Two structures of factor XIa in complex with small synthetic inhibitors are reported. These are the first structures of factor XIa complexed to small non-basic inhibitors.
Keywords: factor XIa, inhibitors, coagulation factors
Abstract
Factor XI (FXI) is a key enzyme in the coagulation pathway and an attractive target for the development of anticoagulant drugs. A small number of high-resolution crystal structures of FXIa in complex with small synthetic inhibitors have been published to date. All of these ligands have a basic P1 group and bind exclusively in the nonprime side of the active site of FXIa. Here, two structures of FXIa in complex with nonbasic inhibitors that occupy both the prime and nonprime sides of the active site are presented. These new structures could be valuable in the design and optimization of new FXIa synthethic inhibitors.
1. Introduction
Thromboembolic diseases are the primary cause of cardiovascular morbidity and mortality in developed countries. Conventional antithrombotic therapy is based on intravenous administration of heparin followed by oral treatment with warfarin. Although this is effective for the treatment and prevention of thromboembolic diseases, it has a number of limitations, such as a lack of reversibility and a small therapeutic window, which put patients at risk of excessive bleeding (Hirsh & Raschke, 2004 ▶). In recent years, a number of inhibitors have been designed to target specific enzymes in the coagulation pathway, such as thrombin and factor Xa (Zikria & Ansell, 2009 ▶; Pinto et al., 2010 ▶). Dabigatran, a thrombin inhibitor, has recently been approved for the treatment of patients with atrial fibrillation at risk of embolism or stroke (Connolly et al., 2009 ▶). Rivaroxaban, a factor Xa inhibitor, has been approved for the prevention of deep-vein thrombosis in patients with knee or hip replacement surgery (Lassen et al., 2008 ▶). However, antithrombotic agents still suffer from significant safety issues (Fiessinger et al., 2005 ▶).
Coagulation factor XI (FXI; EC 3.4.21.27) is a trypsin-like serine protease which plays a major role in the coagulation pathway (Gailani & Smith, 2009 ▶). FXI is cleaved by FXIIa, resulting in a heavy chain and a light chain, which then activates factor IX. Inhibition of the FXIa light chain may result in the formation of smaller thrombi without inhibiting clot formation (Schumacher et al., 2010 ▶). Thus, FXIa inhibitors could potentially offer an improved safety profile as anticoagulation agents.
The first crystal structure of the light chain FXIa in complex with a peptide protease inhibitor was reported by Jin, Pandey, Babine, Gorga et al. (2005 ▶). Initially, crystals of wild-type FXIa in complex with small-molecule inhibitors could not be obtained. However, it was found that by introducing four mutations in its solvent-exposed surface FXIa could be crystallized in complex with benzamidine (Jin, Pandey, Babine, Weaver et al., 2005 ▶). Since then, additional structures of FXIa in complex with a number of covalent and noncovalent synthetic inhibitors have been disclosed (Deng et al., 2006 ▶; Lazarova et al., 2006 ▶; Lin et al., 2006 ▶; Buchanan et al., 2008 ▶; Hanessian et al., 2010 ▶). In all of these complexes the ligand binds in the nonprime side of the active site. Another feature common to all of the inhibitors is a basic P1 group (guanidine or benzamidine) which interacts with Asp189 in FXIa1. Basic groups are often associated with problems such as high protein binding and poor systemic distribution; therefore, inhibitors with nonbasic P1 groups could be more promising as starting points for the development of anticoagulant drugs (Pinto et al., 2007 ▶, 2008 ▶; Pinto, 2008 ▶; Corte et al., 2009 ▶).
In this paper, we present crystal structures of FXIa in complex with two inhibitors with a nonbasic chlorophenyl-tetrazole P1 group. Ligands with this scaffold have previously been crystallized in complex with thrombin (Howard et al., 2006 ▶), but this is the first report of nonbasic ligands in complex with FXIa. These are also the first structures of FXIa in complex with nonpeptidic inhibitors that occupy both the prime and nonprime sides of the active site.
2. Materials and methods
2.1. Protein expression and purification
The catalytic domain of human factor XIa (UniProt code P03951; residues 370–607) mutant C500S was expressed as a secreted protein in the methylotropic yeast Pichia pastoris as described previously for the wild-type protein (Jin, Pandey, Babine, Gorga et al., 2005 ▶). Conditioned medium was concentrated using a 10 kDa cutoff TFF membrane (Vivascience) and dialysed against 20 mM Tris pH 7.4. Recombinant FXIa was initially purified on Zn2+-chelating Sepharose FF. After treatment of the enzyme with Endo Hf (New England Biolabs) at pH 6.0, the protein was further purified by cation-exchange chromatography (SP Sepharose FF, GE Healthcare) and size-exclusion chromatography (Superdex 75 26/60, GE Healthcare).
2.2. Crystallization
Factor XIa (54.7 mg ml−1) was diluted with storage buffer (20 mM Tris–HCl pH 7.5, 75 mM NaCl) to a final concentration of 25 mg ml−1. The inhibitor (ligand 1 or ligand 2) (50 mM in 20 mM Tris–HCl pH 7.5, 75 mM NaCl, 50% DMSO) was added to the protein (2 mM final concentration). Hanging-drop crystallizations were set up by mixing equal volumes of the protein solution and mother liquor (0.1 M citrate pH 4.7–5.2, 20–26% PEG 4K). Crystallization was initiated by inoculating the crystallization drops with microseeds of previously grown FXIa crystals. Crystals appeared after overnight incubation at 293 K.
2.3. Data collection and processing
Crystals were transferred to a general cryosolution (25% glycerol in mother liquor) for a few seconds and flash-cooled in the nitrogen cryostream of the X-ray generator. The crystals diffracted to about 2.2 Å resolution or better. Data collection was performed on a Rigaku MicroMax-007 HF X-ray generator equipped with dual R-AXIS IV++ image-plate detectors and Varimax optics. We collected 125 and 180 images from crystals of FXIa in complex with ligands 1 and 2, respectively.
Diffraction data for the two complexes were integrated and scaled using the CrystalClear processing suite (Rigaku, 1997 ▶). Each structure was solved by rigid-body refinement of an in-house structure with the same space group and similar unit-cell parameters using REFMAC (Murshudov et al., 2011 ▶). Each model was then subjected to a number of cycles of restrained refinement with REFMAC, using Coot (Emsley & Cowtan, 2004 ▶) to rebuild the models at each stage and adding the ligand, water and additional compounds in the crystallization solution. Statistics for the two models are listed in Table 2 ▶. Coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 3sor and 3sos)
Table 2. Data-collection and refinement statistics.
A cutoff of 2.0 in I/σ(I) was used. Values in parentheses are for the outer shell.
| PDB code | 3sos | 3sor |
|---|---|---|
| Ligand | 1 | 2 |
| No. of images | 125 | 180 |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 59.114, b = 59.548, c = 66.755 | a = 59.988, b = 60.06, c = 67.512 |
| Molecules per unit cell | 1 | 1 |
| Matthews coefficient (Å3 Da−1) | 2.01 | 2.04 |
| Resolution range (Å) | 66.75–2.58 (2.95–2.58) | 44.88–2.38 (2.73–2.38) |
| Total No. of reflections | 36355 (12050) | 68740 (22632) |
| No. of unique reflections | 7852 (2551) | 10088 (3291) |
| Average multiplicity | 4.63 (4.71) | 6.76 (6.81) |
| Completeness (%) | 100.0 (100) | 99.7 (99.6) |
| No. of reflections used in refinement | 7446 | 9654 |
| Rmerge | 0.127 (0.345) | 0.097 (0.263) |
| 〈I/σ(I)〉 | 3.9 (3.6) | 11.2 (5.3) |
| Unaveraged I/σ(I) | 7.7 (2.0) | 5.1 (2.4) |
| R factor/Rfree | 0.203/0.278 | 0.222/0.296 |
| No. of chains | 1 | 1 |
| Sequence stretches not built because of absence of electron density | 244–245 | 244–245 |
| Protein atoms | 1928 | 1899 |
| Ligand atoms | 39 | 38 |
| Water molecules | 82 | 174 |
| Other molecules | 1 citrate, 1 2-amino-2-hydroxymethyl-propane-1,3-diol | 1 citrate |
| R.m.s.d. bonds (Å) | 0.021 | 0.013 |
| R.m.s.d. angles (°) | 1.9 | 1.5 |
| Wilson B factor (Å2) | 17.5 | 31.7 |
| Average B factor (Å2) | ||
| Protein | 38.0 | 34.7 |
| Ligand | 43.0 | 42.2 |
| Solvent | 41.2 | 39.2 |
3. Results and discussion
3.1. Overall architecture of FXIa
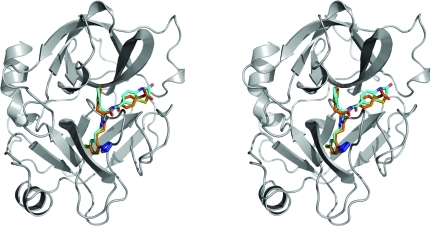
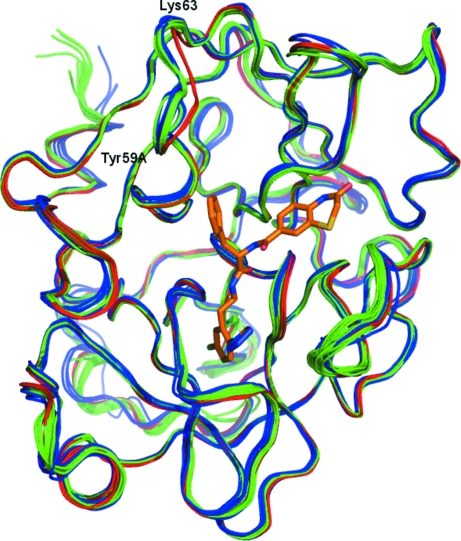
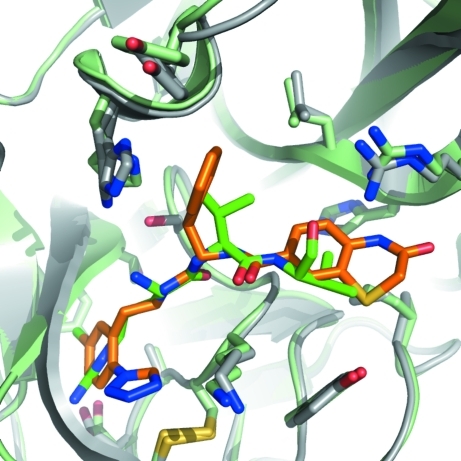
The main structural features of FXIa are two β-barrels facing each other with the catalytic triad (Ser195–His57–Asp102) in between them. A number of loops and two helical features also contribute to define the overall structure of FXIa. Fig. 1 ▶ shows the secondary structure of FXIa in complex with ligands 1 and 2 (see Fig. 2 ▶). The protein structures of the two complexes are very similar (the Cα r.m.s.d. between them is ∼0.2 Å). Fig. 3 ▶ shows an overlay of the Cα traces of the complexes reported in this paper with those of previous FXIa structures. Again, the structure of FXIa appears to be very similar in all of the complexes. The only significant difference is in a short loop comprising residues 59A–63, which is in a slightly different conformation compared with the other structures in the PDB. We have noticed the same conformation in our own structures of FXIa in complex with unrelated ligands, so it is unlikely that this is a ligand-induced effect. Rather, it might be a consequence of the fact that the combination of space group P212121 and unit-cell parameters of ∼60 × 60 × 67 µm in our crystals differs from all of the other structures in the PDB. In our structures, residues 59A–63 are exposed to solvent and do not make any crystal contacts to other protein units. In contrast, the FXIa structures in the PDB make extensive contacts in this region either to symmetry-related protein units or to bound peptide inhibitors. This could explain the conformational differences in this loop.
Figure 1.
Stereo diagram of factor XIa bound to ligands 1 (orange) and 2 (cyan). The protein structure is depicted in grey and corresponds to factor XIa bound to ligand 2. The protein structure for the complex with ligand 1 is not shown, but it is visually indistinguishable (the Cα r.m.s.d. between the two structures is 0.21 Å). All structural figures in this paper were generated with PyMOL (DeLano, 2002 ▶).
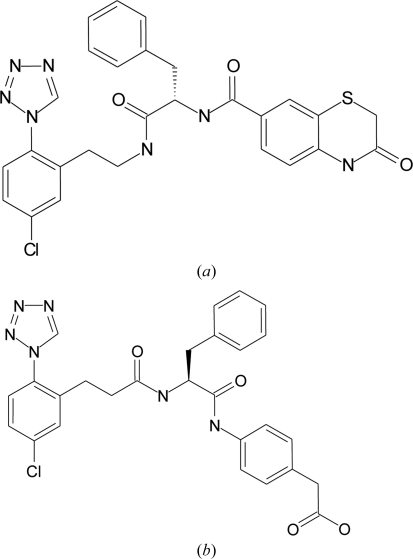
Figure 2.
Chemical structures of (a) ligand 1 and (b) ligand 2.
Figure 3.
Overlay of the Cα traces of all of the structures of activated FXIa that have been deposited in the Protein Data Bank. Complexes of FXIa with a peptide inhibitor are coloured blue, those with a small synthetic inhibitor green and the two complexes reported in this paper red. Bound ligand 1 is shown as orange sticks. For structures with multiple protein chains in the asymmetric unit only the first one is shown.
3.2. Binding mode
The two compounds crystallized in complex with FXIa are depicted in Fig. 2 ▶ and their activities towards FXIa, thrombin and FXa are given in Table 3 ▶. They are potent FXIa inhibitors that are selective against thrombin and FXa (Pinto et al., 2007 ▶, 2008 ▶; Pinto, 2008 ▶; Corte et al., 2009 ▶). The binding modes of the two ligands are very similar, with the chlorophenyl-tetrazole scaffold deep in the S1 pocket and the C—Cl bond perpendicular to the phenol ring of Tyr228 (see Fig. 4 ▶). There are no polar contacts between Asp189 and the chlorophenyl scaffold, although Asp189 and Tyr228 interact through a bridging water. Thermodynamic studies of similar P1 scaffolds in thrombin suggest that the interaction of the Cl atom perpendicularly directed into the centre of the phenol ring can contribute significantly to binding affinity (Baum et al., 2009 ▶). This is consistent with geometrical analyses of similar interactions in the Protein Data Bank and Cambridge Structural Database (Matter et al., 2009 ▶; Imai et al., 2009 ▶). For all of the complexes, the tetrazole ring of the ligand is in close contact with the disulfide bond formed by Cys191 and Cys219, possibly forming an interaction between the π system of the tetrazole ring and the lone pairs of the S atoms. Interestingly, the chlorophenyl-tetrazole fragment and some ligands based around it have been crystallized in complex with thrombin (Howard et al., 2006 ▶; Young et al., 2004 ▶). The comparison with our FXIa structures reveals that the chlorophenyl-tetrazole scaffold has the same binding mode in both proteins. This is not surprising, taking into account that FXIa and thrombin have a high degree of sequence and structural similarity in this region.
Table 3. Ligands and activity data.
Activity data towards FXIa, thrombin and FXa were measured using functional assays with fluorogenic substrates. For FXIa, the activity is given as pEC50. For thrombin and FXa, we could not determine an EC50 for any of the ligands and the percentage inhibition at 10 µM concentration is reported instead. Ligand 1 could not be tested for activity against FXa.
pEC50.
Inhibition at 10 µM.
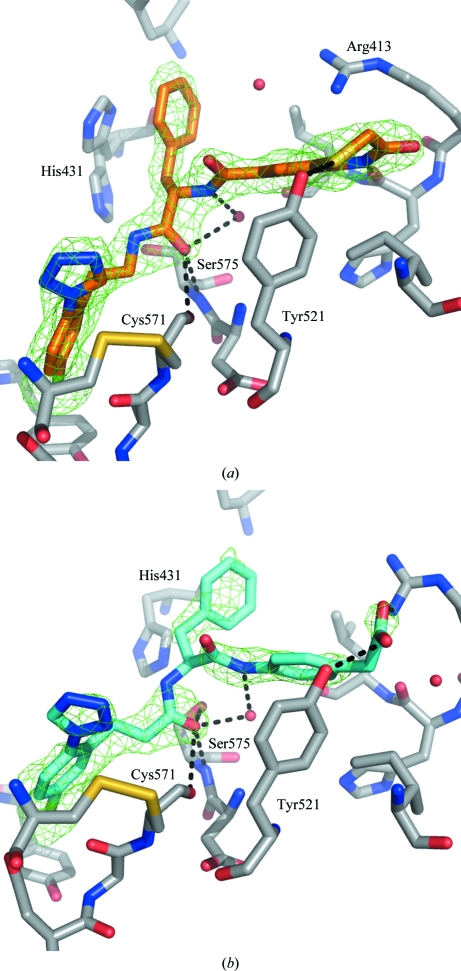
Figure 4.
Schematic diagram of the interactions of ligands 1 (a) and 2 (b) in complex with FXIa. The ligands are depicted in orange (ligand 1) and cyan (ligand 2). Key protein residues are shown in grey and water molecules in the active site are shown as red spheres. His57 of FXIa in complex with ligand 1 has been modelled in two conformations. The OMIT difference density around each ligand (F o − F c map calculated from the final model after removing the ligand) is shown as a green mesh (contoured at 3σ) for the two complexes. Key polar interactions between the ligand and protein or solvent are shown as black dashed lines.
Both ligands have an ethylamide group at position 2 of the phenyl ring, with the amide binding near the catalytic Ser195 and stabilized through hydrogen-bonding contacts to the backbone of Ser195 and Gly193. There is no evidence of protein–ligand covalent bonding in the electron density of either of the two structures. The exit vector of this amide is directed towards the prime side of the pocket, with a benzyl group binding into S1′ and the rest of each ligand into S2′. In both structures the P1′ benzyl ring makes contacts with His57, which is part of the catalytic triad, Tyr59A and Leu39. For ligand 1, the benzyl ring is stacked in a face-to-face interaction with His57, but only for one of the two His57 conformations modelled. For ligand 2, the P1′ benzyl group and His57 are arranged in a displaced edge-to-face arrangement. P1′ is Val in the native substrate and structures from the PDB of FXIa in complex with peptidic inhibitors have either Met or Val in this position (Jin, Pandey, Babine, Gorga et al., 2005 ▶).
Ligand 1 has an amide in the S1′ subpocket that interacts with the backbone of Leu39 through a bridging water molecule. The S2′ site is filled by the benzothiazinone ring, which forms a hydrogen bond to the backbone of His38 and a long-distance (3.5 Å) polar contact between the S atom and the side chain of Tyr143. Lys37D is located ∼3.2 Å above the junction between the phenyl and thiazinone rings, further stabilizing the ligand through a cation–π interaction. P2′ is Val in the native substrate, and peptidic inhibitors cocrystallized with FXIa have either Val or Ala in this position, with the side chain in approximately the same location as occupied by the aromatic ring of the benzothiazinone scaffold. Fig. 5 ▶ shows an overlay of ligand 1 and the P1–P1–P2′ residues of the ECOTINP peptide2 in complex with FXIa. This illustrates the equivalence between the P1, P1′ and P2′ residues of the peptide and the chloroaryl, benzyl and benzothiazinone ring systems of the ligand. The overlay also highlights some relatively minor conformational changes in some of the key residues in the active site between the two complexes: Arg37D, His57, Tyr59A and Lys192.
Figure 5.
Binding mode of ligand 1 in complex with FXIa compared with the ECOTINP peptide mutant. The FXIa–ligand 1 complex is depicted with the protein in grey and the ligand in orange, while the FXIa–ECOTINP complex is depcicted in pale green and green for the protein and peptide, respectively. Proteins are shown as cartoons, with key residues in the active site as sticks. Only residues Arg84-Val85-Val86 of the peptide are depicted (corresponding to P1–P1′–P2′). The FXIa–ECOTINP mutant structure was obtained from the PDB (PDB entry 1xxf; Jin, Pandey, Babine, Gorga et al., 2005 ▶).
Ligand 2 also has an amide at the junction between S1′ and S2′, but it is reversed compared with ligand 1, with the carbonyl group making a hydrogen bond to a water molecule. The phenyl ring directly attached to the amide binds into S2′ close to the location of the Val residue in the substrate-like peptide inhibitors. The acetate group binds between the side chains of Lys37D and Tyr143, making polar interactions with both residues.
Ligand 2 is selective for the closely related thrombin and factor Xa serine proteases, while ligand 1 was also found to be selective against thrombin but could not be tested against FXa (see Table 3 ▶). The three proteases are very similar in the S1 pocket, and the chlorophenyl-tetrazole fragment has previously been found to be active towards thrombin (Howard et al., 2006 ▶). However, there are some key differences in the prime site of the ligand that can explain the selectivity profiles of ligands 1 and 2. Compared with FXIa, both thrombin and FXa have extensive sequence and structural differences which make the S1′ pocket smaller and could prevent the benzyl moiety from binding there. There are also a number of differences in the S2′ pocket and beyond. Key differences are found at Tyr143, which is replaced by Trp (thrombin) or Arg (FXa), and Lys37D, which is replaced by Glu in both thrombin and FXa. These residues are directly involved in the binding of the benzothiazinone ring of ligand 2 and the acetate of ligand 1, which will not be able to make similarly attractive interactions in the active site of thrombin or FXa.
In summary, we have presented crystal structures of FXIa in complex with two small synthetic inhibitors. These are the first structures of FXIa bound to inhibitors with a neutral P1 group, showing that the binding mode for the chlorophenyl-tetrazole group is the same in thrombin and FXIa. These are also the first FXIa structures with nonpeptidic inhibitors bound in the prime side of the active site. A comparative analysis of the binding mode of the two ligands into the prime side identified key ligand-binding interactions in this pocket and explained the selectivity profile of these ligands towards thrombin and FXa. This information could be useful for the design and optimization of selective FXIa inhibitors.
Supplementary Material
PDB reference: factor XIa–inhibitor complexes, 3sor
PDB reference: 3sos
Table 1. Correspondence between FXIa and trypsin residue numbering for the residue numbers mentioned in this paper.
| Residue | FXIa | Trypsin |
|---|---|---|
| Arg | 413 | 37D |
| His | 414 | 38 |
| Leu | 415 | 39 |
| His | 431 | 57 |
| Tyr | 434 | 59A |
| Lys | 440 | 63 |
| Asp | 480 | 102 |
| Tyr | 521 | 143 |
| Asp | 569 | 189 |
| Cys | 571 | 191 |
| Lys | 572 | 192 |
| Gly | 573 | 193 |
| Ser | 575 | 195 |
| Cys | 599 | 219 |
| Tyr | 608 | 228 |
| Ala | 624 | 244 |
| Val | 625 | 245 |
Footnotes
References
- Baum, B., Mohamed, M., Zayed, M., Gerlach, C., Heine, A., Hangauer, D. & Klebe, G. (2009). J. Mol. Biol. 390, 56–69. [DOI] [PubMed]
- Buchanan, M. S., Carroll, A. R., Wessling, D., Jobling, M., Avery, V. M., Davis, R. A., Feng, Y., Xue, Y., Oster, L., Fex, T., Deinum, J., Hooper, J. N. & Quinn, R. J. (2008). J. Med. Chem. 51, 3583–3587. [DOI] [PubMed]
- Connolly, S. J. et al. (2009). N. Engl. J. Med. 361, 1139–1151.
- Corte, J. R., Hu, Z. & Quan, M. L. (2009). Patent WO 2009/114677 A1.
- DeLano, W. (2002). PyMOL http://www.pymol.org.
- Deng, H. et al. (2006). Bioorg. Med. Chem. Lett. 16, 3049–3054.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fiessinger, J. N., Huisman, M. V., Davidson, B. L., Bounameaux, H., Francis, C. W., Eriksson, H., Lundström, T., Berkowitz, S. D., Nyström, P., Thorsén, M. & Ginsberg, J. S. (2005). JAMA, 293, 681–689. [DOI] [PubMed]
- Gailani, D. & Smith, S. B. (2009). J. Thromb. Haemost. 7, Suppl. 1, 75–78. [DOI] [PMC free article] [PubMed]
- Hanessian, S., Larsson, A., Fex, T., Knecht, W. & Blomberg, N. (2010). Bioorg. Med. Chem. Lett. 20, 6925–6928. [DOI] [PubMed]
- Hirsh, J. & Raschke, R. (2004). Chest, 126, 188S–203S. [DOI] [PubMed]
- Howard, N., Abell, C., Blakemore, W., Chessari, G., Congreve, M., Howard, S., Jhoti, H., Murray, C. W., Seavers, L. C. & van Montfort, R. L. (2006). J. Med. Chem. 49, 1346–1355. [DOI] [PubMed]
- Imai, Y., Inoue, Y., Nakanishi, I. & Kitaura, K. (2009). QSAR Comb. Sci. 28, 869–873.
- Jin, L., Pandey, P., Babine, R. E., Gorga, J. C., Seidl, K. J., Gelfand, E., Weaver, D. T., Abdel-Meguid, S. S. & Strickler, J. E. (2005). J. Biol. Chem. 280, 4704–4712. [DOI] [PubMed]
- Jin, L., Pandey, P., Babine, R. E., Weaver, D. T., Abdel-Meguid, S. S. & Strickler, J. E. (2005). Acta Cryst. D61, 1418–1425. [DOI] [PubMed]
- Lassen, M. R. et al. (2008). N. Engl. J. Med. 358, 2776–2786. [DOI] [PubMed]
- Lazarova, T. I., Jin, L., Rynkiewicz, M., Gorga, J. C., Bibbins, F., Meyers, H. V., Babine, R. & Strickler, J. (2006). Bioorg. Med. Chem. Lett. 16, 5022–5027. [DOI] [PubMed]
- Lin, J. et al. (2006). J. Med. Chem. 49, 7781–7791. [DOI] [PubMed]
- Matter, H., Nazaré, M., Güssregen, S., Will, D. W., Schreuder, H., Bauer, A., Urmann, M., Ritter, K., Wagner, M. and Wehner, V. (2009). Angew. Chem. 121, 2955–2960. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Pinto, D. J. P. (2008). Patent WO 2008/076805 A2.
- Pinto, D. J. P., Quan, M. L., Smith, L. M. II, Orwat, M. J. & Gilligan, P. J. (2008). Patent WO 2008/157162 A1.
- Pinto, D. J. P., Smallheer, J. M., Cheney, D. L., Knabb, R. M. & Wexler, R. R. (2010). J. Med. Chem. 53, 6243–6274. [DOI] [PubMed]
- Pinto, D. J. P., Smallheer, J. M., Corte, J. R., Hu, Z., Cavallaro, C. L., Gilligan, P. J., Quan, M. L. & Smith, L. M. II (2007). Patent WO 2007/070826 A1.
- Rigaku (1997). CrystalClear: An Integrated Program for the Collection and Processing of Area Detector Data Rigaku Corporation, Tokyo, Japan.
- Schumacher, W., Luettgen, J., Quan, M. & Seiffert, D. (2010). Arterioscler. Thromb. Vasc. Biol. 30, 388–392. [DOI] [PubMed]
- Young, M. B. et al. (2004). J. Med. Chem. 47, 2995–3008. [DOI] [PubMed]
- Zikria, J. C. & Ansell, J. (2009). Curr. Opin. Hematol. 16, 347–356. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: factor XIa–inhibitor complexes, 3sor
PDB reference: 3sos