
Methods
Mice.
Clock-deficient (Clock−/−) animals were a gift from S. Reppert (Worcester, MA) (32). Experiments were performed in conformity with the institutional animal care and use committee guidelines at the University of California at Irvine. Mice were fed ad libitum. Heterozygous mating pairs were used for breeding so that WT and Clock−/− littermates could be generated for experiments.
Global Metabolic Profiling.
Adult, age-matched, male WT and Clock−/− mice were used for liver metabolic profiling. ZT0 corresponded to lights on and ZT12 to lights off in the animal facility. Livers were harvested, rinsed in PBS, and rapidly frozen in liquid nitrogen. The metabolic profiling platform used by Metabolon combines three independent platforms: UHPLC/MS/MS optimized for basic species, UHPLC/MS/MS optimized for acidic species, and GC/MS (66). Details of sample preparation and data analysis are described in SI Methods.
Indirect Calorimetry.
Indirect calorimetry was performed using negative-flow CLAMS hardware system cages (Columbus Instruments). VO2, VCO2, and food intake were measured. RER (VCO2/VO2) was calculated with Oxymax software (Columbus Instruments).
Preparation of cDNA and Quantitative Real-Time Reverse Transcription.
RNA was isolated and cDNA prepared as previously described (67). The primers for ChIP, semiquantitative PCR, and real-time qPCR were obtained from refs. 67–71 or designed by Primer 3 (v. 0.4.0) and are listed in SI Methods.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation was performed as described in ref. 72 with some modifications (SI Methods).
CircadiOmics.
We developed a computational framework that integrates the above-described liver metabolome data with the following sources of information: KEGG, Circa and GEO, BioGRID, IntAct and MINT, Bmal1 ChIP-seq, and MotifMap. This integration is described in detail in SI Methods. Data integration was carried out using code written in Python and charts were created using Google Chart API and visualized using Cytoscape (version 2.8) (73). All data are freely available at the CircadiOmics Website (http://circadiomics.igb.uci.edu).
Statistics.
Data represent mean ± SEM. In circadian experiments with two variables, statistical significance was calculated by two-way ANOVA using Bonferroni posttests to compare replicate means (Prism 3.0). A P value of <0.05 was considered statistically significant. Mann-Whitney u tests were used to determine significance between two independent samples. For analysis of rhythmic metabolites, the nonparametric test, JTK_CYCLE, was used incorporating a window of 20–28 h for the determination of circadian periodicity (30). Venn diagrams were made with Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Statistical Analysis for Metabolite Profiling.
Statistical Analysis for Metabolite Profiling appears in SI Methods.
Supplementary Material
Acknowledgments
We thank Clarence Gillett at Metabolon for assisting in the study design; Akiko Kawai for assisting with animal care; P.S.-C. and P.B. laboratory members, Dr. Saurabh Sahar, Dr. Robert Eckel, and Dr. Roberto Coppari for discussions; Jordan Hayes for assisting with the Web site creation; and Dr. Karl Kornacker for assistance using the JTK_CYCLE R script. Monetary support includes the following grants: National Institutes of Health (NIH)/National Research Service Award F32 DK083881 (to K.L.E.M.), NIH GM081634 and AG033888 (to P.S.-C.), and Sirtris Pharmaceuticals SP-48984 (to P.S.-C.). The work of V.R.P. and P.B. is supported by the following grants: National Science Foundation IIS-0513376, NIH LM010235, and NIH/National Library of Medicine T15 LM07443 (to P.B.).
Footnotes
Conflict of interest statement: R.P.M. and K.S.V. are current and past employees of Metabolon, respectively.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118726109/-/DCSupplemental.
References
- 1.Froy O. Metabolism and circadian rhythms—implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 2.Gerstner JR, et al. Cycling behavior and memory formation. J Neurosci. 2009;29:12824–12830. doi: 10.1523/JNEUROSCI.3353-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamura H. Suprachiasmatic nucleus clock time in the mammalian circadian system. Cold Spring Harb Symp Quant Biol. 2007;72:551–556. doi: 10.1101/sqb.2007.72.033. [DOI] [PubMed] [Google Scholar]
- 5.Sharifian A, Farahani S, Pasalar P, Gharavi M, Aminian O. Shift work as an oxidative stressor. J Circadian Rhythms. 2005;3:15. doi: 10.1186/1740-3391-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawachi I, et al. Prospective study of shift work and risk of coronary heart disease in women. Circulation. 1995;92:3178–3182. doi: 10.1161/01.cir.92.11.3178. [DOI] [PubMed] [Google Scholar]
- 7.Parkes KR. Shift work and age as interactive predictors of body mass index among offshore workers. Scand J Work Environ Health. 2002;28:64–71. doi: 10.5271/sjweh.648. [DOI] [PubMed] [Google Scholar]
- 8.Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53:103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 9.Antunes LC, Levandovski R, Dantas G, Caumo W, Hidalgo MP. Obesity and shift work: Chronobiological aspects. Nutr Res Rev. 2010;23:155–168. doi: 10.1017/S0954422410000016. [DOI] [PubMed] [Google Scholar]
- 10.Fonken LK, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci USA. 2010;107:18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oishi K, et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 14.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 16.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 18.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 19.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arias I, et al. The Liver: Biology and Pathobiology. New York: Wiley; 2009. [Google Scholar]
- 21.Destici E, Oklejewicz M, Nijman R, Tamanini F, van der Horst GT. Impact of the circadian clock on in vitro genotoxic risk assessment assays. Mutat Res. 2009;680:87–94. doi: 10.1016/j.mrgentox.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 22.O'Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love J, Dodd AN, Webb AA. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell. 2004;16:956–966. doi: 10.1105/tpc.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 30.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 33.DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 34.Le Minh N, Damiola F, Tronche F, Schütz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber AM, et al. Molecular clock is involved in predictive circadian adjustment of renal function. Proc Natl Acad Sci USA. 2009;106:16523–16528. doi: 10.1073/pnas.0904890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes M, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark C, et al. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aranda B, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ceol A, et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2009;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatanaka F, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daily K, Patel VR, Rigor P, Xie X, Baldi P. MotifMap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics. 2011;12:495. doi: 10.1186/1471-2105-12-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie X, Rigor P, Baldi P. MotifMap: A human genome-wide map of candidate regulatory motif sites. Bioinformatics. 2009;25:167–174. doi: 10.1093/bioinformatics/btn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao D, Pizzorno G. Uridine phosophorylase: An important enzyme in pyrimidine metabolism and fluoropyrimidine activation. Drugs Today (Barc) 2004;40:431–443. doi: 10.1358/dot.2004.40.5.850491. [DOI] [PubMed] [Google Scholar]
- 48.Cappiello M, Mascia L, Scolozzi C, Giorgelli F, Ipata PL. In vitro assessment of salvage pathways for pyrimidine bases in rat liver and brain. Biochim Biophys Acta. 1998;1425:273–281. doi: 10.1016/s0304-4165(98)00071-3. [DOI] [PubMed] [Google Scholar]
- 49.Pizzorno G, et al. Homeostatic control of uridine and the role of uridine phosphorylase: A biological and clinical update. Biochim Biophys Acta. 2002;1587:133–144. doi: 10.1016/s0925-4439(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 50.Gasser T, Moyer JD, Handschumacher RE. Novel single-pass exchange of circulating uridine in rat liver. Science. 1981;213:777–778. doi: 10.1126/science.7256279. [DOI] [PubMed] [Google Scholar]
- 51.Yang X, Lamia KA, Evans RM. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- 52.Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol. 2007;28:61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism: The epigenetic link. J Cell Sci. 2010;123:3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcheva B, Ramsey KM, Affinati A, Bass J. Clock genes and metabolic disease. J Appl Physiol. 2009;107:1638–1646. doi: 10.1152/japplphysiol.00698.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 58.O'Reardon JP, et al. Circadian eating and sleeping patterns in the night eating syndrome. Obes Res. 2004;12:1789–1796. doi: 10.1038/oby.2004.222. [DOI] [PubMed] [Google Scholar]
- 59.Goel N, et al. Circadian rhythm profiles in women with night eating syndrome. J Biol Rhythms. 2009;24:85–94. doi: 10.1177/0748730408328914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Reardon JP, Peshek A, Allison KC. Night eating syndrome: Diagnosis, epidemiology and management. CNS Drugs. 2005;19:997–1008. doi: 10.2165/00023210-200519120-00003. [DOI] [PubMed] [Google Scholar]
- 61.Piccione G, Caola G, Refinetti R. Daily rhythms of liver-function indicators in rabbits. J Physiol Sci. 2007;57:101–105. doi: 10.2170/physiolsci.RP014606. [DOI] [PubMed] [Google Scholar]
- 62.Koudela K, Sova Z, Orlík K. Circadian rhythmicity of urea concentration in the liver and blood serum of cockerels. Vet Med (Praha) 1977;22:177–182. [PubMed] [Google Scholar]
- 63.Assenza A, Fazio F, Marcenò G, Piccione G, Caola G. Daily rhythms of serum and salivary parameters in goats. Aust Vet J. 2009;87:397–401. doi: 10.1111/j.1751-0813.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- 64.Piccione G, Caola G, Refinetti R. Annual rhythmicity and maturation of physiological parameters in goats. Res Vet Sci. 2007;83:239–243. doi: 10.1016/j.rvsc.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 65.Piccione G, Foà A, Bertolucci C, Caola G. Daily rhythm of salivary and serum urea concentration in sheep. J Circadian Rhythms. 2006;4:16. doi: 10.1186/1740-3391-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reitman ZJ, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Im YS, et al. Enhanced cytotoxicity of 5-FU by bFGF through up-regulation of uridine phosphorylase 1. Mol Cell. 2009;28:119–124. doi: 10.1007/s10059-009-0116-x. [DOI] [PubMed] [Google Scholar]
- 69.Honda A, et al. Attenuation of cadmium-induced testicular injury in metallothionein-III null mice. Life Sci. 2010;87:545–550. doi: 10.1016/j.lfs.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 70.Pattyn F, Speleman F, De Paepe A, Vandesompele J. RTPrimerDB: The real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 72.Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol. 2008;22:78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.