Interleukin-23 (IL-23), a member of the IL-12 family, is a heterodimeric cytokine composed of p19 and p40 subunits. Human p19 and p40 subunits were cloned and coexpressed in N-acetylglucosaminyltransferase I-negative 293S cells. The glycosylated human IL-23 was purified and crystallized by the hanging-drop vapour-diffusion method.
Keywords: interleukin-23, cytokines, glycosylation
Abstract
Interleukin-23 (IL-23), a member of the IL-12 family, is a heterodimeric cytokine composed of p19 and p40 subunits. IL-23 plays crucial roles in the activation, proliferation and survival of IL-17-producing helper T cells which induce various autoimmune diseases. Human p19 and p40 subunits were cloned and coexpressed in N-acetylglucosaminyltransferase I-negative 293S cells, which produce high-mannose-type glycosylated proteins in order to diminish the heterogeneity of modified N-linked glycans. The glycosylated human IL-23 was purified and crystallized by the hanging-drop vapour-diffusion method. X-ray diffraction data were then collected to 2.6 Å resolution. The crystal belonged to space group P61 or P65, with unit-cell parameters a = b = 108.94, c = 83.79 Å, γ = 120°. Assuming that the crystal contains one molecule per asymmetric unit, the calculated Matthews coefficient was 2.69 Å3 Da−1, with a solvent content of 54.2%. The structure was determined by the molecular-replacement method, with an initial R factor of 52.6%. After subsequent rigid-body and positional refinement, the R work and R free values decreased to 31.4% and 38.7%, respectively.
1. Introduction
The IL-12 cytokine family are key regulators of the activation, differentiation, proliferation and survival of T cells and dysregulation causes susceptibility to pathogen infections, autoimmune diseases, chronic inflammation and tumour growth (Trinchieri, 2003 ▶; Hunter, 2005 ▶; Kastelein et al., 2007 ▶). This family differs from other members of the helical cytokines in consisting of heterodimeric proteins (Kaiser et al., 2004 ▶; Goriely et al., 2008 ▶).
IL-23, a member of the IL-12 cytokine family, is composed of a p40 subunit (Genbank accession No. NM_002187), which is shared with IL-12, and a p19 subunit (Genbank accession No. NM_016584), with an approximate molecular weight of 60 kDa (Oppmann et al., 2000 ▶). p19 was computationally identified as a member of the IL-6 family, which have an up–up–down–down four-helix bundle topology. In contrast, the p40 subunit consists of one immunoglobulin-like domain and a cytokine-binding homology region. These subunits are linked by a disulfide bridge, similar to the p35 and p40 subunits of IL-12. IL-23 is produced by activated dendritic cells and macrophages in response to microorganisms (Smits et al., 2004 ▶; Middleton et al., 2006 ▶). The secreted IL-23 binds to IL-23 receptor (IL-23R) and IL-12 receptor β1 (IL-12Rβ1) on T cells and subsequently activates the Janus kinase (JAK)/signalling transducer and activator of transcription (STAT) pathway in the intracellular region (Parham et al., 2002 ▶). After the phosphorylated STATs form homodimers or heterodimers in the cytoplasm, they translocate to the nucleus and modulate the expression of target genes (Ihle et al., 1997 ▶; Heim, 1999 ▶; Kisseleva et al., 2002 ▶).
Initially, IL-23 was considered to be required for differentiation of IL-17-producing helper T (Th17) cells from naïve T cells (Aggarwal et al., 2003 ▶; Harrington et al., 2005 ▶). However, further investigations revealed that IL-23 is essential for proliferation and survival rather than differentiation of Th17 cells (Veldhoen et al., 2006 ▶; Mangan et al., 2006 ▶; Bettelli et al., 2006 ▶; Zhou et al., 2007 ▶). In experiments using knockout mice deficient in either the p19 or p40 subunit, it was elucidated that IL-23 is involved in several autoimmune diseases such as experimental autoimmune encephalomyelitis (EAE), collagen-induced arthritis (CIA) and inflammatory bowel disease (IBD) (Gyülvészi et al., 2009 ▶; Murphy et al., 2003 ▶; Ahern et al., 2008 ▶). In addition, IL-23 negatively regulates cytotoxic T-cell infiltration into malignant lesions, so that tumour cells escape immune surveillance and elimination (Langowski et al., 2006 ▶). Furthermore, IL-23 induces the production of IL-1β and tumour necrosis factor (TNF), which are important regulatory cytokines for innate immune responses by peritoneal macrophages (Cua et al., 2003 ▶). Taken together, IL-23 plays pivotal roles in both innate and adaptive immunity and is a promising therapeutic target for various autoimmune inflammatory diseases and tumours.
Almost all of the key molecules in the immune system are glycosylated. Eukaryotic cells are useful tools for the expression of such mammalian proteins which are post-transcriptionally modified. However, it is difficult to obtain crystals from glycosylated proteins with diffraction quality suitable for X-ray structural analysis owing to the heterogeneity of the modified glycans. Therefore, the sugar moieties are usually removed from the protein for structural analysis. Although crystal structures of IL-23 have been solved previously by two groups, the modified glycans of these proteins were enzymatically or genetically removed (Lupardus & Garcia, 2008 ▶; Beyer et al., 2008 ▶).
Here, we report the expression in mammalian cells, purification, crystallization and data collection to 2.6 Å resolution of the glycosylated form of human IL-23 (hIL-23).
2. Experimental methods
2.1. Cloning
Genes encoding the full-length human p19 (1–170) and p40 (1–306) subunits with their own signal peptides were amplified by PCR using Human Leukocyte QUICK-Clone cDNA (Clontech) with appropriate primers (5′-ATGCTGGGGAGCAGAGCTGTAATGC-3′ and 5′-GCCTTTAGGGACTCAGGGTTGCTG-3′ for p19 and 5′-AAGATGTGTCACCAGCAGTTGGTCATCTC-3′ and 5′-ATCAGAACCTAACTGCAGGGCACAGATG-3′ followed by 5′-TGTTTCAGGGCCATTGGACTCTCCGTC-3′ and 5′-GCCCATGGCAACTTGAGAGCTGGAAAATC-3′ for p40) and the PCR products were cloned into pCR4Blunt-TOPO vector (Invitrogen). To introduce a C-terminal hexahistidine (His6) tag, p19 was amplified by PCR (5′-TATATAGGTACCATGCTGGGGAGCAGAG-3′ and 5′-ATATATCTCGAGGGGACTCAGGGTTGCTG-3′) and the PCR product was subcloned into the KpnI/XhoI restriction-enzyme sites of pET30b(+) vector (Novagen). For construction of the mammalian cell expression plasmid, the p19-His and p40 genes were amplified by PCR (the sense primer that was used to introduce the His6 tag and 5′-TATATATGCGGCCGCTCAGTGGTGGTG-3′ for p19-His, 5′-GTACATGGTACCATGTGTCACCAGCAGTTG-3′ and 5′-TATATATGCGGCCGCTCAGTGGTGGTG-3′ for p40) and each PCR product was subcloned into the KpnI/NotI restriction-enzyme sites of pLEXm vector (Aricescu et al., 2006 ▶). The final clones were verified by sequencing.
2.2. Expression and purification
N-Acetylglucosaminyltransferase I-negative (GnTI−) 293S cells (Reeves et al., 2002 ▶) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM low glucose, Wako) supplemented with 10% FBS (Wako) at 310 K using 150 mm tissue-culture dishes. These cells are able to modify high-mannose-type glycans instead of the complex forms which interfere in the formation of diffraction-quality glycoprotein crystals (Crispin et al., 2006 ▶; Chang et al., 2007 ▶). For large-scale protein expression, 293S GnTI− cells were cultured in 1750 cm2 polystyrene roller bottles (Corning) in 350 ml DMEM supplemented with 10% FBS. The cells grown in two 150 mm dishes were transferred into one roller bottle. Transfection was performed using polyethyleneimine (PEI; Sigma–Aldrich) with 250 µg of each plasmid when the cells reached about 90% confluency (Aricescu et al., 2006 ▶). 250 µg of each plasmid was added to 50 ml serum-free medium and mixed briefly by vortexing after the addition of 1 ml PEI (1 mg ml−1, pH 7.0). This mixture was incubated for 30 min at 310 K, poured into a roller bottle and incubated at 310 K. 36 h later, the culture medium was collected, centrifuged and filtrated using a filter paper to remove cell debris. The filtrated medium was applied onto an Ni–NTA Superflow column (Qiagen). The column was washed with 25 column volumes of washing buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl). The protein was eluted with 1–2 column volumes of elution buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 100 mM imidazole). The eluate containing p19 and p40 (IL-23) was concentrated to 5 ml and then diluted sevenfold with dilution buffer (20 mM Tris–HCl pH 8.0). hIL-23 was then applied onto a RESOURCE Q anion-exchange column (GE Healthcare). The proteins were eluted between 170 and 200 mM NaCl using a 0–220 mM linear gradient over 22 column volumes. For further purification, the eluate was concentrated and applied onto a HiLoad 16/60 Superdex 200 preparation-grade column (GE Healthcare) equilibrated with equilibration buffer (20 mM HEPES pH 7.4, 150 mM NaCl). The eluted hIL-23 had a molecular weight approximately corresponding to that of a heterodimer consisting of p19 and p40 (∼50 kDa). Purified proteins were collected and concentrated to 5 mg ml−1 for crystallization. 1.4 mg IL-23 was yielded from 1 l culture.
2.3. Crystallization
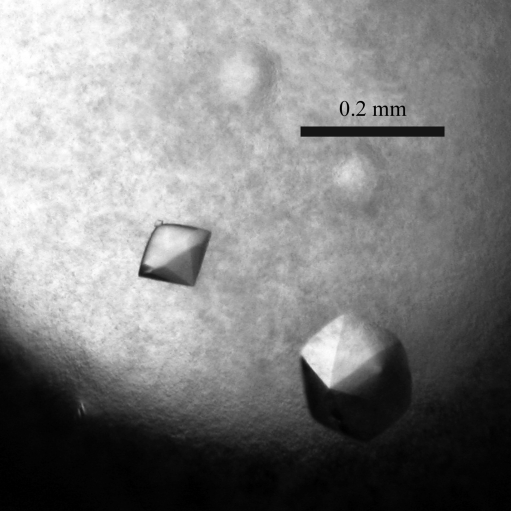
hIL-23 was initially crystallized at 293 K by the hanging-drop vapour-diffusion method using the JCSG+ Suite (Qiagen). Droplets were formed by mixing equal volumes (0.5 µl) of protein solution and reservoir solution. After several days, small crystals appeared using 20%(w/v) PEG 1000, 0.2 M lithium sulfate, 0.1 M phosphate–citrate pH 4.2. Based on these conditions, the pH and the concentrations of protein and PEG were optimized and the droplet size was increased (2 µl protein solution and the same volume of reservoir solution). As a result, large good-shaped crystals suitable for X-ray diffraction experiments were obtained using 6 mg ml−1 hIL-23 and 18%(w/v) PEG 1000, 0.2 M lithium sulfate, 0.1 M phosphate–citrate pH 4.2 (Fig. 1 ▶).
Figure 1.
Crystals of the glycosylated form of hIL-23 (p19/p40). The dimensions of the crystals are 0.15 × 0.15 × 0.15 mm.
2.4. Diffraction data collection and processing
The crystals were transferred into a cryoprotectant composed of reservoir solution with 30%(v/v) glycerol and flash-cooled in a nitrogen-gas stream at 100 K. Diffraction data collection was performed on beamline NW12A of Photon Factory Advanced Ring (PF-AR; Tsukuba, Japan) at 100 K. The best data were collected to a resolution of 2.6 Å using X-rays of wavelength 1.000 Å. All data were processed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The statistics of data collection are shown in Table 1 ▶. The structure was solved by the molecular-replacement method using the program MOLREP (Vagin & Teplyakov, 2010 ▶) with the deglycosylated form of hIL-23 (molecules A and C of PDB entry 3duh; Lupardus & Garcia, 2008 ▶) as a search model.
Table 1. Data-collection statistics.
Values in parentheses are for the outer shell.
| Beamline | PF-AR NW12A |
| Wavelength (Å) | 1.000 |
| Detector | ADSC Quantum 210 CCD |
| Exposure time (s) | 10 |
| Crystal-to-detector distance (mm) | 301 |
| Oscillation angle (°) | 1.0 |
| Sweep angle (°) | 0–360 |
| Temperature (K) | 100 |
| Resolution (Å) | 30.0–2.6 (2.69–2.60) |
| Space group | P61 or P65 |
| Unit-cell parameters (Å, °) | a = 108.94, b = 108.94, c = 83.79, γ = 120 |
| No. of molecules in unit cell (Z) | 6 |
| Matthews coefficient VM (Å3 Da−1) | 2.69 |
| Solvent content (%) | 54.24 |
| No. of observed reflections | 262611 |
| No. of unique reflections | 16401 (1217) |
| Multiplicity | 16.0 (5.5) |
| Completeness (%) | 93.5 (70.0) |
| Rmerge† (%) | 6.8 (45.3) |
| 〈I/σ(I)〉 | 44.3 (1.9) |
R
merge = 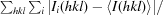
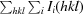 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the mean value of Ii(hkl).
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the mean value of Ii(hkl).
3. Results and discussion
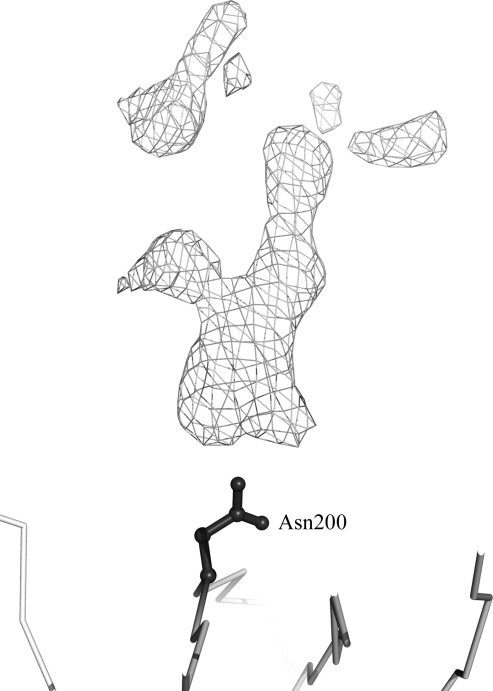
We cloned the p19 and p40 subunits of hIL-23 and coexpressed both subunits in 293S GnTI− cells. The protein was purified using an Ni–NTA Superflow column (Qiagen), a RESOURCE Q anion-exchange column (GE Healthcare) and a HiLoad 16/60 Superdex 200 preparation-grade column (GE Healthcare) and then concentrated to 6 mg ml−1 for crystallization. Crystals were obtained using 18%(w/v) PEG 1000, 0.2 M lithium sulfate, 0.1 M phosphate–citrate pH 4.2 and diffracted to 2.6 Å resolution. The hIL-23 crystal belonged to space group P61 or P65, with unit-cell parameters a = b = 108.94, c = 83.79 Å, γ = 120°. The calculated Matthews coefficient (Matthews, 1968 ▶) was 2.69 Å3 Da−1, with a solvent content of 54.2%, for one molecule in the asymmetric unit. The deglycosylated form of hIL-23 (Lupardus & Garcia, 2008 ▶) was used as a search model for molecular replacement using the program MOLREP (Vagin & Teplyakov, 2010 ▶) and provided a solution with an initial R factor of 52.6%. Subsequent rigid-body refinement was carried out with four rigid groups (p19 and three domains of the p40 subunit) and the individual atom positions of hIL-23 were then refined using the program CNS (Brünger et al., 1998 ▶). The R work and R free values after initial minimization refinement were 31.4% and 38.7%, respectively. Electron density for the modified sugars was observed around Asn200 of the p40 subunit (Fig. 2 ▶). It is likely that the sugar moiety effects the domain movements which we have seen in our structure. Further refinement of the structure is in progress and will be published elsewhere.
Figure 2.
Electron density of the modified glycan at Asn200. The Cα backbone atoms and Asn200 are represented in grey and as a black ball-and-stick representation, respectively. A difference Fourier map contoured at the 2.5σ level for the modified glycan at Asn200 is shown as a grey mesh.
Acknowledgments
We thank Drs Matsugaki and Yamada and Professor Wakatsuki for data collection at the Photon Factory. This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sport and Technology of Japan.
References
- Aggarwal, S., Ghilardi, N., Xie, M.-H., de Sauvage, F. J. & Gurney, A. L. (2003). J. Biol. Chem. 278, 1910–1914. [DOI] [PubMed]
- Ahern, P. P., Izcue, A., Maloy, K. J. & Powrie, F. (2008). Immunol. Rev. 226, 147–159. [DOI] [PubMed]
- Aricescu, A. R., Lu, W. & Jones, E. Y. (2006). Acta Cryst. D62, 1243–1250. [DOI] [PubMed]
- Bettelli, E., Carrier, Y., Gao, W., Korn, T., Strom, T. B., Oukka, M., Weiner, H. L. & Kuchroo, V. K. (2006). Nature (London), 441, 235–238. [DOI] [PubMed]
- Beyer, B. M., Ingram, R., Ramanathan, L., Reichert, P., Le, H. V., Madison, V. & Orth, P. (2008). J. Mol. Biol. 382, 942–955. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chang, V. T., Crispin, M., Aricescu, A. R., Harvey, D. J., Nettleship, J. E., Fennelly, J. A., Yu, C., Boles, K. S., Evans, E. J., Stuart, D. I., Dwek, R. A., Jones, E. Y., Owens, R. J. & Davis, S. J. (2007). Structure, 15, 267–273. [DOI] [PMC free article] [PubMed]
- Crispin, M., Harvey, D. J., Chang, V. T., Yu, C., Aricescu, A. R., Jones, E. Y., Davis, S. J., Dwek, R. A. & Rudd, P. M. (2006). Glycobiology, 16, 748–756. [DOI] [PubMed]
- Cua, D. J. et al. (2003). Nature (London), 421, 744–748.
- Goriely, S., Neurath, M. F. & Goldman, M. (2008). Nature Rev. Immunol. 8, 81–86. [DOI] [PubMed]
- Gyülvészi, G., Haak, S. & Becher, B. (2009). Eur. J. Immunol. 39, 1864–1869. [DOI] [PubMed]
- Harrington, L. E., Hatton, R. D., Mangan, P. R., Turner, H., Murphy, T. L., Murphy, K. M. & Weaver, C. T. (2005). Nature Immunol. 6, 1123–1132. [DOI] [PubMed]
- Heim, M. H. (1999). J. Recept. Signal Transduct. Res. 19, 75–120. [DOI] [PubMed]
- Hunter, C. A. (2005). Nature Rev. Immunol. 5, 521–531. [DOI] [PubMed]
- Ihle, J. N., Nosaka, T., Thierfelder, W., Quelle, F. W. & Shimoda, K. (1997). Stem Cells, 15(S2), 105–112. [DOI] [PubMed]
- Kaiser, P., Rothwell, L., Avery, S. & Balu, S. (2004). Dev. Comput. Immunol. 28, 375–394. [DOI] [PubMed]
- Kastelein, R. A., Hunter, C. A. & Cua, D. J. (2007). Annu. Rev. Immunol. 25, 221–242. [DOI] [PubMed]
- Kisseleva, T., Bhattacharya, S., Braunstein, J. & Schindler, C. W. (2002). Gene, 285, 1–24. [DOI] [PubMed]
- Langowski, J. L., Zhang, X., Wu, L., Mattson, J. D., Chen, T., Smith, K., Basham, B., McClanahan, T., Kastelein, R. A. & Oft, M. (2006). Nature (London), 442, 461–465. [DOI] [PubMed]
- Lupardus, P. J. & Garcia, K. C. (2008). J. Mol. Biol. 382, 931–941. [DOI] [PMC free article] [PubMed]
- Mangan, P. R., Harrington, L. E., O’Quinn, D. B., Helms, W. S., Bullard, D. C., Elson, C. O., Hatton, R. D., Wahl, S. M., Schoeb, T. R. & Weaver, C. T. (2006). Nature (London), 441, 231–234. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 499–501. [DOI] [PubMed]
- Middleton, M. K., Rubinstein, T. & Puré, E. (2006). J. Immunol. 176, 265–274. [DOI] [PubMed]
- Murphy, C. A., Langrish, C. L., Chen, Y., Blumenschein, W., McClanahan, T., Kastelein, R. A., Sedgwick, J. D. & Cua, D. J. (2003). J. Exp. Med. 198, 1951–1957. [DOI] [PMC free article] [PubMed]
- Oppmann, B. et al. (2000). Immunity, 13, 715–725. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Parham, C. et al. (2002). J. Immunol. 168, 5699–5708. [DOI] [PubMed]
- Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. (2002). Proc. Natl Acad. Sci. USA, 99, 13419–13424. [DOI] [PMC free article] [PubMed]
- Smits, H. H., van Beelen, A. J., Hessle, C., Westland, R., de Jong, E., Soeteman, E., Wold, A., Wierenga, E. A. & Kapsenberg, M. L. (2004). Eur. J. Immunol. 34, 1371–1380. [DOI] [PubMed]
- Trinchieri, G. (2003). Nature Rev. Immunol. 3, 133–146. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Veldhoen, M., Hocking, R. J., Atkins, C. J., Locksley, R. M. & Stockinger, B. (2006). Immunity, 24, 179–189. [DOI] [PubMed]
- Zhou, L., Ivanov, I. I., Spolski, R., Min, R., Shenderov, K., Egawa, T., Levy, D. E., Leonard, W. J. & Littman, D. R. (2007). Nature Immunol. 8, 967–974. [DOI] [PubMed]