Abstract
Plasmids are self-replicating genetic elements capable of mobilization between different hosts. Plasmids often serve as mediators of lateral gene transfer, a process considered to be a strong and sculpting evolutionary force in microbial environments. Our aim was to characterize the overall plasmid population in the environment of the bovine rumen, which houses a complex and dense microbiota that holds enormous significance for humans. We developed a procedure for the isolation of total rumen plasmid DNA, termed rumen plasmidome, and subjected it to deep sequencing using the Illumina paired-end protocol and analysis using public and custom-made bioinformatics tools. A large number of plasmidome contigs aligned with plasmids of rumen bacteria isolated from different locations and at various time points, suggesting that not only the bacterial taxa, but also their plasmids, are defined by the ecological niche. The bacterial phylum distribution of the plasmidome was different from that of the rumen bacterial taxa. Nevertheless, both shared a dominance of the phyla Firmicutes, Bacteroidetes, and Proteobacteria. Evidently, the rumen plasmidome is of a highly mosaic nature that can cross phyla. Interestingly, when we compared the functional profile of the rumen plasmidome to two plasmid databases and two recently published rumen metagenomes, it became apparent that the rumen plasmidome codes for functions, which are enriched in the rumen ecological niche and could confer advantages to their hosts, suggesting that the functional profiles of mobile genetic elements are associated with their environment, as has been previously implied for viruses.
Keywords: metagenomics, microbial ecology, rumen microbiology
Wide analysis of prokaryotic genomes has revealed that lateral gene transfer (LGT) greatly accelerates the rate of novel gene introduction into these genomes (1). Hence, LGT significantly contributes to prokaryotic genome novelty and evolution (2). The driving force for LGT is mobile genetic elements that serve as vehicles in the communal gene pool (3). Plasmids, as mobile genetic elements, are self-replicating, extrachromosomal genetic elements that operate as “gene ferries” (4). Plasmids have been considered key vectors of genetic exchange between bacterial chromosomes, both recently and in the past (5). Furthermore, plasmids are thought to have major ecological functions because they are found with high abundance in bacterial populations of many habitats (6). Plasmids usually carry conserved backbone functions of DNA replication and mobilization, important for maintenance within their host and transfer among hosts. In addition, plasmids carry a variable assortment of accessory genes, which make them an important evolutionary force, as these functions often contribute to their host's phenotypic diversity. Plasmids isolated thus far from different ecological niches encode a versatile array of accessory functions, ranging from antibiotic resistance to nitrogen fixation. These functions may confer an advantage to their host in its ecological niche, making the burden of carrying the plasmid worthwhile (3, 6–10). Therefore, accessory functions carried by plasmids in a specific ecological niche may be enriched by requirements that are relevant for that niche. Because of their important role in shaping microbial habitats, understanding and characterizing plasmids is of high importance to understanding microbial environments. To date, several studies have attempted to look at uncultured plasmid communities, overcoming host dependency, albeit allowing the isolation of only a small number of plasmids or resulting with chromosomal DNA contamination (11–13).
The reticulorumen environment, which is part of the ruminant digestive system, is a highly intriguing environmental niche that accommodates complex and important microbial populations responsible for the ruminants’ remarkable ability to convert indigestible plant mass into food products (14). The high microbial density in the rumen (1010–1011/mL) favors LGT among rumen microorganisms (15). Therefore, understanding the reticulorumen microbial niche is tightly linked to an understanding of LGT and the plasmids residing within this niche.
There have been several reports on the isolation of plasmids from rumen bacterial isolates and the transfer of genes between rumen microbes (16–24). Plasmids isolated from rumen bacteria have been characterized as having replication and mobilization functions, and some contain accessory genes (25–28). As most reticulorumen microorganisms cannot be cultured, the ability to study their resident plasmids is limited by the constraints of traditional microbiology methods. A metagenomic approach, enabling the sequencing and study of rumen plasmids as a whole, might offer a solution to this problem. Nevertheless, to date, no broad metagenomic study has been performed on rumen plasmids, possibly because of challenging limitations, such as difficulty distinguishing them from chromosomal DNA, their low copy number, and very small fraction of the microbial DNA content.
In this study, we characterize the rumen plasmidome (overall rumen plasmid population) for its potential hosts, functions, and mosaic nature using a deep-sequencing approach. We developed a plasmid-extraction procedure that overcomes the aforementioned challenges, and extracted plasmid DNA from 16 cows. The rumen plasmid DNA was deep-sequenced via an Illumina paired-end protocol and analyzed using a public and custom-developed bioinformatics pipeline. This pipeline, named Blast Report Plasmid Aggregator (BRPA), is capable of classifying and sorting the different generated contigs according to the localization of their annotated functions. We present a characterization of the rumen plasmidome for the mobility of its genes, potential hosts, and coding functions.
Results and Discussion
Rumen Sampling, Extraction, and Sequencing of Plasmid DNA.
Traditionally, the study of plasmids and mobile genetic elements from the environment has been restricted to culture-dependent methods or molecular PCR-based screens of known plasmid sequences. Here we sought to overcome the culture dependency by using a recently standardized procedure that uses the ability of an exonuclease (plasmid-safe DNase) to digest chromosomal DNA. The chromosomal DNA is sheared during the extraction procedure and its linearized form is then exposed to exonuclease digestion, leaving the plasmid DNA intact. The circular plasmid DNA is then selectively amplified. During this procedure, contamination with chromosomal DNA is monitored by PCR of the 16S rRNA gene. We used this procedure to amplify the total rumen plasmidome. To achieve maximum plasmidome coverage, 16 animals were sampled for their rumen fluid and total rumen microbial populations were isolated from these samples, as previously described (29). To purify the most divergent plasmid DNA from rumen bacteria, we used three different plasmid-purification methods that vary in their lysis abilities and are applicable to both Gram-negative and Gram-positive bacteria (30–32), with minor adjustments. The purified DNA of the 16 different samples was pooled together, in equal ratios (Fig. S1B), and subjected to digestion by plasmid-safe DNase. The purified plasmids were amplified with phi29 DNA polymerase (Fig. S1C), previously used for viral DNA amplification (33, 34). This process was carried out in parallel with an Escherichia coli plasmid (8 kb) as a positive control. The phi29 amplification product was digested with a unique enzyme to give its designative length (Fig. S1A). After verifying that our amplified DNA samples were free of genomic DNA contamination (Fig. S1D), they were subjected to deep sequencing via the Illumina paired-end protocol to enhance the de novo assembly process.
Roughly 34 million reads were generated and subjected to de novo assembly, using two assembly programs: Velvet (35) and SOAP de novo (SDN) (36). Whereas Velvet yielded a higher number of contigs, SDN yielded a higher mean contig length (the two assembly outputs are summarized in Table S1). Furthermore, when aligning the two sets of contigs, almost all of the Velvet contigs aligned with SDN contigs (98%), whereas only 90% of the SDN contigs aligned with the Velvet contigs. Given the longer contig lengths, we selected the contig set generated by SDN for further analyses. Overall, the SDN contig set included 5,771 contigs with a mean contig length of 469 bp and GC content of 47.4%. This relatively short contig length may suggest that the rumen plasmidome has extensive diversity, and that even 34 million reads do not cover a significant part of it.
Phylogenetic Analysis of the Rumen Plasmidome.
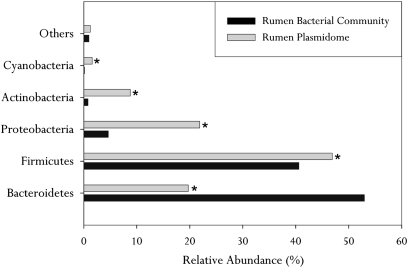
We investigated the potential hosts of the plasmid-originated contigs using their phylogenetic assignment by the SEED database (using a maximum E-value of ≤10−5). Most of the contigs were assigned to the domain Bacteria, with minor representation of the Archaea and Eukarya (Fig. S2). We selected the Bacteria domain for further analysis. The distribution of the dominant bacterial phyla within the rumen plasmidome was: Firmicutes (47%), Bacteroidetes (22%), Proteobacteria (20%), and Actinobacteria (9%) (Fig. 1). This distribution was significantly different from the phylum distribution of all plasmids available at the Integrated Microbial Genome (IMG) and ACLAME (A Classification of Mobile Genetic Elements) databases: notably, Bacteroidetes and Firmicutes were more abundant in the rumen plasmidome and the phylum Proteobacteria was more abundant in the plasmid databases (Fig. S3). Next, we sought to evaluate the phylogenetic distribution of the rumen plasmidome compared with the rumen microbiome. We analyzed metagenomic DNA extracted from the same rumen samples using 16S rRNA gene amplicon pyrosequencing. The Quantitative Insights Into Microbial Ecology (QIIME) pipeline was used to analyze the 172,000 quality-filtered reads generated, revealing a significantly different phylum distribution of Firmicutes (44%), Bacteroidetes (50%), and Proteobacteria (5%) (Fig. 1). This difference was also apparent at lower taxonomical levels (Fig. S4).
Fig. 1.
Phylogenetic analysis of the rumen plasmidome. The relative abundance of each bacterial phylum (established for the same 16 cows) is shown for the bovine rumen plasmidome, as revealed by MG-RAST analysis using similarity to the SEED database with a maximum E-value of ≤10−5, and for the bacterial populations in the same samples as revealed by amplicon pyrosequencing of the V2 and V3 regions of 16S rRNA gene sequences. All phyla that were less abundant than 1% in both datasets are included in the “Others” category. Differences in the relative abundance of each of the phyla were measured using χ2 statistics. *P < 0.05.
Functional Analysis.
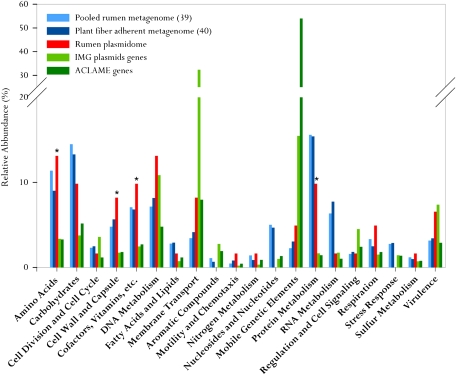
Using a maximum E-value of ≤10−5, the functional SEED assignment of the rumen plasmidome revealed the most highly represented subsystems to be “Amino Acids” and “DNA Metabolism.” The next most represented subsystems were “Cofactors, Vitamins, etc.,” “Carbohydrates,” and “Protein Metabolism.” These subsystems were more abundant than “Virulence,” which is considered to be highly represented in plasmids. High representation was also observed for the “Cell Wall and Capsule” subsystem (Fig. 2). We further analyzed the functional distribution of the rumen plasmidome in comparison with known plasmid databases (37, 38) and previously published rumen metagenomes (39, 40), to determine whether the rumen plasmidome harbors functions relevant to the rumen ecological niche. We assumed that features specific to the rumen niche would be abundant in the rumen metagenomes and plasmidome relative to the plasmid databases. Indeed, the functional distribution of the rumen plasmidome shared some similarities with the rumen metagenomes and exhibited a significantly higher (P < 0.05) proportion of representation of certain functional subsystems compared with the plasmid databases (Table S2). These subsystems included “Cofactors, Vitamins, etc.,” “Cell Wall and Capsule,” “Carbohydrates,” “Respiration,” “Amino Acids,” and “Protein Metabolism.” Overall, the rumen plasmidome contains a unique distribution of functions that does not match the plasmids, which have been isolated and sequenced to date. This distribution profile represents a mixture of plasmid-associated functions and functions unique to the rumen ecological niche.
Fig. 2.
Comparative functional analysis of the rumen plasmidome vs. rumen metagenomes and plasmid databases. The plasmid databases (IMG and ACLAME) and the rumen plasmidome and metagenomes (39, 40) were compared with the SEED database for relative abundance of each of the major SEED subsystems by using a maximum E-value of ≤10−5. SEED subsystems that were less abundant than 1% in all five datasets analyzed are not shown. Differences in the abundances of plasmidome-enriched SEED subsystems compared with the plasmid databases were tested using χ2 statistics. *P < 0.05.
Proximity of Plasmidome Functions on the Contigs.
As the SEED functional assignment does not refer to the arrangement of functions within each contig, we devised a bioinformatics pipeline that enables the analysis of functions carried by each of the contigs and their proximity to each other. This pipeline, termed BRPA, screens generated BLAST reports and maps the assigned functions according to their coordination within the contigs. Next, using sets of search strings and rules, BRPA aggregates contigs according to their assignment (a general scheme of the pipeline is given in Fig. S5). A similar bioinformatics approach was recently used to study metagenomics of microbial traits within oceanic viral communities (41).
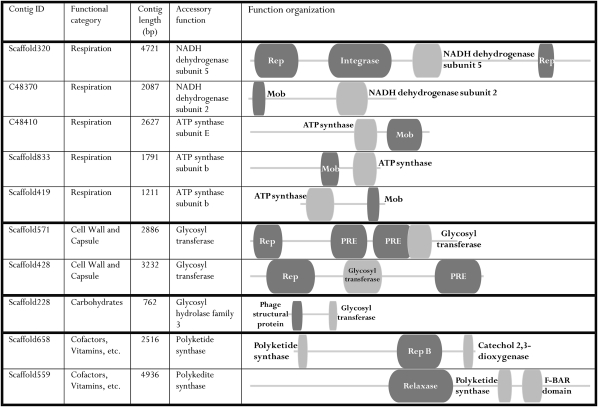
Briefly, we compared our data, using a maximum E-value of ≤10−3, to the National Center for Biotechnology Information-protein nonredundant (NCBI-NR), NCBI-nucleotide (NT), Conserved Domain Database (CDD), ACLAME, and IMG databases (only genes of ACLAME and IMG plasmids were used). In this step, 40% of the contigs had at least one hit against one of the databases, leaving roughly 60% of the data unannotated. This finding is consistent with the findings in virome studies that used the same E-value cutoff where the percentage of annotated contigs varied between 1 and 30 (42–44). The proportion of contigs annotated by the different databases is listed in Table S3. Manual inspection of the annotated contigs revealed the presence of plasmids previously extracted from rumen bacterial isolates (a list of these plasmids is given in Tables S4 and S5). We used BRPA to explore the datasets for contigs that carry a plasmid backbone function in proximity to accessory rumen-associated functions, as revealed by the rumen functional analysis. The rumen-enriched subsystems and plasmid origin of each contig were determined using specific search strings (Materials and Methods). In this classification, 86% of the annotated contig dataset received a plasmid assignment, verifying their plasmid nature. Using this approach, we were also able to locate contigs carrying functions related to these rumen-enriched subsystems carried in proximity to the plasmid backbone genes (Fig. 3).
Fig. 3.
Selected contigs carrying plasmid elements together with rumen-enriched functions. The plasmidome contigs were compared with five different databases with a maximum E-value of ≤ 10−3. The BLAST reports were combined using BRPA, which mapped the hits to each of the contigs. Contigs carrying plasmid backbone functions (shown in dark gray) with selected accessory functions (shown in light gray) of the rumen-enriched SEED subsystems (Materials and Methods) are shown. The BRPA best-hit selection process allows for overlapping hits (illustrated by overlapping boxes) to coexist only if the reading frames or strands are not shared among the two. The lengths of the gray line and boxes represent their relative proportions. PRE, plasmid recombination enzyme.
Mosaic Nature of the Rumen Plasmidome.
To assess the extent of mosaicism of the rumen plasmidome, we determined the phylogenetic association of ORFs on contigs carrying more than one ORF. Their analysis was carried by the Phylogenie pipeline (45) for automated phylome generation and analysis implemented with the maximum likelihood RXaML program (46). We could confidently determine the mosaic nature of 84 of the 220 contigs that carried at list two ORFs with annotations in the NCBI-NR. This proportion was because of unclear determination of the phylogeny association by the Phylogenie pipeline (Materials and Methods). This analysis revealed 74% of the contigs to be mosaic within the phyla level and 14.3% exhibited cross-phylum mosaicism (Table S6).
Discussion
In this study, we were able to distinguish and amplify a large number of high-quality, pure rumen plasmid DNA using exonuclease treatment, phi29 DNA polymerase amplification and genomic DNA monitoring through 16S rRNA PCR. We used this procedure to extract plasmid DNA from the rumen of 16 animals, thus allowing for high plasmid diversity because of the possibility that different plasmids reside in different individuals. We also used three different plasmid-purification methods, each with a different lysis procedure, to allow for maximum diversity and quantity of plasmid DNA.
Most of the hosts of the rumen plasmidome could be attributed to three bacterial phyla (Firmicutes 47%, Bacteroidetes 20%, and Proteobacteria 22%). However, when comparing our data to that generated for the same cows using 16S amplicon pyrosequencing, the proportions of the different phyla varied significantly. These differences in proportions could be explained by variations in plasmid-bearing bacteria from each of the phyla: it is possible that in the rumen, phyla such as Proteobacteria and Actinobacteria host more plasmids than Bacteroidetes. Another possibility is database bias, supported by the scarce representation of Bacteroidetes and the high representation of Proteobacteria in the plasmid databases (Fig. S3), or technical bias created by the plasmid-extraction methods. The BRPA computational pipeline designed for this study provided the ability to focus on the arrangement of the annotated functions within the plasmid-originated contigs: over 86% of our annotated contigs were determined to be of plasmid origin, reinforcing the validity of the plasmid purification and amplification method. Interestingly, 338 of the plasmid-encoded contigs aligned with rumen plasmids previously isolated from different locations and at different time points, whereas rumen plasmidome alignment with other datasets of plasmids isolated from terrestrial and aquatic environments was significantly lower (P < 0.05) (Tables S4 and S5). This finding suggests that not only are bacterial taxa selected by the ecological niche, but the plasmids that they carry are as well. Furthermore, this finding implies that the rumen plasmidome is a recurring outcome of natural selection and evolution with a certain degree of heredity: the rumen plasmidome occurs again and again, with variations, but also remarkable regularities in its genetic content. Inheritance of the rumen plasmidome between animals is likely carried out by a set of hosting bacteria, although the exact source of the rumen plasmidome and what reconstitutes it within each cow remain elusive. Nevertheless, previous studies have speculated that the origin of rumen bacteria stems from contact between the newborn and its mother (47). The remaining contigs, not classified as “plasmid” by BRPA, might still be of plasmid origin but might lack the plasmid features recognized by BRPA.
Whereas most studies examining the identity and extent of laterally transferred genes focus on identifying the tracks of such events in bacterial genomes, this study presents the opportunity to examine the identity of such genes and the phylogenetic barriers they cross on the transfer “vehicles.” By assigning the phylogenetic association of ORFs residing in the same contig using the Phylogenie pipeline, we determined that 74% of the examined contigs are mosaic within the phylum level, which is consistent with a recent report of LGT frequency at different phylogenetic levels (48). Contigs exhibiting cross-phyla mosaicism amounted to 14.3%, possibly implying that in a dense microbial habitat, such as the rumen, gene mobility across phyla commonly occurs (Tables S6 and S7).This theory was also suggested in a recent study where gene-sharing events among species from different taxa were observed to be more frequent in the same habitat (49), which could provide an additional explanation for the differences in the proportion of bacterial phyla in the rumen revealed by the 16S rRNA gene versus the plasmid contigs, as it suggests that some plasmids might be carried by very different host phyla and therefore have more than one phylum designation.
Evidently, the rumen plasmidome encodes a wide array of gene functions (Fig. 2), strongly supporting the notion that most bacterial genes are capable of LGT (50). For functions to be fixated on plasmids they ought to confer their host with an advantage in the ecological niche. Interestingly, when we compared the functional profile of the rumen plasmidome to those of the rumen metagenomes and plasmid databases, it became apparent that except for the intrinsic plasmid-coding functions, there are functions that are significantly enriched in the rumen plasmidome compared with the plasmid databases (P < 0.05). Using BRPA, we mapped some of these rumen-enriched functions in proximity to plasmid backbone functions, such as glycosyltransferase of the “Cell Wall and Capsule” subsystem (Fig. 3). This enzyme and other members of this subsystem have been recently reported to be laterally transferred in the human gut from bacteria into archaeal genomes (51). It was speculated that these genes give their host an advantage in the gut niche, enabling it to vary surface structures, especially capsular polysaccharides, in vivo. Other enriched subsystems included “Amino Acids” and “Protein Metabolism,” which were predominantly constituted of biosynthetic pathway functions, which may grant their hosts an advantage in the rumen, as recently suggested (51). On the other hand, the “Carbohydrates” subsystem was composed mainly of sugars using enzymes that might give their host an advantage in the carbohydrate-rich rumen environment. Overall, these findings support the notion that the mobile genetic elements show distinct profiles that are associated with their environment, something that has been previously shown for viruses and phages (41, 52, 53). Furthermore, by isolating and examining the plasmid population of the rumen environmental niche we could capture the trafficking of plasmid gene mobilization. The validity of the reported results and findings for plasmids in other ecological niches remains to be confirmed. The plasmids’ heredity and distribution between animals, as well as the functions they carry in the rumen environment, also warrant further investigation under changing conditions, such as diet and age. These questions can now be addressed using the unique procedures and computational pipeline developed here.
Materials and Methods
Sample Preparation.
The experimental procedures used in this study were approved by the Faculty Animal Policy and Welfare Committee at the Agricultural Research Organization's (ARO) Volcani Center and were in accordance with the guidelines of the Israel Council of Animal Care.
Israeli Holstein cows (n = 16) were housed at the ARO dairy farm in one shaded corral with free access to water and to a diet consisting of 70% concentrated food, mineral, and vitamin mix and 30% roughage, 17% of which was dietary natural detergent fiber. Samples were taken 1 h after the morning feeding: 500 mL of ruminal contents were collected via the cow's mouth using a stainless-steel stomach tube with a rumen vacuum sampler. Samples were transferred to CO2-containing centrifuge bottles to maintain anaerobic conditions and kept on ice. The ruminal samples were processed immediately after collection.
Microbes were isolated from the rumen samples using a protocol previously described by Stevenson and Weimer (29): following 2 min of homogenization using an ice-cold blender, the homogenate was centrifuged at 10,000 × g and the pellet was dissolved in ice-cold extraction buffer (100 mM Tris-HCl, 10 mM EDTA, 0.15 M NaCl pH 8.0); 1 g of the pellet was dissolved in 4 mL buffer and incubated at 4 °C for 1 h. The suspension was then centrifuged gently at 500 × g for 15 min at 4 °C to remove ruptured plant particles while keeping the bacterial cells in suspension. The supernatant was then passed through four layers of cheesecloth, centrifuged (10,000 × g, 25 min, 4 °C), and the pellet was kept at −20 °C until plasmid DNA extraction.
Plasmids were purified from the bacterial pellet by three different methods, with some minor adjustments, to maximize lysis as different bacteria require different lysis methods (30–32). Equal amounts of the DNA purified from the 16 rumen samples were pooled together and 10 μg of the pooled, purified DNA was subjected to Plasmid Safe DNase (Epicentre) digestion. The reactions were incubated overnight at 37 °C, following DNase inactivation at 70 °C for 30 min, and chilled on ice as previously described (54). The presence of genomic DNA was tested by PCR using 16S rRNA universal primers (BAC338F: 5′-ACTCCTACGGGAGGCAG-3′ and BAC805R: 5′-GACTACCAGGGTATCTAATCC-3′) (29). When 16S rRNA PCR product was visible on an electrophoretic DNA gel, another overnight digestion reaction was performed until 16S rRNA PCR product could no longer be visualized.
To selectively amplify circular DNA in the plasmid samples, phi29 DNA polymerase (New England Biolabs) was used. Reactions contained: 5 μL of the plasmid DNA as template, 1.5 μL of 10 μM exonuclease-resistant random hexamers (Fermentas), 3 μL phi29 DNA Polymerase Reaction Buffer (New England Biolabs), and 15.7 μL double-distilled water. Reactions were incubated at 95 °C for 5 min and immediately chilled on ice. Following 5 min on ice, 1.6 μL phi29 DNA polymerase was added along with 0.2 μL pyrophosphatase, inorganic (yeast) (New England Biolabs) and 3 μL dNTPs (10 mM). Reactions were incubated at 30 °C for 16 h. Finally, 3 μL from each reaction was loaded onto a 1% agarose gel with ethidium bromide staining for analysis. Following the amplification, the reaction product was used as the template for another 16S rRNA PCR to determine genomic DNA contamination. Only samples which failed to produce a 16S rRNA PCR product were sequenced.
Sequencing and Bioinformatics.
The pure amplified plasmid DNA was subjected to deep sequencing via Illumina (Illumina GAIIX sequencer). To account for gaps that might occur during the assembly process because of repetitive DNA motifs, such as insertion sequence elements, we used the paired-end protocol, which increases the odds of overcoming such gaps during de novo assembly. The reads were assembled using two different assembly programs: Velvet (35) and SDN (36). The two sets of assembled contigs were compared using the BWA program (with standard parameters) (55).
We analyzed the taxonomic and functional compositions of our dataset with the MG-RAST server (56) using similarity to the SEED database with a maximum E-value of ≤10−5 (57). The relative abundance of the SEED subsystems in our data were compared with that in two previously published rumen metagenomes (39, 40), as well as to that in the IMG and ACLAME (37, 38) plasmid databases using the same method. For the data published by Brulc et al. (39), we pooled all four metagenomic samples together and used them as a whole metagenome.
Contig Function Localization, Classification, and Sorting.
The SDN-generated contig set was compared with five different databases: NCBI-NR (BLASTX), CDD (RPS-BLAST), NCBI-NT, ACLAME, and IMG (TBLASTX) using an E-value cutoff of ≤10−3. The BRPA pipeline was designed to map and arrange functional annotations on the contigs and sort them according to function. The essence of BRPA is to inspect and integrate the hits generated by the different BLAST reports into the different contigs and divide them into selected categories according to key words or specified parameters. BRPA is trained using the custom-made “hifdisk.pl” tool that searches for characteristic high-frequency keywords in a specific database, compared with a general database. For our purposes, the comparison was between the ACLAME plasmid database and NCBI-NR database. Of the main suggested keywords, we manually chose the most plasmid-oriented ones and added the “plasmid” or “conj” strings, as mentioned below. Because overlap between hits is expected, BRPA was directed to choose the best hit from all overlapping hits when the overlapping regions shared the same frame; other cases were considered nonoverlapping to enable the detection of two genes on opposite strands or different frames. Plasmid functions were defined by BRPA using the following rules: (i) a hit is generated by similarity to one of the plasmid databases (ACLAME or IMG plasmids); (ii) The description of the hit contains one of the following strings that we produced and added with the “hifdisk.pl” tool: “rep,” “mob,” “tra,” “plasmid,” or “conj” (some of the shorter strings contained a space character, filtering out false-positives). The functions of the rumen-enriched subsystems were defined by BRPA using the following strings: “NADH” (58), “ATP synthase” (59–61), “glycosyltransferase” (51), “glycosyl hydrolase” (40). Representatives of other subsystems could not be found in proximity to plasmid backbone functions using BRPA. Finally, the general collection of contigs carrying plasmid backbone functions near accessory functions was manually inspected to validate their plasmid origin and the integrity of their annotation. Following this manual inspection, we were also able to locate functions of the “Cofactors, Vitamins, etc.” subsystem, which are represented by polyketide synthase function.
Phylogenetic Assignment of ORFs.
For the phylogenetic assignment of ORFs we used the previously described Phylogenie pipeline (45), which was implemented with the maximum likelihood RXaML program (46) using NCBI-NR as database and 100 bootstrap replicates. We analyzed 220 contigs that had at least two ORFs with NCBI-NR annotations. The taxonomic assignment of each ORF was determined using bootstrap support of 80% or higher. We excluded contigs that failed to generate a tree, as well as those carrying ORFs with bootstrap support of less than 80% (30% of the contigs) or ORFs, the taxonomic assignments of which, were not in agreement (17% of the contigs, which were considered mosaic).
454 Tag Amplicon Pyrosequencing and Data Analyses.
Samples from the same 16 cows were also used for metagenomic DNA extraction as previously described (29). 454 amplicon pyrosequencing of metagenomic DNA was performed by the Research and Testing Laboratory (Lubbock, TX) using primers covering the 103- to 530-bp region of the 16S rRNA gene sequence, corresponding to regions V2 and V3 (107 F: 5′-GGCGVACGGGTGAGTAA-3′ and 530 R: 5′-CCGCNGCNGCTGGCAC-3′). The tagging and sequencing protocol was as described previously (62). Data quality control and analyses were mostly performed using the QIIME pipeline (63). First, reads were assigned to their designated rumen sample using the split_library.py script, which also performs quality filtering based on length (<200 bp) and quality of the reads. The Uclust default method was used and the degree of similarity between sequences was defined as ≥97% to obtain operational taxonomic unit (OTU) identity at the species level. OTUs that clustered single reads were manually removed. After constructing an OTU table, taxonomy was assigned using the BLAST algorithm and the reference database, found at http://blog.qiime.org, designated “most recent Greengenes OTUs.”
Supplementary Material
Acknowledgments
We thank Dr. Nitzan Kol and Dr. Eran Halperin for the help with the assembly process, Prof. Tal Pupko and Ofir Cohen for their suggestions on the phylogeny analysis, and Dr. Uri Gophna for his comments and suggestions regarding this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the MG-RAST database, http://metagenomics.anl.gov/metagenomics.cgi?page=Home (accession nos. 4460391.3 and 4483775.3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116410109/-/DCSupplemental.
References
- 1.Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol. 2003;20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Wolf YI. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 4.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci USA. 2010;107:127–132. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heuer H, Abdo Z, Smalla K. Patchy distribution of flexible genetic elements in bacterial populations mediates robustness to environmental uncertainty. FEMS Microbiol Ecol. 2008;65:361–371. doi: 10.1111/j.1574-6941.2008.00539.x. [DOI] [PubMed] [Google Scholar]
- 7.Novick RP. Mobile genetic elements and bacterial toxinoses: The superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/s0147-619x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 8.Crossman LC. Plasmid replicons of Rhizobium. Biochem Soc Trans. 2005;33:157–158. doi: 10.1042/BST0330157. [DOI] [PubMed] [Google Scholar]
- 9.van der Meer JR, Sentchilo V. Genomic islands and the evolution of catabolic pathways in bacteria. Curr Opin Biotechnol. 2003;14:248–254. doi: 10.1016/s0958-1669(03)00058-2. [DOI] [PubMed] [Google Scholar]
- 10.Gil R, Sabater-Muñoz B, Perez-Brocal V, Silva FJ, Latorre A. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene. 2006;370:17–25. doi: 10.1016/j.gene.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 11.Zhang T, Zhang XX, Ye L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE. 2011;6:e26041. doi: 10.1371/journal.pone.0026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bekele AZ, Koike S, Kobayashi Y. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol Lett. 2010;305:49–57. doi: 10.1111/j.1574-6968.2010.01911.x. [DOI] [PubMed] [Google Scholar]
- 13.Smalla K, et al. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl Environ Microbiol. 2000;66:4854–4862. doi: 10.1128/aem.66.11.4854-4862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint HJ. The rumen microbial ecosystem—Some recent developments. Trends Microbiol. 1997;5:483–488. doi: 10.1016/S0966-842X(97)01159-1. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Vallvé S, Romeu A, Palau J. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol Biol Evol. 2000;17:352–361. doi: 10.1093/oxfordjournals.molbev.a026315. [DOI] [PubMed] [Google Scholar]
- 16.Tóthová T, Pristas P, Javorský P. Mercuric reductase gene transfer from soil to rumen bacteria. Folia Microbiol (Praha) 2006;51:317–319. doi: 10.1007/BF02931823. [DOI] [PubMed] [Google Scholar]
- 17.Flint HJ, Thomson AM, Bisset J. Plasmid-associated transfer of tetracycline resistance in Bacteroides ruminicola. Appl Environ Microbiol. 1988;54:855–860. doi: 10.1128/aem.54.4.855-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fliegerová K, Benada O, Flint HJ. Large plasmids in ruminal strains of Selenomonas ruminantium. Lett Appl Microbiol. 1998;26:243–247. doi: 10.1046/j.1472-765x.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- 19.Flint HJ, McPherson CA, Martin J. Expression of two xylanase genes from the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 cloned in pUC13. J Gen Microbiol. 1991;137:123–129. doi: 10.1099/00221287-137-1-123. [DOI] [PubMed] [Google Scholar]
- 20.Gilmour M, Mitchell WJ, Flint HJ. Genetic transfer of lactate-utilizing ability in the rumen bacterium Selenomonas ruminantium. Lett Appl Microbiol. 1996;22:52–56. doi: 10.1111/j.1472-765x.1996.tb01107.x. [DOI] [PubMed] [Google Scholar]
- 21.Kazimierczak KA, Flint HJ, Scott KP. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob Agents Chemother. 2006;50:2632–2639. doi: 10.1128/AAC.01587-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melville CM, Scott KP, Mercer DK, Flint HJ. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob Agents Chemother. 2001;45:3246–3249. doi: 10.1128/AAC.45.11.3246-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer DK, Melville CM, Scott KP, Flint HJ. Natural genetic transformation in the rumen bacterium Streptococcus bovis JB1. FEMS Microbiol Lett. 1999;179:485–490. doi: 10.1111/j.1574-6968.1999.tb08767.x. [DOI] [PubMed] [Google Scholar]
- 24.Mercer DK, Patel S, Flint HJ. Sequence analysis of the plasmid pRRI2 from the rumen bacterium Prevotella ruminicola 223/M2/7 and the use of pRRI2 in Prevotella/Bacteroides shuttle vectors. Plasmid. 2001;45:227–232. doi: 10.1006/plas.2000.1515. [DOI] [PubMed] [Google Scholar]
- 25.Al-Khaldi SF, Evans JD, Martin SA. Complete nucleotide sequence of a cryptic plasmid from the ruminal bacterium Selenomonas ruminantium HD4 and identification of two predicted open reading frames. Plasmid. 1999;42:45–52. doi: 10.1006/plas.1999.1405. [DOI] [PubMed] [Google Scholar]
- 26.May T, Kocherginskaya SA, Mackie RI, Vercoe PE, White BA. Complete nucleotide sequence of a cryptic plasmid, pBAW301, from the ruminal anaerobe Ruminococcus flavefaciens R13e2. FEMS Microbiol Lett. 1996;144:221–227. doi: 10.1111/j.1574-6968.1996.tb08534.x. [DOI] [PubMed] [Google Scholar]
- 27.Ohara H, et al. Structural analysis of a new cryptic plasmid pAR67 isolated from Ruminococcus albus AR67. Plasmid. 1998;39:84–88. doi: 10.1006/plas.1997.1324. [DOI] [PubMed] [Google Scholar]
- 28.Ogata K, et al. Structural organization of pRAM4, a cryptic plasmid from Prevotella ruminicola. Plasmid. 1996;35:91–97. doi: 10.1006/plas.1996.0011. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 30.Hansen JB, Olsen RH. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978;135:227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson DG, McKay LL. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 34.Johne R, Müller H, Rector A, van Ranst M, Stevens H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009;17:205–211. doi: 10.1016/j.tim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 37.Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: A CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32(Database issue):D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz VM, et al. The integrated microbial genomes system: An expanding comparative analysis resource. Nucleic Acids Res. 2010;38(Database issue):D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brulc JM, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 41.Sharon I, et al. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 2011;5:1178–1190. doi: 10.1038/ismej.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg Miller ME, et al. Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol. 2012;14:207–227. doi: 10.1111/j.1462-2920.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- 43.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 45.Frickey T, Lupas AN. PhyloGenie: Automated phylome generation and analysis. Nucleic Acids Res. 2004;32:5231–5238. doi: 10.1093/nar/gkh867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 47.Fonty G, Gouet P, Jouany J-P, Senaud J. Establishment of the microflora and anaerobic fungi in the rumen of lambs. J Gen Microbiol. 1987;133:1835–1843. [Google Scholar]
- 48.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–1074. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorek R, et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 51.Lurie-Weinberger MN, Peeri M, Gophna U. Contribution of lateral gene transfer to the gene repertoire of a gut-adapted methanogen. Genomics. 2012;99:52–58. doi: 10.1016/j.ygeno.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 53.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 54.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer F, et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hino T, Russell JB. Effect of reducing-equivalent disposal and NADH/NAD on deamination of amino acids by intact rumen microorganisms and their cell extracts. Appl Environ Microbiol. 1985;50:1368–1374. doi: 10.1128/aem.50.6.1368-1374.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell JB, Strobel HJ. ATPase-dependent energy spilling by the ruminal bacterium, Streptococcus bovis. Arch Microbiol. 1990;153:378–383. doi: 10.1007/BF00249009. [DOI] [PubMed] [Google Scholar]
- 60.Miwa T, Esaki H, Umemori J, Hino T. Activity of H(+)-ATPase in ruminal bacteria with special reference to acid tolerance. Appl Environ Microbiol. 1997;63:2155–2158. doi: 10.1128/aem.63.6.2155-2158.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miwa T, Abe T, Fukuda S, Ohkawara S, Hino T. Regulation of H(+)-ATPase synthesis in response to reduced pH in ruminal bacteria. Curr Microbiol. 2001;42:106–110. doi: 10.1007/s002840010187. [DOI] [PubMed] [Google Scholar]
- 62.Dowd SE, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.