Abstract
Endosymbiotic acquisition of bacteria by a protist, with subsequent evolution of the bacteria into mitochondria and plastids, had a transformative impact on eukaryotic biology. Reconstructing events that created a stable association between endosymbiont and host during the process of organellogenesis—including establishment of regulated protein import into nascent organelles—is difficult because they date back more than 1 billion years. The amoeba Paulinella chromatophora contains nascent photosynthetic organelles of more recent evolutionary origin (∼60 Mya) termed chromatophores (CRs). After the initial endosymbiotic event, the CR genome was reduced to approximately 30% of its presumed original size and more than 30 expressed genes were transferred from the CR to the amoebal nuclear genome. Three transferred genes—psaE, psaK1, and psaK2—encode subunits of photosystem I. Here we report biochemical evidence that PsaE, PsaK1, and PsaK2 are synthesized in the amoeba cytoplasm and traffic into CRs, where they assemble with CR-encoded subunits into photosystem I complexes. Additionally, our data suggest that proteins routed to CRs pass through the Golgi apparatus. Whereas genome reduction and transfer of genes from bacterial to host genome have been reported to occur in other obligate bacterial endosymbioses, this report outlines the import of proteins encoded by such transferred genes into the compartment derived from the bacterial endosymbiont. Our study showcases P. chromatophora as an exceptional model in which to study early events in organellogenesis, and suggests that protein import into bacterial endosymbionts might be a phenomenon much more widespread than currently assumed.
Keywords: endosymbiosis, gene transfer, plastid evolution, photosynthesis
Mitochondria and primary plastids were established more than 1 billion years ago in the eukaryotic kingdom by endosymbiosis from free-living bacteria. This process of organellogenesis, a consequence of the merger of two organisms into a single chimeric organism with new biochemical capabilities, had a transformative impact on eukaryotic evolution. Organellogenesis was accompanied by a massive size reduction of the endosymbiont genome with the transfer of many endosymbiont genes to the host nuclear genome by a process designated as endosymbiotic gene transfer (EGT). Critical for integrating host–endosymbiont metabolic and regulatory networks was the evolution of mechanisms for trafficking nuclear-encoded proteins into the endosymbiont-derived compartment, an evolutionary advance often considered as defining the transition from endosymbiont to genetically integrated organelle. Today, the vast majority of organellar proteins is encoded on the nuclear genome and posttranslationally targeted to mitochondria or plastids by cleavable N-terminal targeting presequences. These presequences are recognized by specialized complexes (translocon of inner and outer mitochondrial or chloroplast membranes, or Tim/Tom or Tic/Toc, respectively) that facilitate translocation of the protein across the double envelope membranes of the organelle (1). Presequences that localize cytoplasmically synthesized proteins to plastids are called transit peptides (TPs).
A central question crucial for understanding organellogenesis concerns the evolution of the path for importing proteins into nascent organelles. The recent discovery of a Tic/Toc-independent targeting mechanism for primary plastids that depends on protein trafficking through endoplasmic reticulum (ER) and Golgi, led to the idea that organelle-targeted proteins were initially imported through the endomembrane system and that this system was gradually replaced by translocon-dependent import (2–4). Other authors propose that ER/Golgi-mediated targeting represents a derived evolutionary feature, and that the translocon complexes evolved gradually from simpler transporters that already existed on bacterial genomes (5). As the acquisition of organelles from free-living bacteria has remained an extremely rare process, it has been difficult to develop an experimental approach to test these hypotheses. Therefore, it is of great biological interest to identify organisms in which organelle establishment is at an earlier stage; such organisms might provide a window through which we can begin to decipher the series of events critical for plastid evolution.
Recently, the cercozoan amoeba Paulinella chromatophora was shown to have an organelle in an early stage of evolutionary development. P. chromatophora harbors two photosynthetic compartments of cyanobacterial origin, termed chromatophores (CRs), that are delineated from the “host cell” cytoplasm by a double envelope membrane and peptidoglycan layer. Recent evidence suggests that CRs evolved independently of plastids (6–9) and experienced a major genome size reduction [∼3.0 to ∼1.0 Mb (10, 11)], and that more than 30 expressed nuclear genes are the result of EGT from the CR (11–13). Interestingly, most of the transferred genes encode small proteins predicted to have a function associated with photosynthesis and light-acclimation. In particular, the nuclear genes psaE, psaK1, and psaK2 encode low molecular mass subunits of photosystem I (PSI), which is one of the two reaction centers critical for oxygenic photosynthesis. The other nine PSI subunits are encoded on the CR genome.
As a number of proteins encoded by EGT genes of P. chromatophora likely function in thylakoid membranes, we speculated that a subset of proteins synthesized in the P. chromatophora cytoplasm would be imported into the CR (13). In this study, we show that the nuclear-encoded PsaE, PsaK1, and PsaK2 polypeptides are synthesized in the host cell cytoplasm, imported into the CR, and associate with CR-encoded PSI subunits in functional PSI complexes. Additionally, our analyses suggest that the Golgi apparatus might function as an intermediate in trafficking of cytoplasmically synthesized proteins into CRs.
Results
Nuclear-Encoded PsaE Protein Is Localized to CRs.
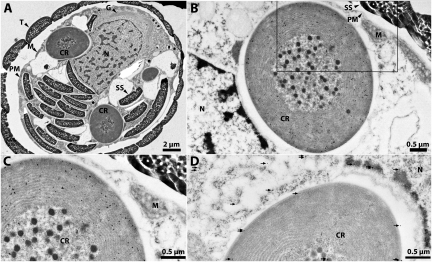
To determine if the nuclear-encoded PSI subunit PsaE is synthesized in P. chromatophora and if the mature protein localizes to the CR, we generated peptide antibodies against P. chromatophora PsaE and established its subcellular localization by using Immunogold EM on thin-sectioned cells. EM images of P. chromatophora cells are shown in Fig. 1, with a transect through the cell in Fig. 1A. The cell is delineated by a cell wall or theca composed of silica scales (SSs) and a plasma membrane adjacent to the wall. Visible in the cell interior are CRs, and within CRs are carboxysomes (hexagonal bodies in the center of CR) that are surrounded by thylakoid membranes. Also apparent in this image is the centrally localized nucleus, cytoplasmically localized SSs used for construction of cell walls of daughter cells upon cell division, mitochondria, and the Golgi apparatus. Fig. 1 B and C show details of an EM image of a P. chromatophora cell Immunogold-labeled with affinity-purified α-PsaEpepC, which shows avid and specific binding to PsaE in Western blots (Fig. S1). Most of the gold particles are located over the CR, with very few particles over the cytoplasm and nucleus. Within the CR, thylakoid membranes are densely decorated with gold particles, whereas very few localize over the carboxysomes. When preimmune serum is used as a control, gold particles appear randomly distributed over cells (Fig. 1D, and as detailed later). These results demonstrate that, in P. chromatophora, nuclear-encoded PsaE protein becomes localized to the CR, where it is likely associated with thylakoid membranes.
Fig. 1.
Immunogold EM of sectioned P. chromatophora cells. (A) Cell cross-section. (B) Detailed cross-section of CR labeled with α-PsaEpepC and α-rabbit-IgG-15 nm gold. (C) Close-up of area highlighted by the rectangle in B. Note dense decoration of thylakoids with gold particles (crisp black dots). (D) Cross-sectioned CR labeled with preimmune serum and α-rabbit-IgG-15 nm gold. Black arrows highlight gold particles. M, mitochondria; N, nucleus; PM, plasma membrane; SS, silica scales; T, theca.
PsaE, PsaK1, and PsaK2 Associate with PSI.
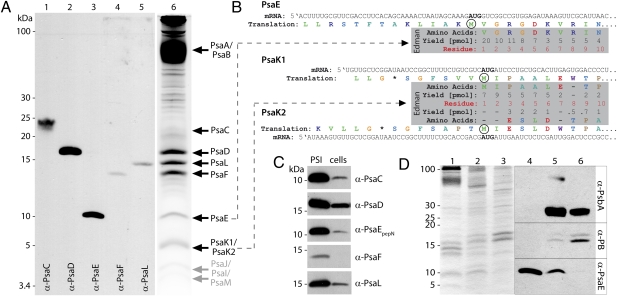
To determine whether PsaE (and the other nuclear-encoded PSI subunits, PsaK1 and PsaK2) in the CRs is associated with PSI (i.e., site of function), trimeric PSI was isolated from P. chromatophora cells and its various subunits identified. PSI subunits were resolved by SDS/PAGE, and the identity of many of the subunits was determined by immunoblot analysis using monospecific, polyclonal antibodies raised to cyanobacterial PSI subunits (α-PsaC, α-PsaD, α-PsaF, α-PsaL) and α-PsaEpepN raised to P. chromatophora PsaE (Fig. 2A). PsaC formed a sharp band that colocalized with PsaE in 16% Schägger gels without urea (Fig. 2C). To resolve PsaE and PsaC, 7 M urea was added to the resolving gel and the polyacrylamide concentration was increased to 18%; these modifications caused PsaC to migrate much more slowly than PsaE, and as a more diffuse, poorly staining band (Fig. 2A). PsaE identity was confirmed by N-terminal sequencing (Fig. 2B). Other PSI polypeptides, for which no specific antibodies were available, were identified by MS (PsaA/PsaB; Table S1) or N-terminal sequencing (PsaK1/PsaK2). The band at approximately 5 kDa contained predominantly PsaK1 with a lower amount of PsaK2, for which not all amino acids were recovered (Fig. 2B). N-terminal sequences defined for PsaE, PsaK1, and PsaK2 demonstrated that the mature polypeptides had the translation start site predicted from cDNA sequences, although the initiator methionine was removed from PsaE (Fig. 2B). Additional low molecular mass polypeptides (<5 kDa), likely PsaJ, PsaI, and PsaM, were observed when highly concentrated protein samples were analyzed. Furthermore, several minor polypeptides (between 30 and >100 kDa) of unknown identity were consistently observed in isolated PSI complexes (Fig. 2A, lane 6); whether they are associated with PSI or just copurify with the complex is not known.
Fig. 2.
Characterization of PSI isolated from P. chromatophora. (A) Fifteen micrograms of protein of PSI was resolved by SDS/PAGE on an 18% Schägger gel containing 7 M urea. Polypeptides were transferred to PVDF membranes for Western blot analysis using antibodies to various PSI subunits as indicated (lanes 1–5). Additionally, 140 μg protein of isolated PSI was resolved in the same gel system and stained with Sypro Ruby (lane 6); individual polypeptides were excised from the gels for MS analysis (PsaA/PsaB band) or transferred to PVDF membranes for N-terminal sequencing by Edman degradation (PsaE and PsaK1/PsaK2 band). (B) Edman degradation. Sequences shown represent the beginning of the full-length mRNA and their deduced translation products, with predicted initiator methionines encircled and stop codons designated by asterisks. Within the gray box are identities and quantities of the first 10 N-terminal amino acid residues, determined by Edman degradation. (C) Equal concentrations of protein (15 μg per lane) from isolated PSI complexes (left lanes) and from whole-cell extracts (right lanes) were resolved by SDS/PAGE, transferred to PVDF membranes, and subjected to immunoblot analyses using α-PsaC, α-PsaD, α-PsaEpepN, α-PsaF, and α-PsaL. (D) Equal amounts of protein (15 μg per lane) from PSI complexes (lanes 1 and 4), thylakoid membranes (lanes 2 and 5), and whole-cell extracts (lanes 3 and 6) were resolved by SDS/PAGE, stained by CBB (lanes 1–3), or transferred to PVDF membranes and subjected to immunoblot analysis by using α-PsbA, α-PB, and α-PsaEpepN. (lanes 4–6).
To determine if PsaE, PsaK1, and PsaK2 are specifically associated with PSI, we had to determine the purity of the isolated PSI complex. To this end, the abundance of photosystem II (PSII) polypeptides and phycobiliproteins in isolated PSI complexes were assessed by immunoblot analysis by using α-PsbA as a PSII marker and α-PB as a phycobilisome marker (Fig. 2D). Proteins of PSII and light-harvesting phycobilisomes were strongly depleted in isolated PSI complexes. In the thylakoid fraction PSII proteins appear enriched, whereas the soluble phycobilisomes are depleted, as expected. Furthermore, abundances of individual PSI polypeptides within the isolated PSI complex were highly enriched relative to their abundance in total protein extracts (Fig. 2C). Together, these results demonstrate that the isolated PSI complex is relatively pure, and notably, that the nuclear-encoded PsaE, PsaK1, and PsaK2 polypeptides are associated with this complex.
Nuclear-Encoded PSI Subunits Are Synthesized on 80S Ribosomes.
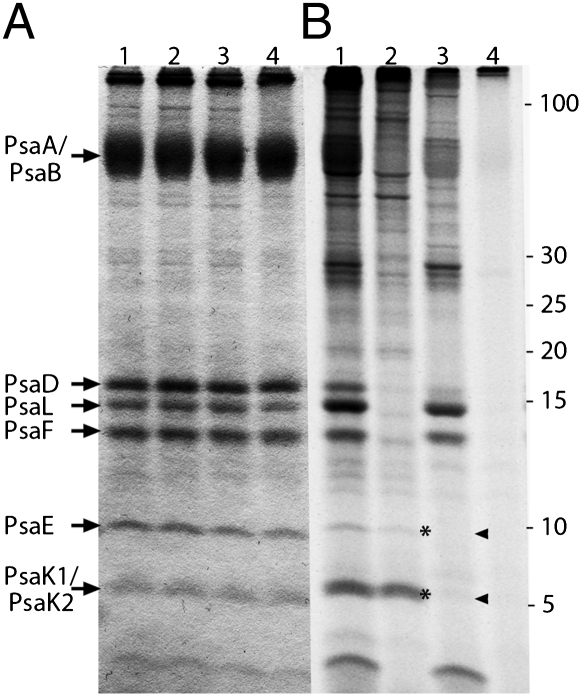
To establish the site of translation of the nuclear-encoded PsaE, PsaK1, and PsaK2 polypeptides, P. chromatophora proteins were radiolabeled with NaH14CO3 in the absence of translational inhibitors or in the presence of chloramphenicol (which inhibits translation on 70S CR ribosomes; Fig. S2), cycloheximide (which inhibits translation on 80S cytoplasmic ribosomes; Fig. S2), or both. Labeling of polypeptides with 35SO42− was precluded because PsaE has a single sulfur-containing amino acid (initiator methionine) that is cleaved from the nascent polypeptide, as revealed by N-terminal sequencing (Fig. 2B). As shown in Fig. 3, all components of PSI incorporate 14C when no inhibitor of translation is added (Fig. 3B, lane 1). The simultaneous addition of both inhibitors blocks incorporation of nearly all radioactivity into protein (Fig. 3B, lane 4). However, if cytoplasmic translation is blocked selectively by cycloheximide, incorporation of label into PsaE and PsaK1/PsaK2 polypeptides is completely abolished (Fig. 3, arrowheads), with some other polypeptides (e.g., PsaD, PsaA/PsaB) having somewhat less incorporation relative to the no-inhibitor control (Fig. 3B, lane 1 vs. lane 3). If translation in CRs is blocked selectively by chloramphenicol, PsaE and PsaK1/PsaK2 still become labeled (Fig. 3, asterisks), whereas accumulation of label in the other PSI polypeptides (PsaA/PsaB, PsaD, PsaL, and PsaF) is markedly reduced (Fig. 3B, lane 2). The experiment was repeated twice with little variation in the results.
Fig. 3.
Translation of PsaE, PsaK1, and PsaK2 on 80S ribosomes. Equal amounts (45 μg protein per lane) of P. chromatophora PSI labeled in vivo with NaH14CO3 without translation inhibitors (lane 1, A and B), in the presence of chloramphenicol (lane 2, A and B), cycloheximide (lane 3, A and B), or both (lane 4, A and B) were resolved by SDS/PAGE on an 18% polyacrylamide, 7 M urea Schägger gel. Resolved polypeptides were stained with CBB (A) and the 14C signal visualized using a PhosphorImager (B). Asterisks and arrowheads highlight presence or absence (respectively) of radiolabeled PsaE and PsaK1/PsaK2.
PsaE Is Likely Routed into CRs via Secretory Pathway.
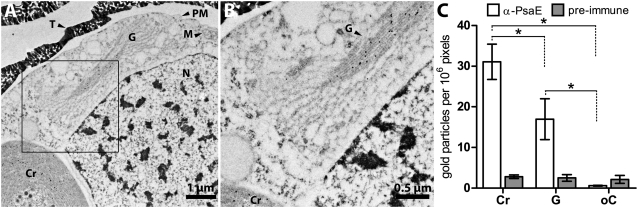
Interestingly, Immunogold studies with affinity-purified α-PsaEpepC showed specific labeling not only over CRs but also over the Golgi (labeled G in Fig. 4 A and B). No accumulation of gold particles over the Golgi was observed when preimmune serum was used in the procedure (Fig. 4C). These findings suggest that PsaE is transported into CRs via the secretory pathway. We did attempt to determine whether Brefeldin A, a Golgi-disrupting agent, impacts trafficking of the nuclear-encoded PSI polypeptides. Although this reagent at a dosage of 1 μg mL−1 3 h−1 strongly inhibited translation, as revealed by decreased incorporation of radioactive label into protein, there was no clear effect on Golgi morphology (as observed by EM; Fig. S3).
Fig. 4.
Immunogold EM of various compartments of P. chromatophora cells. (A) Labeling with α-PsaEpepC and α-rabbit-IgG-15 nm gold. (B) Close up of Golgi in same section (area highlighted by black rectangle in A). Note the decoration of Golgi with gold particles (crisp black dots). (C) Statistical analysis of gold particle densities over CRs (Cr), Golgi (G), and other cell compartments (oC) in cells Immunogold-labeled with α-PsaEpepC or with preimmune serum. Displayed are mean and SD; n = 6. One-way ANOVA with repeated measures for antibody treatment revealed differences for mean gold particle densities in CR, Golgi, and other cell compartments [F(2,5) = 129.6; P < 0.001]; for preimmune treatment, no significant differences were found [F(2,5) = 1.282; P = 0.32]. (*P < 0.001, Holm–Sidak test.) M, mitochondria; N, nucleus; PM, plasma membrane; T, theca.
Discussion
Protein Import into CRs.
By applying (i) Immunogold EM on thin-sectioned P. chromatophora cells using α-PsaE antibodies, (ii) isolation and analysis of PSI complexes, (iii) and in vivo radiolabeling of proteins in the presence of translational inhibitors, we demonstrated that some nuclear EGT genes encode proteins synthesized in the cytoplasm and then transported into P. chromatophora CRs. When they are in CRs, the imported proteins assemble with proteins synthesized within the organelle into multiprotein complexes. To our knowledge, this is the first report of the integration of bacterial endosymbiont–host genetic and biosynthetic machineries (i.e., EGT combined with import of encoded proteins) outside of that established for mitochondria and plastids.
Previously, we (and others) hypothesized that various host-derived solute transporters were inserted into CR envelope membranes (10, 11), as membrane transport systems encoded on the CR genome are extremely limited and cannot account for the expected metabolite exchange between CR and host cell. Transcriptional/translational control of these solute transporters would allow the host to impact CR growth and performance. Both our recent data (13), and data presented in this study, have revealed that, in addition to host control of metabolite fluxes, the host may exert direct control over CR activity by regulating the abundance of nuclear-encoded proteins involved in physiological processes that occur within CRs.
Nuclear-Encoded PSI Subunits.
PsaE is a stromal subunit of PSI that facilitates the interaction between ferredoxin and PSI; residues R9 and R39 of PsaE of Synechocystis sp. PCC 6803 are directly involved in this interaction (14). In contrast, the function of PsaK, an integral membrane protein of PSI, is not known (15). Our observations imply that PsaE and PsaK1/PsaK2 are functional within CRs. First, all three subunits are assembled into PSI complexes. Second, the functionally important PsaE residues R9 and R39 of PCC6803 are conserved in P. chromatophora.
It was previously shown that there can be some decline in the levels of newly synthesized plastid-encoded polypeptides of a plastid protein complex when an inhibitor is used to specifically block the synthesis of nuclear-encoded subunits of that complex (16, 17). The potential reason for this phenomenon is that, when one subunit of a complex is not synthesized, other subunits may be poorly integrated into the complex (even if they are synthesized), as integration may depend on the interaction of the newly synthesized subunit with a subunit already in the complex. Furthermore, when polypeptides of a complex do not efficiently assemble, specific regulatory mechanisms may limit the synthesis of other subunits of that complex (18). Overall, these findings could explain the observed decrease of newly synthesized PsaA/PsaB and PsaD polypeptides in isolated PSI complexes labeled in the presence of cycloheximide (Fig. 3).
A fascinating question concerns the identification of factors that govern which endosymbiont genes first become stable, functional entities in the genome of the host organism. Sequence analysis of the CR genome and a set of more than 3,500 nuclear expressed sequence tags of a second Paulinella strain, Paulinella FK01, revealed differential gene transfer between the two Paulinella strains (11, 12). Although psaE was transferred to the host genome in both strains, psaK still resides on the CR genome of Paulinella FK01. In contrast, although the psaI gene (encoding another low molecular mass PSI subunit) of Paulinella FK01 was transferred to the nuclear genome, it resides on the CR genome of P. chromatophora CCAC 0185. Furthermore, PsaE and PsaK are both nuclear-encoded in the Viridiplantae whereas they are plastid-encoded in red algae. These findings suggest that there is no pronounced functional pressure to specifically move psaE or psaK genes into the host nucleus. However, we note that only low molecular mass PSI subunits have been transferred to the nucleus, suggesting, as previously noted (13), a bias in P. chromatophora for EGT of small genes. It is unclear if this bias originates from size restrictions imposed by the protein import mechanism or it reflects a decreased probability of large gene sequences exiting the CR and arriving in the nucleus physically intact.
Trafficking Through Golgi.
Movement of proteins from the P. chromatophora cytoplasm into CRs requires that the proteins traverse two membranes, the outer one most likely derived from the host phagocytotic membrane and the inner one homologous to the cyanobacterial inner membrane (10). Although we were unable to use Brefeldin A to study protein trafficking in detail because it did not disrupt Golgi morphology in P. chromatophora, Immunogold labeling with α-PsaEpepC revealed significant accumulation of gold particles over Golgi membranes, raising the possibility that trafficking of PsaE into CRs might involve vesicular transport through the Golgi. The high specificity of the affinity purified α-PsaEpepC observed in Western blots, and the fact that no accumulation of signal over the Golgi was noted with the preimmune serum control, strongly suggest that the increased number of gold particles over Golgi membranes reflects PsaE accumulation in that compartment.
Nuclear-encoded proteins imported into plastids surrounded by three envelope membranes [i.e., complex plastids of euglenophytes and peridinin-containing dinoflagellates that are derived from algae with primary plastids through secondary endosymbioses (19)] were shown to passage through the Golgi (20, 21). Plastid proteins synthesized in the cytoplasm of these algal cells have N-terminal, bipartite presequences composed of an ER-targeting signal [signal peptide (SP)] and a TP-like sequence. The discovery that more than 8% of the Arabidopsis plastid proteome have SPs rather than TPs revealed that a secretory system-dependent pathway for protein import is not restricted to complex plastids (22). Although some of these plant proteins appear to be attached to the outer envelope membrane (23), for others, Golgi-mediated plastid import was biochemically demonstrated (4, 24).
Another intriguing example of protein targeting is represented by the nonobligate endosymbiotic nitrogen-fixing bacterium Rhizobium. Rhizobia within host-derived membrane vesicles in nodules of the legume Medicago truncatula take up peptides that evolved from effectors of the plant's innate immune system (25). These peptides have N-terminal SPs and are imported through the secretory pathway into the bacteria, where they trigger dramatic morphological and physiological changes. Details of the transport mechanism have not been established.
Targeting Signals.
Even though import of cytoplasmically synthesized proteins through the Golgi occurs in many biological systems, it is surprising that the PsaE, PsaK1, and PsaK2 polypeptides (deduced from full-length mRNA sequences) of P. chromatophora do not appear to have SPs, even if noncanonical start codons are considered (discussed in detail in refs. 13, 26; Fig. 2B). In addition, N-terminal sequencing of PsaE, PsaK1, and PsaK2 performed in this study demonstrated that the start of the mature protein was predicted correctly and that no cleavable SP occurs downstream of this start. Thus, the mechanism for targeting PsaE, PsaK1, and PsaK2 does not appear to depend on SPs. However, there are various presequence-independent protein import pathways that depend on internal targeting signals; such signals have been identified for mitochondrial membrane proteins (27). Furthermore, nuclear-encoded proteins without SPs or TPs have been identified in proteomes of primary and complex plastids (22, 28). The mechanism for importing these plastidal proteins is not known. Interestingly, for “tail-anchored” proteins (i.e., proteins anchored to a membrane by a single C-terminal transmembrane domain), SP-independent insertion into the ER has been observed; a posttranslational, partner protein-dependent mechanism of targeting and insertion has recently been described (29).
PsaK1 and PsaK2 have a C-terminal transmembrane domain that might function as an internal cryptic targeting signal. However, PsaE is a soluble protein that associates with PSI subunits and does not have any known sequence that likely functions in targeting. Clearly, additional information is required to elucidate mechanisms involved in targeting proteins to CRs.
Relevance to Other Endosymbiotic Systems.
Numerous obligate bacterial endosymbionts, widespread over the eukaryotic tree of life, provide their host with a range of physiological functions (30, 31). Common to these systems is a reduction in the size of the endosymbiont genome (30) which, in extreme cases, may be as little as 0.16 Mb (32). There is also increasing documentation of the occurrence of EGT in these symbiotic interactions (33), although the import of host-synthesized EGT proteins into resident “endosymbionts” has not been demonstrated. The reduced genomes of many of these endosymbionts do not encode many essential genes, and endosymbiont survival within host tissue appears to depend on metabolite transfer from the host to the endosymbiont. In some cases, the genome reduction has been accompanied by a loss of genes involved in DNA replication, transcription, and translation, i.e., functions required by endosymbionts that are not easily compensated for at the metabolite level (34, 35). These findings make it almost certain that host-encoded proteins are delivered to the endosymbiont, and that this phenomenon is widespread in nature. An interesting question that arises from the observed import of host peptides in rhizobial endosymbionts and from the finding that many host-derived transporters are likely to be inserted into CR envelope membranes, is whether host-originating rather than endosymbiont-originating proteins (encoded by EGT sequences) are the first targeted to the endosymbiont/nascent organelle. A scenario in which host proteins are readily targeted to and function within the endosymbiont would explain the loss of endosymbiont independence in many developing associations, and an evolutionary path in which the host organism can coopt endosymbiont functionalities.
Conclusion
With a growing knowledge of endosymbiotic systems, the border between organelles and endosymbionts has been blurred. The import of host-encoded proteins into many bacterial endosymbionts must occur based on the limited repertoire of genes on endosymbiont genomes. We present evidence that EGT proteins of P. chromatophora are synthesized on cytoplasmic ribosomes and transported into the CR, and assemble into functional complexes. Trafficking does not appear to depend on a cleavable presequence, although our initial data suggest that proteins routed to the CR pass through the Golgi. Our characterizations identify P. chromatophora as a potentially powerful model for empirically elucidating evolutionary aspects of organelle establishment, maintenance, and maturation. A comprehensive identification of imported CR proteins and their functional features and phylogenetic origins, and the establishment of targeting and import pathway(s) for cytoplasmically synthesized proteins will provide new insights into mechanisms that host cells use to “enslave” bacterial symbionts and provide general information concerning the way in which a homeostatic, synergistic association between the biochemical capacities of an endosymbiont and its host cell can develop during the process of organellogenesis.
Materials and Methods
Strain and Growth Conditions.
P. chromatophora strain CCAC 0185 [a subisolate of strain M0880/a (10)] was grown at 17 °C in Fernbach flasks containing 0.75 L Waris-H culture medium (36) supplemented with 1.5 mM Na2SiO3. Cultures were maintained in a light/dark cycle (14/10 h) at an intensity of approximately 10 μmol photon m−2 s−1.
Primary Antibodies.
α-PsaEpepN and α-PsaEpepC, raised against synthetic peptides corresponding to N- and C-terminal sequences of PsaE from P. chromatophora, were purified by (NH4)2SO4 precipitation followed by dead Sephadex column chromatography or affinity-purified. Antibodies against other cyanobacterial PSI subunits were provided by Gaozhong Shen (Pennsylvania State University, University Park, PA) (α-PsaC and α-PsaD) and Wade Johnson (Susquehanna University, Selinsgrove, PA) (α-PsaF and α-PsaL), whereas antibodies against the PSII D1 protein (α-PsbA) were obtained from Agrisera (AS05-084). The α-PB is a mixture of antibodies to phycocyanin and allophycocyanin subunits generated in the Stanford Animal Facility (by A.R.G.). All primary antibodies were polyclonal and raised in rabbits.
Postembedding Immunogold EM.
Ultrathin sections (∼60 nm) of P. chromatophora cells embedded in LR White resin (Polysciences) were Immunogold-labeled with affinity-purified α-PsaEpepC (1:5) or, as a control, preimmune serum (1:500; justification of this dilution is shown in Fig. S4). Primary antibodies were reacted with goat α-rabbit IgGs conjugated with 15-nm gold particles. Samples were contrasted with uranyl acetate and lead citrate (37). Micrographs were taken with a Jeol JEM-1400 transmission electron microscope. For statistical analysis of gold particle distribution, montage images of whole-cell sections were created at a magnification of 5,000× by using SerialEM software (38); images were subdivided into different cell compartments, and gold particles over these compartments were automatically counted by using ImageJ software (http://rsb.info.nih.gov/ij/).
Isolation of PSI.
Trimeric PSI was isolated from P. chromatophora essentially as described for Synechococcus sp. PCC7002 (39). Protein concentration of PSI samples was determined in triplicate by using the BCA assay (BioRad). SDS/PAGE 4× loading buffer [150 mM Tris-HCl, pH 7.0, 12% (wt/vol) SDS, 30% (vol/vol) glycerol, 6% (vol/vol) β-mercaptoethanol, 0.05% Coomassie brilliant blue G-250 (CBB; Serva)] was added to the samples (three volumes of sample to one volume of loading buffer), which were then mixed, snap-frozen in liquid nitrogen, and stored at −80 °C until they were used.
SDS/PAGE and Protein Identification.
Proteins of isolated PSI were solubilized by boiling for 1 min in loading buffer and resolved by SDS/PAGE on 16% polyacrylamide gels or 18% polyacrylamide, 7-M urea gels with a discontinuous Tris-Tricine buffer system [i.e., Schägger system (40)]. For Western blots, proteins were transferred to PVDF membranes and analyzed by using an enhanced chemiluminescence assay (GE Healthcare) after incubation with primary antibodies. For mapping peptide masses, PSI polypeptides resolved by SDS/PAGE on an 18% Schägger gel containing 7 M urea and stained with SYPRO Ruby (BioRad) were excised from gel, and MS was performed at the protein facility of Stanford University. For N-terminal sequencing, PSI polypeptides were resolved in the same way, but with 2 mM mercaptoacetic acid in the cathode buffer to scavenge N-blocking free radicals. Protein bands were blotted onto PVDF membrane, stained with CBB acid-free, destained in 50% methanol/10% acetic acid, and rinsed with dH2O. N-terminal sequences of purified polypeptides were determined by Edman chemistry on a Procise liquid-pulse protein sequenator (Applied Biosystems) at the protein facility of Stanford University.
In Vivo Radiolabeling.
P. chromatophora cells were radiolabeled over a period of 6 h by using 5 μCi/mL NaH14CO3. For inhibition of translation, cells were incubated with chloramphenicol (100 μg/mL) and/or cycloheximide (1 μg/mL) during the labeling period. PSI complexes were isolated as described earlier, the polypeptide subunits resolved by SDS/PAGE, and the label in the polypeptides visualized by using a Typhoon scanner (Amersham Biosciences). More details of study protocols are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Michael Melkonian for both intellectual and financial (antibody generation) support, Jon Mulholland and John Perrino for help with Immunogold EM, and Richard Winant for help with protein identification. This study was supported by Deutsche Forschungsgemeinschaft Grant NO 888/1-1 (to E.C.M.N.) and National Science Foundation Grant MCB-0951094 (to A.R.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.A. is a guest editor invited by the Editorial Board.
See Commentary on page 5142.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118800109/-/DCSupplemental.
References
- 1.Schleiff E, Becker T. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya D, Archibald JM, Weber APM, Reyes-Prieto A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays. 2007;29:1239–1246. doi: 10.1002/bies.20671. [DOI] [PubMed] [Google Scholar]
- 3.Radhamony RN, Theg SM. Evidence for an ER to Golgi to chloroplast protein transport pathway. Trends Cell Biol. 2006;16:385–387. doi: 10.1016/j.tcb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Villarejo A, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- 5.Bodył A, Mackiewicz P, Stiller JW. Early steps in plastid evolution: Current ideas and controversies. Bioessays. 2009;31:1219–1232. doi: 10.1002/bies.200900073. [DOI] [PubMed] [Google Scholar]
- 6.Marin B, Nowack ECM, Glöckner G, Melkonian M. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol Biol. 2007;7:85. doi: 10.1186/1471-2148-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin B, Nowack ECM, Melkonian M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist. 2005;156:425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Yoon HS, et al. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol Biol. 2009;9:11. doi: 10.1186/1471-2148-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon HS, Reyes-Prieto A, Melkonian M, Bhattacharya D. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr Biol. 2006;16:R670–R672. doi: 10.1016/j.cub.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Nowack ECM, Melkonian M, Glöckner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- 11.Reyes-Prieto A, et al. Differential gene retention in plastids of common recent origin. Mol Biol Evol. 2010;27:1530–1537. doi: 10.1093/molbev/msq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama T, Ishida K. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr Biol. 2009;19:R284–R285. doi: 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 13.Nowack ECM, et al. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol Biol Evol. 2011;28:407–422. doi: 10.1093/molbev/msq209. [DOI] [PubMed] [Google Scholar]
- 14.Xu W, Tang H, Wang Y, Chitnis PR. Proteins of the cyanobacterial photosystem I. Biochim Biophys Acta Bioenergetics. 2001;1507:32–40. doi: 10.1016/s0005-2728(01)00208-0. [DOI] [PubMed] [Google Scholar]
- 15.Naithani S, Hou JM, Chitnis PR. Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res. 2000;63:225–236. doi: 10.1023/A:1006463932538. [DOI] [PubMed] [Google Scholar]
- 16.Chua NH, Gillham NW. The sites of synthesis of the principal thylakoid membrane polypeptides in Chlamydomonas reinhardtii. J Cell Biol. 1977;74:441–452. doi: 10.1083/jcb.74.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egelhoff T, Grossman A. Cytoplasmic and chloroplast synthesis of phycobilisome polypeptides. Proc Natl Acad Sci USA. 1983;80:3339–3343. doi: 10.1073/pnas.80.11.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choquet Y, Wollman FA. Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 2002;529:39–42. doi: 10.1016/s0014-5793(02)03260-x. [DOI] [PubMed] [Google Scholar]
- 19.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 20.Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- 21.Sulli C, Fang Z, Muchhal U, Schwartzbach SD. Topology of Euglena chloroplast protein precursors within endoplasmic reticulum to Golgi to chloroplast transport vesicles. J Biol Chem. 1999;274:457–463. doi: 10.1074/jbc.274.1.457. [DOI] [PubMed] [Google Scholar]
- 22.Kleffmann T, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 23.Armbruster U, et al. Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol Plant. 2009;2:1325–1335. doi: 10.1093/mp/ssp082. [DOI] [PubMed] [Google Scholar]
- 24.Nanjo Y, et al. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell. 2006;18:2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 26.Mackiewicz P, Bodył A, Gagat P. Possible import routes of proteins into the cyanobacterial endosymbionts/plastids of Paulinella chromatophora. Theory Biosci. 2011 doi: 10.1007/s12064-011-0147-7. 10.1007/s12064-011-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal S, Striepen B. More membranes, more proteins: Complex protein import mechanisms into secondary plastids. Protist. 2010;161:672–687. doi: 10.1016/j.protis.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moya A, Peretó J, Gil R, Latorre A. Learning how to live together: Genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 31.Nowack ECM, Melkonian M. Endosymbiotic associations within protists. Philos Trans R Soc Lond B Biol Sci. 2010;365:699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 33.Dunning Hotopp JC. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011;27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamames J, et al. The frontier between cell and organelle: Genome analysis of Candidatus Carsonella ruddii. BMC Evol Biol. 2007;7:7. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McFadden GI, Melkonian M. Use of hepes buffer for microalgal culture media and fixation for electron-microscopy. Phycologia. 1986;25:551–557. [Google Scholar]
- 37.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Shen G, et al. Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J Biol Chem. 2002;277:20343–20354. doi: 10.1074/jbc.M201103200. [DOI] [PubMed] [Google Scholar]
- 40.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.