The crystallization of the ligand-binding domain of the methyl-accepting chemotaxis protein chemoreceptor McpS (McpS-LBD) is reported.
Keywords: methyl-accepting chemotaxis proteins, ligand-binding domains, Pseudomonas putida KT2440
Abstract
Methyl-accepting chemotaxis proteins (MCPs) are transmembrane proteins that sense changes in environmental signals, generating a chemotactic response and regulating other cellular processes. MCPs are composed of two main domains: a ligand-binding domain (LBD) and a cytosolic signalling domain (CSD). Here, the crystallization of the LBD of the chemoreceptor McpS (McpS-LBD) is reported. McpS-LBD is responsible for sensing most of the TCA-cycle intermediates in the soil bacterium Pseudomonas putida KT2440. McpS-LBD was expressed, purified and crystallized in complex with two of its natural ligands (malate and succinate). Crystals were obtained by both the counter-diffusion and the hanging-drop vapour-diffusion techniques after pre-incubation of McpS-LBD with the ligands. The crystals were isomorphous and belonged to space group C2, with two molecules per asymmetric unit. Diffraction data were collected at the ESRF synchrotron X-ray source to resolutions of 1.8 and 1.9 Å for the malate and succinate complexes, respectively.
1. Introduction
Bacteria need to constantly sense changes in environmental signals and to adapt their metabolism and behaviour accordingly to guarantee survival. These regulatory processes are primarily mediated by members of several large protein families, namely two-component systems, methyl-accepting chemotaxis proteins (MCPs), Ser/Thr/Tyr protein kinases, diguanylate cyclases, cyclic di-GMP-specific phosphodiesterases and adenylate cyclases (Galperin, 2005 ▶). These proteins all recognize environmental signals and generate regulatory responses.
MCPs are typically transmembrane proteins composed of a periplasmic ligand-binding domain and a cytosolic signalling domain which forms a ternary complex with the CheA autokinase and the CheW coupling protein (Hazelbauer et al., 2008 ▶). Many chemoreceptors mediate bacterial chemotaxis, whereas others have been shown to be involved in the regulation of different cellular processes such as modulating the level of the second-messenger cyclic di-GMP (Hickman et al., 2005 ▶).
Bioinformatic analyses have revealed that bacterial chemoreceptors can be classified according to the size of their ligand-binding domain (LBD). Cluster I receptors (54% of the receptors) possess an LBD of 120–210 amino acids, whereas cluster II receptors (39% of the receptors) have an LBD which contains 220–300 amino acids (Lacal, García-Fontana et al., 2010 ▶). There is abundant structural information available on cluster I LBDs, of which the best characterized are TarH, PAS and GAF domains (Pokkuluri et al., 2008 ▶; Podust et al., 2008 ▶; Milburn et al., 1991 ▶). In marked contrast to cluster I LBDs, no published structural information is available on the larger cluster II domains. One other chemoreceptor for TCA-cycle intermediates has been identified, which is the TCP receptor of Salmonella typhimurium (Iwama et al., 2006 ▶). However, sequence analysis of this protein revealed a cluster I LBD, indicating that there are at least two different domains which sense this group of ligands in the context of a chemotactic response.
The McpS chemoreceptor of Pseudomonas putida KT2440 has been found to mediate a specific chemotactic response towards six TCA-cycle intermediates (succinate, malate, fumarate, oxaloactetate, citrate and isocitrate) as well as towards butyrate (Lacal, Alfonso et al., 2010 ▶; Lacal et al., 2011 ▶). The LBD of this receptor consists of 257 amino acids and therefore corresponds to a cluster II domain. This domain was produced as an individual recombinant protein (McpS-LBD) and submitted to biophysical analyses. Analytical ultracentrifugation studies have shown that there is a dynamic equilibrium between monomers and dimers and that ligand binding stabilizes the protein dimer (Lacal, Alfonso et al., 2010 ▶). Isothermal titration calorimetry studies revealed that the seven ligands mentioned above bind to McpS with affinities in the range 8–300 µM (Lacal, Alfonso et al., 2010 ▶; Lacal et al., 2011 ▶). The two ligands which caused the strongest chemotactic response in vivo were malate and succinate (Lacal, Alfonso et al., 2010 ▶). These compounds bound to McpS-LBD with dissociation constants of 8 and 82 µM, respectively.
McpS has been predicted to be composed of six helices (two long and four short helices). This prediction has not been verified since structural data are only available for the smaller cluster I domains. Here, we report the crystallization and preliminary X-ray crystallographic analysis of McpS-LBD in complex with malate and succinate. The resulting structures will contribute to compensate for the existing lack of structural information on bacterial cluster II sensor domains.
2. Materials and methods
2.1. Overexpression and purification
The plasmid pETMcpS was constructed for expression of the McpS ligand-binding domain (Gly47–Ser283) fused to an N-terminal polyhistidine tag. The construction of this plasmid as well as protein expression and purification have been described previously (Lacal, García-Fontana et al., 2010 ▶). In summary, Escherichia coli BL21 (DE3) cells containing pETMcpS were grown at 310 K until the culture reached an OD600 of 0.6 and were subsequently induced with 0.1 mM IPTG. Growth was then continued at 289 K overnight. Cells were harvested by centrifugation and subsequently resuspended in buffer. Cell disruption was performed using a French press. After a centrifugation step, the supernatant was loaded onto a HisTrap HP column (GE Healthcare) and eluted with an imidazole gradient. Protein-containing fractions were pooled, concentrated to 5 ml, dialyzed against 50 mM Tris–HCl, 0.5 M NaCl pH 8.0 and loaded onto a HiPrep 26/60 Sephacryl S200 gel-filtration column (GE Healthcare). The protein was eluted isocratically (1 ml min−1) with the same buffer. Coomassie-stained SDS–PAGE gels of pure McpS-LBD showed a highly pure sample. The protein was concentrated to 20 mg ml−1 and submitted to crystallization trials. Selenomethionine-derivatized McpS-LBD was produced as described by Doublié (1997 ▶) and purified using the above-described protocol.
2.2. Crystallization
McpS-LBD in purification buffer was subjected to buffer-exchange with analysis buffer (5 mM Tris–HCl, 5 mM HEPES, 5 mM MES pH 8) using 0.5 ml concentration units (Amicon). McpS-LBD at a concentration of 0.6 mM (15 mg ml−1) was then incubated with either 30 mM malate or 100 mM succinate for 30 min on ice. Subsequently, excess malate and succinate were removed by buffer-exchange with the same buffer.
The initial crystallization screen was performed with freshly purified McpS-LBD using the GCB-CSK-24 (Triana Science and Technology) counter-diffusion screening kit with capillaries of 0.1 mm inner diameter (CP-01-50; Triana Science and Technology). The screen was conducted in the absence and presence of ligands, but crystals were exclusively observed in ligand-containing samples. Initial conditions were further improved by the vapour-diffusion method using a hanging-drop configuration. Plate-shaped crystals were obtained by mixing 1 µl protein solution (18 mg ml−1) and 1 µl reservoir solution and equilibrating against 500 µl reservoir solution. Crystals were cryoprotected in mother liquor supplemented with 20% PEG 400 and flash-cooled in liquid nitrogen.
2.3. Data collection and structure determination
Data were collected from native and selenomethionine-derivatized crystals on ESRF beamlines BM16 and ID14-4 using an ADSC Quantum 4 CCD detector and were processed with XDS (Kabsch, 2010 ▶). F A values (structure-factor amplitudes of the heavy-atom model) were calculated using SHELXC (Sheldrick, 2008 ▶). Based on initial analysis of the data, the maximum resolution for substructure determination and initial phase calculation was set to 2.5 Å. 15 Se atoms from a maximum number of 16 heavy atoms requested in the search were found using SHELXD (Sheldrick, 2008 ▶). The correct hand of the substructure was determined using ABS (Hao, 2004 ▶) and SHELX (Sheldrick, 2008 ▶). The twofold noncrystallographic symmetry (NCS) operator was found using RESOLVE (Terwilliger, 2000 ▶). Density modification, phase extension and NCS averaging were performed using DM (Cowtan, 1994 ▶). 81% of the model was built using ARP/wARP (Perrakis et al., 1999 ▶; Morris et al., 2004 ▶). Refinement is in progress using REFMAC (Winn et al., 2011 ▶) and Coot (Emsley & Cowtan, 2004 ▶) for manual building.
3. Results
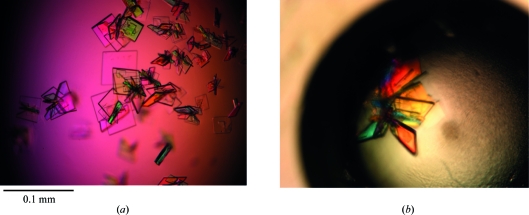
Crystallization of McpS-LBD was first attempted using the expressed and purified domain without any additional ligand. Unfortunately, all attempts to produce crystals failed, probably because a ligand is required for LBD stabilization. It is known that P. putida McpS-LBD can recognize up to seven different ligands which provoke different magnitudes of chemotactic response. Quantitative capillary chemotaxis assays revealed that malate and succinate are amongst the strongest chemoattractants in vivo (Lacal, Alfonso et al., 2010 ▶). In addition, isothermal titration calorimetric studies showed that these ligands bind to the recombinant McpS-LBD with affinities of 8 and 82 µM, respectively. Therefore, both dicarboxylic acids were selected for crystallization assays after incubation with McpS-LBD. Initial crystals of the McpS-LBD–malate complex were obtained by the counter-diffusion technique (García-Ruiz, 2003 ▶) in 0.1 mm inner diameter capillaries in condition No. 18 of the GCB-CSK-24 kit (25% PEG 4000, 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate) at 277 K. Since only one single crystal was observed in the first series of trials, alternative crystallization techniques were employed in the subsequent rounds of crystal improvement. Optimal crystal size was finally obtained using the vapour-diffusion technique (Fig. 1 ▶) in the hanging-drop configuration.
Figure 1.
Crystals of McpS-LBD. (a) Improved McpS-LBD crystals after pre-incubation with 30 mM malic acid. (b) Plate-shaped crystals of McpS-LBD in complex with succinate grown under similar conditions as the malate complex.
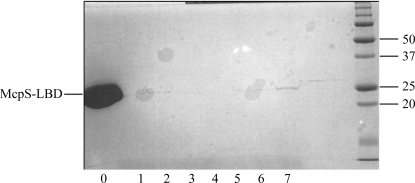
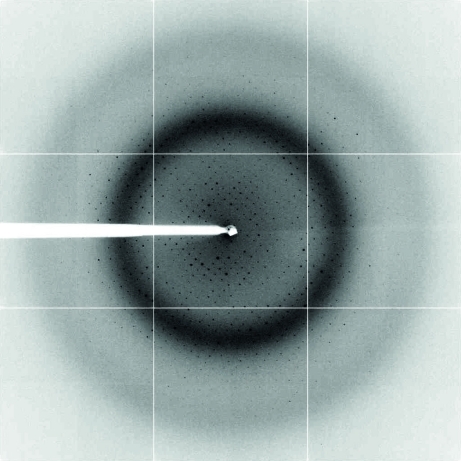
Plate-shaped crystals were grown by mixing 1 µl protein solution (18 mg ml−1) and 1 µl reservoir solution consisting of 20% PEG 4000, 0.25 M ammonium sulfate, 100 mM sodium acetate pH 4.8. Analysis of a dissolved crystal by SDS–PAGE indicated the presence of full-length McpS-LBD (Fig. 2 ▶). The crystals were cryocooled and were subjected to diffraction data collection, showing resolution limits of 1.8 Å (Fig. 3 ▶) for the complex with malate and 1.9 Å for the succinate complex. The lack of a search model for molecular replacement prompted the production of selenomethionine derivatives. The crystallization conditions for McpS-LBD–succinate and the selenomethionine-derivatized protein differed slightly, but the resulting crystals were of similar morphology. One crystal of SeMet-McpS-LBD in complex with malate diffracted X-rays to 2.0 Å resolution, but the initial phase calculation was set to 2.5 Å. 15 of a sequence-derived maximal number of 16 methionine residues (eight per molecule in the asymmetric unit) were detected and used for phasing. Complete data-collection statistics and crystal parameters are shown in Table 1 ▶. Iterative cycles of phase recombination, model building and refinement are being carried out in order to obtain a complete model. The succinate-containing structure will be solved by molecular replacement (MR) using the final malate-containing structure as the search model. Careful analysis of both structures will help us to understand the binding mode of McpS-LBD to the strong chemoattractants succinate and malate.
Figure 2.
SDS–PAGE gel of McpS-LBD incubated with malic acid. Crystals were resuspended in 20 µl reservoir buffer and subjected to several rounds of washing by centrifugation. Lane 0 contains the protein used for the crystallization experiments. Lanes 1–6 contain the supernatant from subsequent washing steps. After the final wash, crystals were resuspended in reservoir buffer followed by addition of 10 µl 1× loading buffer prior to analysis by SDS–PAGE (lane 7).
Figure 3.
X-ray diffraction image of an McpS-LBD–malate crystal. The crystal diffracted to 1.8 Å resolution. This image is representative of a complete data set collected from a native McpS-LBD–malate crystal.
Table 1. Crystal parameters and data-collection statistics.
Values in parentheses are for the last shell.
| Malate complex | |||
|---|---|---|---|
| Native | SeMet, peak | Succinate complex | |
| Crystal parameters | |||
| Space group | C2 | C2 | C2 |
| Unit-cell parameters | |||
| a (Å) | 226.35 | 226.47 | 226.07 |
| b (Å) | 45.85 | 46.06 | 46.04 |
| c (Å) | 51.18 | 51.09 | 50.82 |
| β (°) | 95.812 | 96.085 | 95.93 |
| Data collection | |||
| Temperature | 100 | 100 | 100 |
| X-ray source | BM16 | ID14-4 | ID14-4 |
| Detector | ADSC Quantum 4 CCD | ADSC Quantum 4 CCD | ADSC Quantum 4 CCD |
| Wavelength | 0.98 | 0.98 | 0.98 |
| Resolution (Å) | 25.0–1.8 (1.85–1.80) | 25.0–2.0 (2.05–2.00) | 20.0–1.9 (1.97–1.90) |
| Rmeas (%) | 7.2 (59.2) | 11.7 (62.1) | 13.5 (40.8) |
| 〈I/σ(I)〉 | 13.96 (2.49) | 8.35 (1.94) | 7.0 (2.8) |
| Completeness (%) | 96.8 (94.8) | 97.7 (97.1) | 97.3 (99.5) |
| Multiplicity | 3.8 (3.6) | 1.9 (1.9) | 3.4 (3.3) |
Acknowledgments
This work was financed by grants from the BBVA Foundation, the Andalusian Regional Government (P09-RNM-4509) and FEDER-supported grants from the Spanish Ministry for Science and Innovation (BIO2010-16937 and BIO2010-17227) and FEDER Excelencia Junta de Andalucía (CVI-3010). We would like to thank Cristina García-Fontana for technical assistance. This publication is a product of the ‘Factoría Española de Cristalización’, Consolider-Ingenio 2010 project (MICINN). EP is supported by a ‘Ramón y Cajal’ research contract (MICINN). We are also grateful to the beamline staff at BM16 and ID14-4, ESRF, Grenoble, France for their help during data collection.
References
- Cowtan, K. (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 31, 34–38.
- Doublié, S. (1997). Methods Enzymol. 276, 523–530. [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Galperin, M. Y. (2005). BMC Microbiol. 5, 35. [DOI] [PMC free article] [PubMed]
- García-Ruiz, J. M. (2003). Methods Enzymol. 368, 130–154. [DOI] [PubMed]
- Hao, Q. (2004). J. Appl. Cryst. 37, 498–499.
- Hazelbauer, G. L., Falke, J. J. & Parkinson, J. S. (2008). Trends Biochem. Sci. 33, 9–19. [DOI] [PMC free article] [PubMed]
- Hickman, J. W., Tifrea, D. F. & Harwood, C. S. (2005). Proc. Natl Acad. Sci. USA, 102, 14422–14427. [DOI] [PMC free article] [PubMed]
- Iwama, T., Ito, Y., Aoki, H., Sakamoto, H., Yamagata, S., Kawai, K. & Kawagishi, I. (2006). J. Biol. Chem. 281, 17727–17735. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lacal, J., Alfonso, C., Liu, X., Parales, R. E., Morel, B., Conejero-Lara, F., Rivas, G., Duque, E., Ramos, J. L. & Krell, T. (2010). J. Biol. Chem. 285, 23126–23136. [DOI] [PMC free article] [PubMed]
- Lacal, J., García-Fontana, C., Callejo-García, C., Ramos, J. L. & Krell, T. (2011). J. Mol. Recognit. 24, 378–385. [DOI] [PubMed]
- Lacal, J., García-Fontana, C., Muñoz Martínez, F., Ramos, J. L. & Krell, T. (2010). Environ. Microbiol. 12, 2873–2884. [DOI] [PubMed]
- Milburn, M. V., Privé, G. G., Milligan, D. L., Scott, W. G., Yeh, J., Jancarik, J., Koshland, D. E. & Kim, S.-H. (1991). Science, 254, 1342–1347. [DOI] [PubMed]
- Morris, R. J., Zwart, P. H., Cohen, S., Fernandez, F. J., Kakaris, M., Kirillova, O., Vonrhein, C., Perrakis, A. & Lamzin, V. S. (2004). J. Synchrotron Rad. 11, 56–59. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Podust, L. M., Ioanoviciu, A. & Ortiz de Montellano, P. R. (2008). Biochemistry, 47, 12523–12531. [DOI] [PMC free article] [PubMed]
- Pokkuluri, P. R., Pessanha, M., Londer, Y. Y., Wood, S. J., Duke, N. E., Wilton, R., Catarino, T., Salgueiro, C. A. & Schiffer, M. (2008). J. Mol. Biol. 377, 1498–1517. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.