Abstract
The geographic and temporal origins of Madagascar's biota have long been in the center of debate. We reconstructed a time-tree including nearly all native nonflying and nonmarine vertebrate clades present on the island, from DNA sequences of two single-copy protein-coding nuclear genes (BDNF and RAG1) and a set of congruent time constraints. Reconstructions calculated with autocorrelated or independent substitution rates over clades agreed in placing the origins of the 31 included clades in Cretaceous to Cenozoic times. The two clades with sister groups in South America were the oldest, followed by those of a putative Asian ancestry that were significantly older than the prevalent clades of African ancestry. No colonizations from Asia occurred after the Eocene, suggesting that dispersal and vicariance of Asian/Indian groups were favored over a comparatively short period during, and shortly after, the separation of India and Madagascar. Species richness of clades correlates with their age but those clades that have a large proportion of species diversity in rainforests are significantly more species-rich. This finding suggests an underlying pattern of continuous speciation through time in Madagascar's vertebrates, with accelerated episodes of adaptive diversification in those clades that succeeded radiating into the rainforests.
Keywords: Cretaceous-Tertiary, historical biogeography, lineage diversification, rainforest adaptation, overseas dispersal
Madagascar's unique biodiversity has attracted the interest of evolutionary biologists and biogeographers for a long time. This island was part of the Gondwana supercontinent. As a part of Indo-Madagascar, it separated from Africa 160–130 Mya. The breakup of Indo-Madagascar and northwards drifting of India and the Seychelles started 88 Mya, leaving Madagascar isolated in the Indian Ocean and without subaerial connection to any other landmass for the last 65–80 Myr (1).
The isolation of Madagascar coincided with the end of the Cretaceous, a period of global mass extinction and biotic turnover, probably linked to a major meteorite impact marking the Cretaceous–Tertiary (K–T) boundary at 65.5 Mya (2). In Madagascar, the different composition of the Late Cretaceous vs. the extant vertebrate fauna led to the hypothesis of a major biotic change in deep time (3). The Cretaceous fauna included lungfishes, gars, nonranoid giant frogs, dinosaurs, and marsupial and gondwanatherian mammals (3–8), whereas the extant vertebrate fauna is composed of mainly percomorph freshwater fishes, ranoid frogs, modern squamate reptiles, lemurs, rodents, carnivores, afrotherian mammals, bats, and numerous families of birds (9).
Reconstructing the temporal pattern of this striking biotic turnover is hampered by the almost complete lack of post-Cretaceous and pre-Pleistocene terrestrial fossil deposits. This fossil gap presents difficulties in understanding how and when the extant clades of vertebrates colonized the island, and how their subsequent diversification took place. In recent times, explicit paleogeographic and paleoclimatic modeling associated with the reconstruction of molecular time-trees have started to resolve the biogeography of Madagascar, previously characterized as one of the greatest mysteries of natural history (10, 11). For numerous Malagasy clades of amphibians, squamates, and mammals, sister-group relationships to African taxa and a Cenozoic age are now well established, suggesting a predominance of Out-of-Africa overseas dispersal favored by oceanic paleocurrents (11–15). However, the exact timing of colonizations, as well as their possible clustering in particular periods, remain unstudied for many vertebrate clades and are contentious for others (11), largely because of the use of different molecular markers and time constraints. In addition, the closest relatives of several other taxa are found in South America or Asia (16, 17). The temporal pattern of vertebrate colonization of Madagascar from these different source continents has not been comprehensively studied to date.
Analyses of molecular data have led to great progress in understanding the timing of vertebrate diversification (18, 19). Here, we generated a comprehensive dataset that uses the same molecular markers and time constraints for nearly all terrestrial and freshwater vertebrate clades occurring on Madagascar and their sister taxa. The selected genes, BDNF (brain-derived neurotrophic factor) and RAG1 (recombination activating gene 1), are single-copy, protein-coding, and universal among gnathostomes. We use multiple cross-validated time constraints in a single time-tree to obtain compatible age estimates across clades (20). Those estimates allow us to assess general biogeographic patterns of Madagascar's colonization by vertebrates, for which we test whether: (i) the majority of extant vertebrate clades colonized Madagascar during or after the K–T boundary, as suggested by the fossil record (7); (ii) colonizations followed different temporal patterns depending on the source continent; (iii) species richness of clades are related to their age in Madagascar; and (iv) clade diversification was influenced by habitat type.
Results
Vertebrate Time-Trees Based on Congruent Time Constraints.
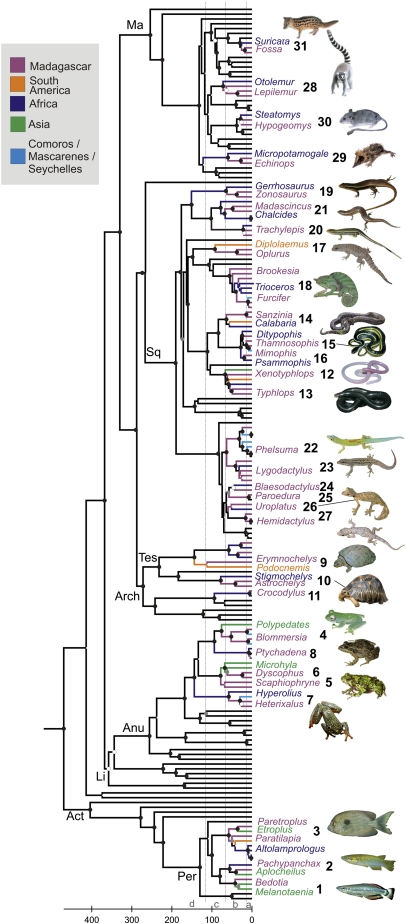
Phylogenetic analyses of the combined 1,747 base pairs of RAG1 and BDNF for 188 taxa representing nonflying and nonmarine Malagasy vertebrate clades, their known non-Malagasy sister-groups, and a set of other vertebrate taxa, recovered most of the generally accepted deep and shallow relationships among vertebrates (Fig. 1). Time-tree reconstruction with 43 selected time constraints was based on two approaches that either allow substitution rates to vary independently over clades (ICR) or in an autocorrelated fashion (ACR) (Materials and Methods). The extremes of the 95% credibility intervals (CrIs) from the two approaches were merged into a single composite CrI (21).
Fig. 1.
ACR vertebrate time-tree based on a 50% majority rule consensus tree from a Bayesian analysis of 1,747-bp DNA sequences of the RAG1 and BDNF genes. Black dots at nodes indicate Bayesian support >0.98, gray dots 0.95–0.98, and white dotted nodes were topology-constrained before the analysis. Colors indicate geographic distribution of taxa and numbers denote clades as listed in Table 1. Only representative taxa of the clades occurring in Madagascar and their sister groups are named (see SI Appendix for tree with names of all terminals); Inset pictures show one representative species for most of the numbered Malagasy clades. Major clade abbreviations: Act, Actinopterygii; Anu, Anura; Arch, Archosauria; Li, Lissamphibia; Ma, Mammalia; Per, Percomorpha; Sq, Squamata; Tes, Testudines. Vertical gray dotted lines separate four main time intervals, at bottom: (a) <15 Mya, corresponding to the period where surface currents did not favor dispersal from Africa; (b) 66–16 Mya, when sea currents favored overseas rafting from Africa; (c) 120–66 Mya, when overland dispersal or short-distance rafting from Asia was possible; (d) >121 Mya, when overland dispersal from Africa and Antarctica might have been possible (15).
A high congruence among most of an initial set of 48 time constraints was obtained in three cross-validation approaches using ACR, here named CV1–CV3 (see SI Appendix for details). In CV1 we assessed statistically the effect of removing single constraints on the overall differences between the constraint ages and molecular age estimates. None of the initial time constraints were highly incongruent, as their removal did not lead to a significant decrease in the variance of the s parameter in one-tailed F-tests (22). In CV2 we performed separate analyses after excluding one constraint in turn, and manually assessed whether the CrI estimated for the respective node was congruent with the original (excluded) constraint. This analysis flagged only three of our initial 48 constraints as incongruent. In CV3 we performed separate analyses after exclusion, in turn, of all but one constraint. The great majority of time-trees recovered correctly most constraints within CrIs. A preferred time-tree was then calculated after exclusion of five constraints, including the three that were most incongruent in CV2 (SI Appendix).
Times and Patterns of the Vertebrate Colonizations of Madagascar.
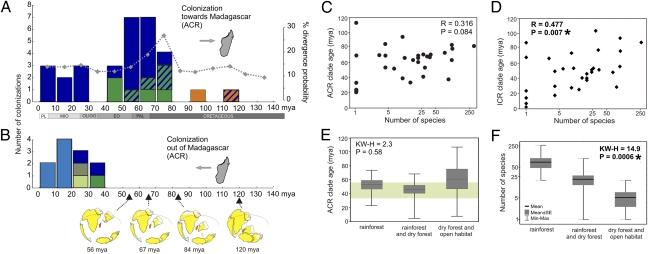
Crown divergence times were not available for all taxa and cannot be obtained for monospecific clades, such as podocnemidid turtles. We thus summarized stem divergence times for the 31 Malagasy clades analyzed (Fig. 1). Although the colonization patterns reconstructed by ACR and ICR at first seem rather different (Fig. 2 and SI Appendix), the ages per clade are correlated (nonparametric correlation, Spearman's R = 0.664, P < 0.001) and concordant regarding the general temporal framework of colonization and differences among source regions. The two analyses agree in a predominant Cenozoic age of origin of the extant Malagasy vertebrates, with ages of 25 (ACR) and 23 (ICR) of 31 clades reconstructed at ≤65.5 Mya (Table 1). The composite CrIs exceeded 125 Mya only in three cases (podocnemidid and testudinid turtles, and tenrecs), confirming that the majority of extant vertebrates must have colonized Madagascar after its separation from Africa at 130 Mya. Additionally, the analyses display clade-age differences depending on their geographic origin. In both analyses, the two clades with South American origins, iguanids and podocnemidid turtles, have Mesozoic stem divergences. In addition, there is a trend for clades with assumed Asian or Asian/African ancestry being older than those originating from Africa: 60 Mya (77–41 Mya) vs. 43 Mya (79–4 Mya) in ACR analyses, 66 Mya (92–39 Mya) vs. 38 Mya (101–1 Mya) in ICR; this result is significant for the ICR estimations (U test: Z = 2.88, P = 0.003), and nearly so for the ACR estimations (Z = 1.73; P = 0.08). No colonization of unambiguous Asian origin was recovered as younger than 39 Mya by any analysis.
Fig. 2.
Age distribution of Malagasy vertebrate clades and species-richness correlates. (A) Histogram of ACR stem divergence ages for vertebrate clades occurring in Madagascar derived from the time-tree in Fig. 1, color-coded depending on the distribution of their sister group. Rhomboids connected by dotted line show the probability of clade origins derived from a model assuming 80% dispersal and 20% vicariance (11). (B) ACR age histogram for clades of assumed out-of-Madagascar dispersal. Inset maps (1) show reconstructions of continental shorelines. (C and D) Scatterplots of species richness (logarithmic scale) of endemic Madagascar clades vs. stem age of the ACR and ICR analyses, respectively. (E and F) Plots of species richness (logarithmic scale) and stem age of endemic Madagascar clades with highest species diversity distribued in rainforest (>75% of species), in rainforest as well as dry forest, or predominantly (>75%) in dry forest (and deforested areas within humid bioclimates). The light green bar in E marks the Eocene period during which the origin of Madagascar's rainforest has been hypothesized (26).
Table 1.
Vertebrate clades endemic to Madagascar and their estimated stem-based age and species richness
| Clade | ACR stem age | ICR stem age | Species |
| Actinopterygii | |||
| 1 Bedotiidae – As | 42 (24-68) | 51 (26–82) | 26 |
| 2 Aplocheilidae – As | 41 (24-65) | 53 (28–82) | 6 |
| 3 Cichlidae – Af | 58 (38-86) | 76 (46–103) | 33 |
| Lissamphibia | |||
| 4 Mantellidae – As | 76 (50–108) | 87 (55–122) | 266 |
| 5 Microhylidae: Cophylinae/ Scaphiophryninae– As/Af | 77 (49–114) | 92 (63–123) | 95 |
| 6 Microhylidae: Dyscophinae – As | 62 (37–94) | 65 (38–95) | 3 |
| 7 Hyperoliidae – Af | 57 (30–94) | 53 (25–85) | 10 |
| 8 Ptychadenidae – Af | 8 (2–20) | 13 (4–24) | 1 |
| Archosauria | |||
| 9 Podocnemididae – SA/Af | 112 (73–159) | 87 (65–111) | 1 |
| 10 Testudinidae – Af | 79 (33–134) | 16 (6–30) | 4 |
| 11 Crocodylidae – Af | 5 (0–20) | 1 (0–2) | 1 |
| Squamata | |||
| 12 Xenotyphlopidae – As/Af | 61 (42–82) | 66 (43–90) | 1 |
| 13 Typhlopidae – As/Af | 54 (36–74) | 39 (21–59) | 15 |
| 14 Boidae – Af | 61 (48–75) | 47 (22–74) | 4 |
| 15 Lamprophiidae: Pseudoxyrhophiinae – Af | 24 (14–37) | 28 (15–41) | 80 |
| 16 Lamprophiidae: Psammophiinae – Af | 19 (10–31) | 22 (10–37) | 1 |
| 17 Opluridae – SA | 90 (62–120) | 72 (32–117) | 9 |
| 18 Chamaeleonidae – Af | 54 (34–77) | 54 (34–75) | 94 |
| 19 Gerrhosauridae – Af | 61 (32–96) | 37 (17–63) | 19 |
| 20 Scincidae: Trachylepis – Af | 19 (9–37) | 24 (10–39) | 13 |
| 21 Scincidae: Scincinae – Af | 65 (39–96) | 47 (23–70) | 67 |
| 22 Gekkonidae: Phelsuma – Af | 62 (39–91) | 49 (34–65) | 34 |
| 23 Gekkonidae: Lygodactylus – Af | 62 (39–91) | 49 (34–65) | 24 |
| 24 Gekkonidae: Blaesodactylus – Af | 42 (23–68) | 27 (10–45) | 4 |
| 25 Gekkonidae: Paroedura – Af | 57 (34–86) | 43 (27–60) | 19 |
| 26 Gekkonidae: Uroplatus – Af | 51 (29–78) | 38 (20–58) | 19 |
| 27 Gekkonidae: Hemidactylus – Af | 4 (1–10) | 6 (1–11) | 1 |
| Mammalia | |||
| 28 Strepsirhini (lemurs) – As/Af | 71 (51–94) | 73 (46–103) | 96 |
| 29 Tenrecidae – Af | 60 (40–84) | 101 (62–141) | 29 |
| 30 Muridae: Nesomyinae – Af | 28 (17–42) | 47 (26–71) | 24 |
| 31 Eupleridae – Af | 26 (16–38) | 26 (14–39) | 9 |
Clade numbers as in Fig. 1; Af, Africa; As, Asia; SA, South America. Ages are given in Mya.
Prevailing ocean currents were periodically favoring rafting from Africa to Madagascar in the Early Cenozoic, but not after a reconfiguration of surface flows in the mid-Miocene, 20–15 Mya (10). Nevertheless, our analyses suggest three arrivals from Africa after the 15-Mya tipping point: ptychadenid frogs, crocodiles, and Hemidactylus geckos (Fig. 2A and Table 1). All of the 11 out-of-Madagascar dispersals are relatively young (36–3 Mya) and peak after the Oligocene (Fig. 2B). The dispersal of the ancestors of day geckos to the Mascarene Islands located east of Madagascar occurred at 22 Mya, and dispersals to the continental Seychelles, and to Asia and Africa occurred in the Oligocene and Eocene (36–28 Mya). Dispersals to the Comoro islands west of Madagascar were recovered by both analyses post-Oligocene, in agreement with favorable surface currents in this period, a pattern confirmed by the cross-validations also for those Comoro-Malagasy splits used as age constraints in the main analyses.
Clade Age and Rainforest Habitat Influence Species Richness.
Besides the extraordinary degree of endemism at higher taxonomic levels, Madagascar's biota is characterized by a high, although incompletely known, species richness, and by a high proportion of range-restricted species that are microendemic to particular areas of the island (9). This pattern might also be typical for other tropical regions, but Madagascar is a particularly well-suited model region to determine the underlying patterns of biotic diversification (23).
A debated biological question is whether species richness is primarily influenced by clade longevity or diversification rate (24, 25). Among Malagasy vertebrates we expect species richness to be correlated with clade age if their diversification took place following a stochastic process of phylogenetic clade accumulation through time. In contrast, if many of the clades underwent rapid adaptive radiations after reaching the island, we expect this correlation to be weak or absent. Using nonparametric rank correlation analyses, stem age and species richness are significantly correlated based on ICR ages (Spearman's R = 0.477; P = 0.007) and nearly so based on ACR (R = 0.316; P = 0.084) (Fig. 2 C and D).
We defined a number of covariables likely to influence either the diversification process or the species-area relationship of clades (Materials and Methods). An ANCOVA model supports that species numbers increase with ACR clade age, as well as with external fertilization, parental care, terrestrial habits, and occurrence in rainforest, the latter variable being the most significant predictor (SI Appendix, Table S8).
Madagascar is renowned for its high diversity of biomes, ranging from subarid shrublands to humid rainforests (9). Vertebrate clades are unevenly distributed over these biomes, with some clades predominantly found in humid and subhumid forests and others in dry forests and open habitats. The six most species-rich clades in our analysis with >50 species (mantellid and microhylid frogs, pseudoxyrhophiine snakes, chameleons, skinks, and lemurs) are rather variable in age (96–20 Mya) but all have a majority or an important proportion of their species diversity living in rainforests (SI Appendix, Table S7). Among all clades, those with the majority (>75%) of species in rainforests have significantly higher species richness than those distributed in both habitats, and the lowest species richness is found in clades specializing to dry and open biomes (Fig. 2F). These three groups of clades do not differ significantly in clade age (Fig. 2E) but most of the species-rich rainforest clades colonized Madagascar at the onset of the Eocene, just before the putative spread of Madagascar's rainforests (26).
Discussion
This comprehensive assessment of Malagasy vertebrate ages and colonization patterns robustly reconstructs the origins of these organisms in Cretaceous and Cenozoic times. Because we included all clades in a single time-tree and cross-validated all time constraints, we can exclude biases, which could arise if incongruent constraints are used in independent single-clade analyses. Even taking into account the conservative composite CrIs, there are only three estimates >125 Mya. This finding strongly supports a predominant origin of Madagascar's extant vertebrates after Madagascar separated from Africa at 160–130 Mya (15). Most of these clades have African sister groups, confirming a predominance of overseas dispersal from Africa (11), in agreement with clade age estimates previously published for 17 Malagasy clades (15). ACR ages are in 12 clades on average 19% (0–43%) older than the published data and in five clades on average 23% (6–29%) younger (SI Appendix, Table S6). ICR ages are more strongly deviating: eight clades are on average 23% (8–39%) younger, and nine clades on average 35% (2–54%) older. Several of the ICR ages are rather unrealistic, given previous assessments, such as an age of tenrecs as old as 103 Mya and of tortoises as young as 16 Mya. We therefore consider the ACR results to be more reliable. In general, we acknowledge that any single-point estimate in our data might be subject to changes as novel methods, better time constraints, or more comprehensive molecular datasets become available. However, we expect changes not to exceed the composite CrIs obtained here and, rather, to support even younger ages given our conservative selection of age constraints.
Only three literature ages fall outside of the ACR CrIs: cichlids and xenotyphlopids, which were younger, and hyperoliid frogs, which were older in our analyses. Among the biogeographically most-relevant deviations of our data from previous analyses (27, 28) are the younger ages of cichlids and of the other two fish clades. The ACR and ICR analyses agreed in placing the origin of these fishes into the Latest Cretaceous or Early Cenozoic, thus supporting dispersal hypotheses for their origin (29). The origin of Madagascar boas was estimated in the Paleocene–Eocene rather than Cretaceous, suggesting dispersal from Africa rather than from Antarctica, as suggested before (17).
Our results can be directly compared with an explicit model for stem divergence ages for Malagasy vertebrates (11) assuming 80% dispersal and 20% vicariance, which predicts a clear peak at the time of Indo-Madagascar breakup at 88–60 Mya. This finding is in perfect agreement with the results of the ACR analysis (Fig. 2A), whereas the ICR ages suggest a more recent Cenozoic peak and a more regular spacing of ages in the Mesozoic (SI Appendix). Divergences >90 Mya are rare in our analyses but are predicted, albeit at low prevalence, by the model. This discordance confirms paleontological evidence for a massive biotic change around the K–T boundary. It also suggests that this biotic change affected all vertebrates, including small frogs and lizards, for which paleontological evidence is typically scarce.
Only a few taxa are missing from our analysis. Apart from flying vertebrates (bats and birds), these include a genus of cyprinodontiform fishes (Pantanodon), with one representative in Africa, some nonprimary freshwater fish clades, three gecko clades (Geckolepis, Matoatoa, and Paragehyra) with uncertain phylogenetic relationships, and Cryptoblepharus shoreline lizards, which dispersed recently from the Australasian region. Three extinct Pliocene-Pleistocene clades are the nonflying Aepyornis related to the Australian and New Zealand flightless birds, pygmy hippos with clear relationships to Africa, and the Malagasy aardvark Plesiorycteropus, which might belong into the Afrotheria. Most of the bird and bat lineages, as well as hippos and aardvarks, presumably arrived in Madagascar between the Oligocene and present (15), and their inclusion would further reinforce the pattern of Cenozoic origins of most of Madagascar's extant vertebrates. However, bats and birds are independent from ocean currents and thus include numerous post-Oligocene arrivals from Asia (15).
In ACR and ICR analyses, the largest proportion of divergence times at 88–60 Mya corresponds to groups with Asian or ambiguously Asian/African ancestry, fitting the expectations of Indo-Madagascar vicariance. However, the Malagasy mantellid and dyscophine frogs are phylogenetically deeply nested in widespread Asian taxa. This finding suggests at least occasional ancient dispersal from India to Madagascar (30), probably facilitated by discontinuous land bridges and stepping-stones after the connection of India with the Asian plate (1). No colonizations from Asia are estimated after the Eocene, which suggests that colonizations from Asia became severed after India had reached its current position, with a large open-sea distance to Madagascar (15).
The two clades with clear South American relationships among extant taxa, iguanas and podocnemidid turtles, are by far oldest in the ACR analyses and were among the oldest seven in the ICR analysis, confirming previous estimates (17). However, our data indicate that these clades might even be older than previously thought, and could have reached Madagascar in the Early Cretaceous. This theory would be in agreement with recent paleogeographic reconstructions (31, 32) that do not support the previously hypothesized Late Cretaceous connections of Madagascar and South America via Antarctica and the Kerguelen/Gunnerus ridges (16). The third group for which such a connection has been previously invoked, Madagascar boas, turned out to be distinctly younger, which agrees with their recently discovered phylogenetic relationship to the African Calabaria (17). Clades of African ancestry were rather evenly spaced over the Cenozoic, and a few of them even in the period 15–0 Mya, coinciding with an unfavorable pattern of surface currents (10).
The equilibrium model of island biogeography predicts a positive area/species diversity relationship because of balanced rates of colonization and extinction (33). Because Madagascar's biota evolved largely in isolation, with a limited number of colonizations, the assembly of its extant species richness has been mainly a consequence of speciation processes rather than immigration. In such situations, a nonequilibrium model of diversity can be applied and species richness is expected to mainly be influenced by variation in net diversification rates or clade age (34). Our analyses are congruent in supporting clade age (time since colonization) as one predictor of species richness of Madagascar's vertebrates. This analysis suggests, in many but not all clades, a rather regular net diversification rate with time, but the rather low correlation coefficients suggest an influence also of other drivers of species richness. In our analysis, the relative time lags between (i) the separation of the Malagasy clade from its non-Malagasy sister group and (ii) the first speciation events within Madagascar decreases with clade species richness (SI Appendix). This decrease could be explained by species-poor clades having been more strongly affected by past extinctions, also at deep levels, but we hypothesize it indicates lower diversification rates of these clades instead.
One factor explaining such differences in diversification rate might be the ability of a clade to adapt to rainforest conditions. Although the subarid and arid biomes of Madagascar span over wide environmental gradients in altitude, moisture, and soil composition, which especially for squamate reptiles offer numerous opportunities for specialization, mainly species-poor clades are the ones predominantly living in these habitats. In fact, among the clades with fewer than 20 species none has its center of diversity in rainforests. The species-poor clades specialized to dry conditions are either old Gondwanan relicts (e.g., podocnemidid turtles, iguanas), or very recent colonizers. The vast majority of such young clades that arrived in the Miocene or later (e.g., Ptychadena, Crocodylus, Hemidactylus, Trachylepis, Mimophis), contain no or very few strict rainforest specialists but are composed of species adapted to dry conditions or generalists surviving in open landscapes. This finding might reflect plesiomorphic traits favorable for overseas dispersal, and for survival after arriving at Madagascar's dry west coast after a transoceanic rafting from Africa. The ability of a clade to adapt to rainforest might have been influenced by intrinsic morphological or physiological constraints, or by ecological interactions with other organisms: either in terms of predator–prey relationships (e.g., the diversity of pseudoxyrhophiine snakes might be influenced by the earlier diversification of frogs and lizards, which constitute their main prey), or competition (e.g., psammophiine snakes might not have radiated into rainforests because these were already occupied by the earlier pseudoxyrhophiine snake radiation).
In conclusion, clade species richness in Madagascar is influenced by clade age but also by their adaptability to rainforest habitats. In contrast to purely vicariant speciation scenarios that have typically been invoked in Madagascar (23), these results suggest a role for adaptive speciation during at least some episodes of the evolutionary history of species-rich clades, but possibly less so for species-poor clades. We predict that future exploration of this and other evolutionary hypothesis in the Madagascar model system will much benefit from the comprehensive temporal framework provided herein.
Materials and Methods
Total genomic DNA was extracted using standard protocols and a combination of various degenerated primers were used to amplify overlapping fragments for a total length of 535 aa of the RAG1 gene (amino acid positions 467–1001 in human RAG1) and a fragment corresponding to 221 aa of the BDNF gene (see SI Appendix for details). Chromatographs were checked and sequences were aligned using CodonCode Aligner (v. 3.7.1, Codon Code). The alignment of newly determined sequences was complemented with sequences retrieved from GenBank (SI Appendix, Table S2). For BDNF the software Gblocks (35) was used to delete highly divergent regions, which were either not unambiguously aligned or saturated by multiple substitutions. Additionally, all positions with gaps in both genes were excluded from the analyses. The final concatenated alignment was 1,747 bp long.
We conducted partitioned Bayesian inference searches based on the concatenated dataset in MrBayes 3.1.2 (36) with two partitions: first-plus-second positions, and third positions, grouped for both genes. The partition scheme was selected based on a Bayes factor analysis (SI Appendix). Both partitions were assigned to a general time-reversible substitution model with estimated γ-shaped distribution and proportion of invariable sites, as suggested by MrModeltest (37). We performed four independent runs of 20 million generations sampling trees every 1,000 generations. The first four million generations were discarded based on empirical evaluation of convergence (see SI Appendix for methods used). A number of nodes were constrained according to well-established knowledge on vertebrate phylogeny (Fig. 1 and SI Appendix).
We selected 48 age constraints across the vertebrate tree, of which 43 were used for the final analysis (SI Appendix), with a preference for ample, conservative estimates rather than narrow upper and lower constraints or point estimates because fossil uncertainties are prone to lead to pseudoaccuracy. Constraints are coded C1–C48 (missing numbers refer to excluded constraints; details and references, and rationale for exclusion in SI Appendix). Slashes represent the phylogenetic split of one clade from the other; when no slash is given, the age is of the split of the respective clade from their unspecified sister group. C1, diapsids/synapsids, 338–288 Mya; C2, lungfishes/tetrapods, 419–408 Mya; C3, archosaurs/lepidosaurs, 299.8–259.7 Mya; C4, birds/crocodiles, 250–235 Mya; C5, alligators/caimans, 71–66 Mya; C6, Pelomedusa/Pelusios turtles, >25 Mya; C7, Erymnochelys/Podocnemis turtles, >65 Mya; C8, Cryptodira/Pleurodira turtles, >210 Mya; C9, podocnemidid/pelomedusid turtles, >100 Mya; C10, sea turtles, >110 Mya; C11, turtles, >220 Mya; C12, Sphenodon/squamates, >228 Mya; C13, geckos, >55 Mya; C14, amphisbaenian/lacertid lizards, > 64 Mya; C17, Booidea/Caenophidia snakes, >75 Mya; C18, Comoran Furcifer chameleons, <15 Mya; C20, Canary geckos Tarentola boettgeri/delalandi, <14 Mya; C21, Comoran gecko Phelsuma nigristriata, <15 Mya; C22, Comoran Phelsuma comorensis, <15 Mya; C23, Mascarene Phelsuma inexpectata/ornata, <2.1 Mya; C24, Canary skinks Chalcides sexlineatus/viridanus, <14 Mya; C25, Caniformia/Feliformia, 63.8–42.8 Mya; C26, hippomorph/ceratomorph Perissodactyla, 58–54 Mya; C28, Lagomorpha, 61.5–48.6 Mya; C30, Cetartiodactyla, 65–55 Mya; C31, Spanish/Moroccan Discoglossus frogs, >5.3 Mya; C32, Spanish/Moroccan Alytes frogs, >5.3 Mya; C33, Comoran Blommersia frogs, <15 Mya; C34, Comoran Boophis frogs, <15 Mya; C35, frog/salamander, >230 Mya; C36, Calyptocephalella frogs, >53 Mya; C37, pelomedusine/pelodryadine treefrogs, >42 Mya; C38, African/American pipid frogs, >86 Mya; C39, cryptobranchid/hynobiid salamanders, >161 Mya; C40, pipid frogs, >140 Mya; C41, discoglossid frogs, >167 Mya; C42, Lake Tanganyika cichlid fishes, <12 Mya; C43, Lake Malawi cichlids, <2 Mya; C44, cichlids, >45 Mya; C45, Actinopterygii, >392 Mya; C46, Tetraodontiformes, >98 Mya; C47, Ostariophysi, >146 Mya; C48, Elopomorpha, >151 Mya.
Time-trees were reconstructed with two different relaxed-clock methods with uncorrelated and correlated substitution rates, respectively, over clades (here named ICR and ACR), using the computer programs BEAST (38) and Multidivtime (39). For methodological details of analyses, see SI Appendix. Age estimates for stem-based nodes (separation of Malagasy clades from their closest non-Malagasy sister group) as well as stem-based ages of assumed out-of-Madagascar dispersal events were extracted from the time-tree results. Composite 95% CrIs were compiled by combining CrIs from ICR and ACR analyses which in simulation studies (20) cover the true time in >97% of the estimated times.
Species richness of endemic Madagascar clades was compiled from recent publications (complete species list in SI Appendix). Linear models (ANCOVAs) were performed in R (40) on species richness as dependent and a series of factors that might influence species richness as independent variables: clade age, rainforest occurrence, body size, age to maturity, endothermy, trophic position, internal fertilization, parental care, and aquatic vs. terrestrial habits. Factors were deleted sequentially from the full model based on the Akaike Information Criterion.
Supplementary Material
Acknowledgments
We thank A. Bauer, W. R. Branch, M. Casiraghi, U. Fritz, F. Glaw, S. B. Hedges, I. A. Irisarri, U. Joger, B. P. Noonan, A. Ohler, S. Rocha, T. Townsend, and M. Vargas for crucial samples or sequences; G. Keunecke, M. Kondermann, and E. Saxinger for assistance in the laboratory; J. R. Ali for providing digital maps; K. Samonds for fruitful discussion; and the Malagasy authorities for research and export permits. Computational resources were provided by Le Centre de Ressources Informatiques-Lille 1 supported by the Centre National de la Recherche Scientifique and Lille 1 University–Science and Technology. This work was supported by the Volkswagen Foundation and the Deutsche Forschungsgemeinschaft, AmphibiaTree Project National Science Foundation Grant EF-0334939, and Spanish Ministry of Science and Innovation Grant CGL2009-10198 (to D.R.V.); Borsa di Perezionamento all'Estero grant from Università degli Studi di Milano and by Fundação para a Ciência e a Tecnologia Grant SFRH/ BPD/72908/2010 (to A.C.); and a postdoctoral grant from the University of Lille 1 (to C.P.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Genbank database (accession nos. JQ073048–JQ073135, JQ073138–JQ073291). The full alignments reported in this paper have been deposited in the Dryad data repository, datadryad.org (http://dx.doi.org/10.5061/dryad.50r80407).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112487109/-/DCSupplemental.
References
- 1.Ali JR, Aitchison JC. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma) Earth Sci Rev. 2008;88:145–166. [Google Scholar]
- 2.Schulte P, et al. The Chicxulub asteroid impact and mass extinction at the Cretaceous-Paleogene boundary. Science. 2010;327:1214–1218. doi: 10.1126/science.1177265. [DOI] [PubMed] [Google Scholar]
- 3.Krause DW, Hartman JH, Wells NA. In: Natural Change and Human Impact in Madagascar. Goodman SM, Patterson BD, editors. Washington, DC: Smithsonian Institution; 1997a. pp. 3–43. [Google Scholar]
- 4.Krause DW. Fossil molar from a Madagascan marsupial. Nature. 2001;412:497–498. doi: 10.1038/35087649. [DOI] [PubMed] [Google Scholar]
- 5.Krause DW. In: The Natural History of Madagascar. Goodman SM, Benstead JP, editors. Chicago: Univ of Chicago Press; 2003. pp. 40–47. [Google Scholar]
- 6.Krause DW, Prasad GVR, von Koenigswald W, Sahni A, Grine FE. Cosmopolitanism among Gondwanan Late Cretaceous mammals. Nature. 1997b;390:504–507. [Google Scholar]
- 7.Krause DW, et al. The Late Cretaceous vertebrate fauna of Madagascar: Implications for Gondwanan paleobiogeography. GSA Today. 1999;9(8):1–7. [Google Scholar]
- 8.Evans SE, Jones MEH, Krause DW. A giant frog with South American affinities from the Late Cretaceous of Madagascar. Proc Natl Acad Sci USA. 2008;105:2951–2956. doi: 10.1073/pnas.0707599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman S, Benstead J. The Natural History of Madagascar. Chicago: Univ of Chicago Press; 2003. [Google Scholar]
- 10.Ali JR, Huber M. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature. 2010;463:653–656. doi: 10.1038/nature08706. [DOI] [PubMed] [Google Scholar]
- 11.Yoder AD, Nowak M. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu Rev Ecol Evol Syst. 2006;37:405–431. [Google Scholar]
- 12.Poux C, et al. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst Biol. 2005;54:719–730. doi: 10.1080/10635150500234534. [DOI] [PubMed] [Google Scholar]
- 13.Vences M, et al. Multiple overseas dispersal in amphibians. Proc Biol Sci. 2003;270:2435–2442. doi: 10.1098/rspb.2003.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raxworthy CJ, Forstner MRJ, Nussbaum RA. Chameleon radiation by oceanic dispersal. Nature. 2002;415:784–787. doi: 10.1038/415784a. [DOI] [PubMed] [Google Scholar]
- 15.Samonds K, et al. Spatial and temporal arrival patterns of Madagascar's vertebrate fauna explained by distance, ocean currents, and ancestor type. Proc Natl Acad Sci USA. 2012;109:5352–5357. doi: 10.1073/pnas.1113993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren BH, et al. Why does the biota of the Madagascar region have such a strong Asiatic flavour? Cladistics. 2010;26:526–538. doi: 10.1111/j.1096-0031.2009.00300.x. [DOI] [PubMed] [Google Scholar]
- 17.Noonan BP, Chippindale PT. Vicariant origin of malagasy reptiles supports late cretaceous antarctic land bridge. Am Nat. 2006;168:730–741. doi: 10.1086/509052. [DOI] [PubMed] [Google Scholar]
- 18.Hedges SB, Kumar S. The Timetree of Life. New York: Oxford Univ Press; 2009. [Google Scholar]
- 19.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 20.Hugall AF, Foster R, Lee MSY. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
- 21.Battistuzzi FU, Filipski A, Hedges SB, Kumar S. Performance of relaxed-clock methods in estimating evolutionary divergence times and their credibility intervals. Mol Biol Evol. 2010;27:1289–1300. doi: 10.1093/molbev/msq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Near TJ, Meylan PA, Shaffer HB. Assessing concordance of fossil calibration points in molecular clock studies: An example using turtles. Am Nat. 2005;165:137–146. doi: 10.1086/427734. [DOI] [PubMed] [Google Scholar]
- 23.Vences M, Wollenberg KC, Vieites DR, Lees DC. Madagascar as a model region of species diversification. Trends Ecol Evol. 2009;24:456–465. doi: 10.1016/j.tree.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 24.McPeek MA, Brown JM. Clade age and not diversification rate explains species richness among animal taxa. Am Nat. 2007;169:E97–E106. doi: 10.1086/512135. [DOI] [PubMed] [Google Scholar]
- 25.Wiens JJ. The causes of species richness patterns across space, time, and clades and the role of “ecological limits”. Q Rev Biol. 2011;86:75–96. doi: 10.1086/659883. [DOI] [PubMed] [Google Scholar]
- 26.Wells NA. In: The Natural History of Madagascar. Goodman SM, Benstead JP, editors. Chicago: Univ of Chicago Press; 2003. pp. 16–34. [Google Scholar]
- 27.Azuma Y, Kumazawa Y, Miya M, Mabuchi K, Nishida M. Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol Biol. 2008;8:215. doi: 10.1186/1471-2148-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sparks JS, Smith WL. Phylogeny and biogeography of the Malagasy and Australasian rainbowfishes (Teleostei:Melanotaenioidei): Gondwanan vicariance and evolution in freshwater. Mol Phylogenet Evol. 2004;33:719–734. doi: 10.1016/j.ympev.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Vences M, Freyhof J, Sonnenberg R, Kosuch J, Veith M. Reconciling fossils and molecules: Cenozoic divergence of cichlid fishes and the biogeography of Madagascar. J Biog. 2001;28:1091–1099. [Google Scholar]
- 30.van der Meijden A, et al. Nuclear gene phylogeny of narrow-mouthed toads (Family: Microhylidae) and a discussion of competing hypotheses concerning their biogeographical origins. Mol Phylogenet Evol. 2007;44:1017–1030. doi: 10.1016/j.ympev.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Ali JR, Aitchison JC. Kerguelen Plateau and the Late Cretaceous southern-continent bioconnection hypothesis: tales from a topographical ocean. J Biogeogr. 2009;36:1778–1784. [Google Scholar]
- 32.Ali JR, Krause DW. Late Cretaceous bioconnections between Indo-Madagascar and Antarctica: Refutation of the Gunnerus ridge causeway hypothesis. J Biogeogr. 2011;38:1855–1872. [Google Scholar]
- 33.MacArthur RH, Wilson EO. The Theory of Island Biogeography. Princeton: Princeton Univ Press; 1967. [Google Scholar]
- 34.Rabosky DL, Glor RE. Equilibrium speciation dynamics in a model adaptive radiation of island lizards. Proc Natl Acad Sci USA. 2010;107:22178–22183. doi: 10.1073/pnas.1007606107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 36.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 37.Nylander JAA. 2008. MrModeltest v2.3. Program distributed by the author. Available from: http://www.abc.se/∼nylander/. Accessed September 10, 2009.
- 38.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 40.Fox J. 2009. car: Companion to Applied Regression. R package version 1.2-16. http://CRAN.R-project.org/package=car. Accessed July 20, 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.