Crystals of the SCAN domain of Zfp206 are tetragonal, belonging to space group I422 with unit-cell parameters a = 67.57, c = 87.54 Å and one molecule in the asymmetric unit, and diffract to 1.85 Å resolution.
Keywords: Zfp206, SCAN domain, transcription factors
Abstract
Zfp206 (also named Zscan10) is a transcription factor that plays an important role in maintaining the pluripotent state of embryonic stem cells. Zfp206 is a member of the SCAN-domain family of C2H2 zinc-finger transcription factors. The SCAN domain is a highly conserved motif of 84 residues which mediates the self-association of and heterodimerization between SCAN-domain family transcription factors. The SCAN domain may therefore be the key to the selective oligomerization of and may combinatorially enhance the regulatory versatility of C2H2 zinc fingers. This paper describes crystallization attempts with the SCAN domain of Zfp206 (Zfp206SCAN) and optimization strategies to obtain diffraction-quality crystals. The best diffracting crystal was grown in a solution consisting of 0.3 M ammonium sulfate, 0.1 M Tris–HCl pH 8.6, 25% PEG 3350, 0.1 M ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA) using the hanging-drop vapour-diffusion technique. Optimized crystals diffracted to 1.85 Å resolution and belonged to space group I422, with unit-cell parameters a = 67.57, c = 87.54 Å. A Matthews analysis indicated the presence of one Zfp206SCAN molecule per asymmetric unit.
1. Introduction
Zfp206 is a transcription factor that has been reported to be a pluripotency factor (Zhang et al., 2006 ▶; Wang, Kueh et al., 2007 ▶; Yu et al., 2009 ▶). Zfp206 is highly expressed in mouse and human undifferentiated embryonic stem cells (ESCs) but is repressed upon differentiation (Zhang et al., 2006 ▶). Overexpression of Zfp206 in ESCs could assist the cells to resist differentiation. Thus, Zfp206 is known to be a maintainer of pluripotency (Zhang et al., 2006 ▶; Wang, Kueh et al., 2007 ▶). In ESCs, Zfp206 was found to be directly regulated by other key pluripotency transcription factors such as Oct4 and Sox2 (Wang, Teh et al., 2007 ▶; Yu et al., 2009 ▶). The Zfp206 protein contains 14 C2H2 zinc fingers and one SCAN domain near the N-terminus. The SCAN domain of Zfp206 suggests that this protein may self-associate or form heterodimers with other family members.
The SCAN domain is a highly conserved leucine-rich (LeR) motif of 84 residues which was found at the N-terminal end of a subfamily of zinc-finger transcription factors. The SCAN domain was originally identified and named after the first letters of some previously discovered family members [SREZBP, Ctfin51, AW-1 (ZNF174) and Number 18 cDNA; Williams et al., 1995 ▶; Pengue et al., 1994 ▶]. There are 63 human and 40 mouse SCAN-family members and the domain is not found outside the mammalian lineage (Edelstein & Collins, 2005 ▶). The function of the SCAN domain appears to be solely to mediate homodimerization or selective heterodimerization with other SCAN-family members; it has no proven transcriptional activation or repression activity (Schumacher et al., 2000 ▶; Williams et al., 1999 ▶; Stone et al., 2002 ▶). Previous NMR structural studies of the SCAN domains of ZNF174 and MZF1 (Ivanov et al., 2005 ▶; Peterson et al., 2006 ▶) and the crystal structure of the SCAN domain of ZNF24 (PDB entry 3lhr; Center for Eukaryotic Structural Genomics, unpublished work) showed that this domain exists as a domain-swapped homodimer. Based on previous studies, SCAN proteins can be broadly classified as (i) exclusive homodimers, (ii) exclusive heterodimers or (iii) homodimerization and heterodimerization generalists (Schumacher et al., 2000 ▶; Williams et al., 1999 ▶; Stone et al., 2002 ▶). Therefore, homodimeric or heterodimeric structures of SCAN domains will provide insights into the folding topology and specificity determinants for SCAN dimerization. This will aid efforts to understand combinatorial transcriptional networks involving SCAN-domain C2H2 zinc fingers and to modulate their regulatory activity. It was found in our in vitro MBP pull-down assay that the SCAN domain of Zfp206 interacts with the SCAN domain of MZF1 and the SCAN domain of ZNF24, but not with the SCAN domain of ZNF174 (unpublished work). To understand the structural features of the SCAN domain of Zfp206 that favour interaction with its partners, we crystallized and attempted to solve the structure of the SCAN domain of Zfp206.
2. Experimental procedures
2.1. Cloning methods
The SCAN domain of mouse Zfp206 (amino acids 36–128) was amplified from the Zfp206 cDNA (IMAGE clone 30006755) by PCR using the primers 5′-ggggacaagtttgtacaaaaaagcaggcttcgaaaacctgtatttt cagggcAGGCCTAGGCCTGAGGTGGCC-3′ and 5′-ggggaccactttgtacaagaaagctgggttTTACATGTGGCTGATGTCTCTGGG-3′ containing attB sites (underlined) and a tobacco etch virus (TEV) protease recognition and cleavage site (bold) preceding the coding sequence. The PCR product was introduced into the Gateway entry vector pDONR221 (Invitrogen). The resulting entry vector pENTR-Zfp206SCAN was verified by sequencing. The insert was then introduced into the destination vector pDEST-HisMBP (Nallamsetty et al., 2005 ▶) by a Gateway LR reaction (Invitrogen), resulting in the expression plasmid pDEST-HisMBP-Zfp206SCAN encoding Zfp206SCAN with an N-terminal HisMBP tag.
2.2. Protein expression and purification
The pDEST-HisMBP-Zfp206SCAN expression plasmid was transformed into Escherichia coli BL21 (DE3) cells (Invitrogen). The cells transformed with pDEST-HisMBP-Zfp206SCAN were grown overnight in 200 ml Terrific Broth (TB) medium supplemented with 100 µg ml−1 ampicillin at 310 K. The overnight culture was used to inoculate 6 l TB medium containing 100 µg ml−1 ampicillin. The cells were grown at 310 K to an OD600 of 0.6 and isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.5 mM. The cells were grown overnight at 291 K and harvested by centrifugation at 6000g for 5 min. The cell pellet was suspended in lysis buffer (50 mM HEPES pH 7.3, 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol) and sonicated at Amp 30%, pulse 6 s for 20 min (Fisher Scientific Model 500 Sonic Dismembrator). The extract was clarified by centrifugation for 1 h at 25 000 rev min−1 at 277 K. The supernatant was loaded onto 10 ml amylose resin (New England Biolabs). The amylose resin was further washed with 50 ml lysis buffer. The bound HisMBP-Zfp206SCAN protein was eluted with elution buffer (50 mM HEPES pH 7.3, 100 mM NaCl, 10 mM maltose, 1 mM EDTA, 10 mM β-mercaptoethanol). The HisMBP tag was removed from the Zfp206SCAN protein by TEV protease digestion [1:50(w:w) ratio of TEV to HisMBP-Zfp206SCAN] at 277 K overnight. After TEV digestion, a non-native Gly residue was retained on the N-terminus of Zfp206SCAN. The Zfp206SCAN protein was further purified using a 6 ml Resource S cation-exchange column (GE Healthcare). The column was pre-equilibrated with buffer A (50 mM HEPES pH 7.3, 100 mM NaCl) and then eluted by gradually increasing the NaCl concentration from 100 mM to 1 M. Finally, the Zfp206SCAN protein was loaded onto a Sephacryl S-200 16/60 gel-filtration column (GE Healthcare) pre-equilibrated with buffer A. All column-chromatography purifications were performed using an ÄKTAxpress system (GE Healthcare). The fractions containing Zfp206SCAN were pooled and concentrated to 6 mg ml−1 in 50 mM HEPES pH 7.3, 100 mM NaCl for high-throughput crystallization screening or to 15 mg ml−1 for final optimization. The protein was stored at 277 K.
2.3. Crystallization of Zfp206SCAN
The high-throughput crystallization screen was set up with purified Zfp206SCAN (6 mg ml−1) using the sitting-drop vapour-diffusion technique. Crystallization-condition screening was aided by a liquid-dispensing robot (Innovadyne). 200 nl protein solution and 200 nl reservoir solution were mixed and equilibrated over a 50 µl reservoir using commercial crystallization screens from Hampton Research and Qiagen. Crystallization hits were manually optimized in a hanging-drop setting. 1 µl Zfp206SCAN protein (15 mg ml−1) mixed with 1 µl reservoir solution was equilibrated over 600 µl reservoir solution. Optimized crystals were transferred into reservoir buffer containing 10% glycerol as a cryoprotectant before subjecting them to a stream of liquid nitrogen for X-ray diffraction studies. Mercury-derivatived crystals were prepared by soaking crystals in the reservoir solution containing 10 mM HgCl2; they were then soaked in reservoir solution containing 10% glycerol and flash-frozen directly in liquid nitrogen.
2.4. X-ray data collection and processing
Initial X-ray diffraction data collection was performed using a Rigaku FR-E SuperBright X-ray generator with a Cu target, VariMax-HR optics, an R-AXIS IV++ imaging-plate detector and an Xstream 2000 low-temperature system. The data-collection procedure was controlled by CrystalClear software (Rigaku). After further optimization of the crystallization conditions, a native data set was collected to 1.85 Å resolution on the X29 beamline at the Macromolecular Crystallography Research Resource (PXRR, USA). A 2.2 Å resolution data set from a mercury-derivative crystal was collected and processed using the same protocol.
3. Results and discussion
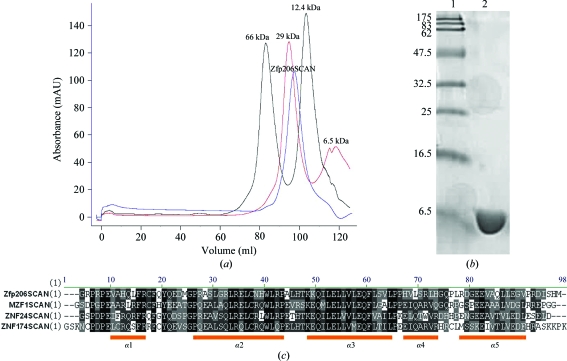
The Zfp206SCAN protein could be expressed and purified from E. coli in a soluble form with a yield of 1 mg pure protein per litre of bacterial expression culture. The Zfp206SCAN protein eluted from the final Sephacryl S-200 gel-filtration column as a single symmetric peak corresponding to the molecular mass of the dimeric form of the protein (∼21 kDa; Fig. 1 ▶ a). SDS–PAGE analysis indicated >95% purity after the final purification step (Fig. 1 ▶ b). Multiple sequence alignments of Zfp206SCAN with ZNF174SCAN (Ivanov et al., 2005 ▶), MZF1SCAN (Peterson et al., 2006 ▶) and ZNF24SCAN (PDB entry 3lhr; Center for Eukaryotic Structural Genomics, unpublished work) were performed using ClustalW (Thompson et al., 1994 ▶; Fig. 1 ▶ c). Helix 2 and helix 3 showed the highest sequence conservation of the five helices. Sequence variations could be observed in helix 1, 4 and 5.
Figure 1.
(a) Elution profile of Zfp206SCAN (indicated by the arrow) from a Sephacryl S-200 gel-filtration column calibrated with molecular-mass standards. Zfp206SCAN elutes as a single symmetric peak corresponding to the molecular mass of the dimeric form of the protein (∼21 kDa; blue). The molecular-mass standards albumin, bovine serum (66 kDa) and carbonic anhydrase from bovine erythrocytes (29 kDa) are shown in black. The molecular-mass standards cytochrome c from horse heart (12.4 kDa) and aprotinin from bovine lung (6.5 kDa) are shown in red. (b) Purified Zfp206SCAN on a 15% SDS–PAGE gel. The 15% SDS–PAGE gel shows purified Zfp206SCAN protein with purity >98% (lane 2) and molecular-mass standards (lane 1; labelled in kDa). (c) Multiple sequence alignments of Zfp206SCAN with other SCAN proteins from the PDB using ClustalW. Secondary-structure elements of SCAN domains are shown under the alignments as orange blocks.
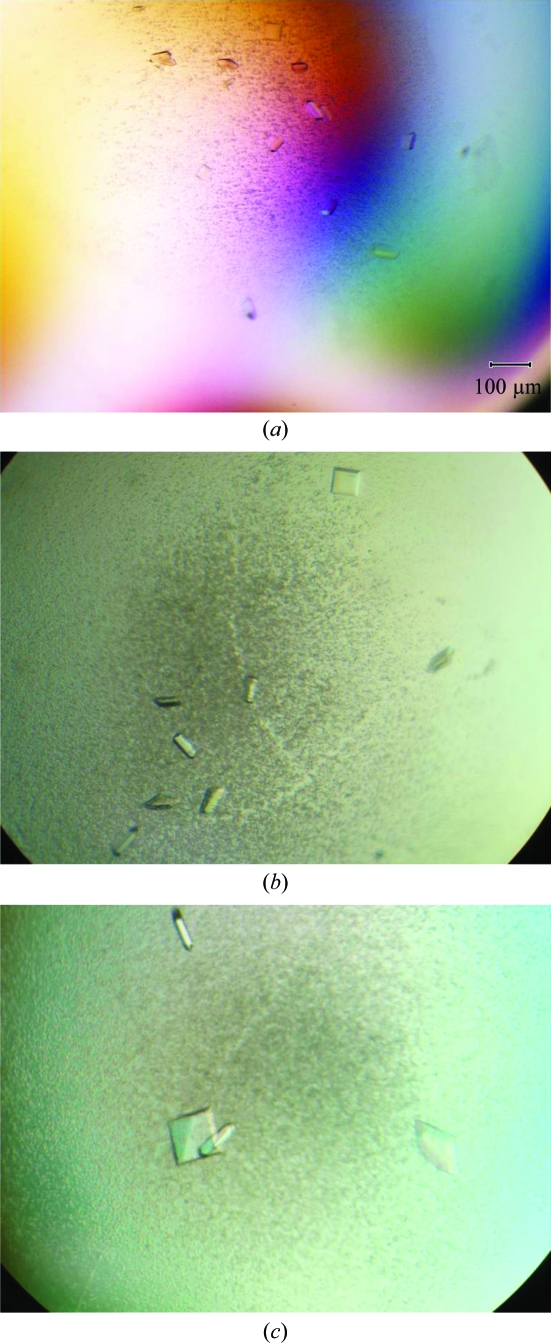
Initial crystal hits were obtained at 291 K with a reservoir buffer consisting of 0.2 M ammonium sulfate, 0.1 M Tris–HCl pH 8.6, 25% PEG 3350 (Fig. 2 ▶ a). The optimized condition was 0.3 M ammonium sulfate, 0.1 M Tris–HCl pH 8.6, 25% PEG 3350 with 15 mg ml−1 Zfp206SCAN protein at 291 K. An additive screen (Hampton Research) was conducted to further improve the crystals. The addition of 0.1 M EDTA was found to improve the crystal growth of Zfp206SCAN. Further optimizations showed that a 2:1:1(v:v:v) ratio of Zfp206SCAN solution, reservoir solution and additive resulted in larger and more homogeneous crystals (Fig. 2 ▶ b).
Figure 2.
Zfp206SCAN crystals. (a) Microcrystals grown using 0.2 M ammonium sulfate, 0.1 M Tris–HCl pH 8.6, 25% PEG 3350 at 291 K. (b) Crystals grown in the presence of 0.3 M ammonium sulfate, 0.1 M Tris–HCl pH 8.6, 25% PEG 3350 supplemented with 0.1 M EDTA at 291 K. (c) Tetragonal crystals obtained by the hanging-drop controlled vapour-diffusion technique using the same condition as in (b) with an oil barrier.
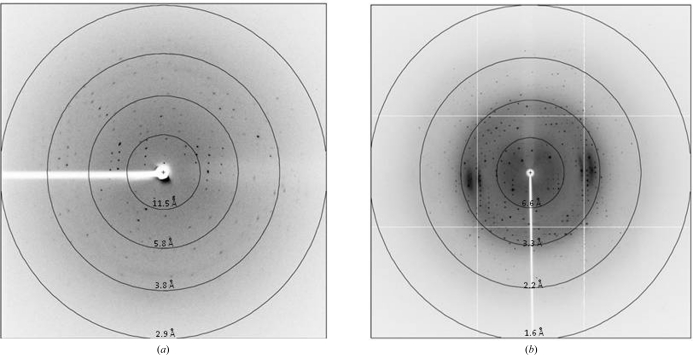
However, crystals of Zfp206SCAN grown in the above conditions only diffracted to 3.2 Å resolution (Fig. 3 ▶ a). Additionally, data processing was hampered by overlapping lattices and high mosaicity. To solve these problems we attempted to control the vapour-diffusion rate using oils (Chayen, 1997 ▶). Using a hanging-drop vapour-diffusion setup for Zfp206SCAN under the optimized condition described previously, various volumes of a mixture of paraffin and silicone oil [50:50%(v/v)] were applied to cover 600 µl reservoir solution (after the protein drop has been mixed with reservoir solution and additive reagent, thus preventing oil from entering the drop). The mixture of paraffin and silicone oil acted as a barrier to vapour diffusion between the reservoir and the drop and thus controlled the rate of vapour diffusion as a function of the thickness of the oil layer. Volumes between 60 and 420 µl of the mixture of paraffin and silicone oil were evaluated. Using an oil volume of 60 µl (10% of the volume of the reservoir solution) single crystals suitable for X-ray diffraction studies were obtained (Fig. 2 ▶ c).
Figure 3.
Diffraction images of Zfp206SCAN crystals. (a) Diffraction image of crystals grown in the presence of 0.3 M ammonium sulfate, 0.1 M Tris–HCl pH 8.6, 25% PEG 3350 supplemented with 0.1 M EDTA at 291 K (3.2 Å). (b) Diffraction image of tetragonal crystals obtained by the hanging-drop controlled vapour-diffusion technique using the same reservoir solution as in (a) with an oil barrier (1.85 Å resolution).
A native data set for Zfp206SCAN to 1.85 Å resolution collected on the X29 beamline was processed using the iMOSFLM software (Battye et al., 2011 ▶; Fig. 3 ▶ b). The data set was complete with good merging statistics. Data processing revealed that the crystal belonged to the I-centred tetragonal space group I422, with unit-cell parameters a = 67.57, c = 87.54 Å. A summary of the data-set statistics is given in Table 1 ▶. The value of the Matthews coefficient (Matthews, 1968 ▶) was 2.32 Å3 Da−1 for one molecule in the asymmetric unit and the estimated solvent content was 48.28%. However, Zfp206SCAN was observed to be a dimer in solution (Fig. 1 ▶). There were no peaks corresponding to noncrystallographic symmetry in the resultant map (data not shown) when a self-rotation was performed. The biological unit is therefore likely to be arranged around a crystallographic twofold axis. Molecular-replacement trials (Chai et al., 2003 ▶) will be used to determine the structure of Zfp206SCAN based on a model derived from the MZF1SCAN structure (PDB entry 2fi2; Peterson et al., 2006 ▶). The mercury derivative belonged to the same space group I422. The statistics for this data set are also included in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Native | Mercury derivative | |
|---|---|---|
| Wavelength (Å) | 1.081 | 1.000 |
| Space group | I422 | I422 |
| Unit-cell parameters (Å) | ||
| a | 67.57 | 67.78 |
| c | 87.54 | 87.46 |
| No. of observed reflections | 136740 | 162986 |
| No. of unique reflections | 8995 | 5459 |
| Resolution of data set (Å) | 50–1.85 (1.92–1.85) | 50–2.20 (2.28–2.20) |
| Multiplicity | 15.2 (14.1) | 29.9 (27.5) |
| Completeness (%) | 100 (100) | 100 (100) |
| Mean I/σ(I) | 47.5 (6.0) | 41.1 (6.7) |
| Rmerge† (%) | 4.5 (46.8) | 8.5 (61.5) |
R
merge = 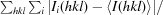
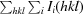 , where Ii(hkl) and 〈I(hkl)〉 are the intensity of measurement i and the mean intensity for the reflection with indices hkl, respectively.
, where Ii(hkl) and 〈I(hkl)〉 are the intensity of measurement i and the mean intensity for the reflection with indices hkl, respectively.
Acknowledgments
This work was supported by the Agency for Science, Technology and Research (A*STAR), Singapore. We are grateful to Dr Robert Robinson (IMCB, Singapore) and Dr Wang Chern Hoe for helping us with diffraction data collection. We also thank Dr Howard Robinson for access to the X29 beamline at the Macromolecular Crystallography Research Resource (PXRR, USA) and for providing assistance in data collection. The PXRR is supported by the NIH’s National Center for Research Resources and the DOE Office of Biological and Environmental Research. We thank the National Synchrotron Light Source (NSLS) for access to the X29 beamline. We would also like to thank Dr Ralf Jauch for critical comments on the manuscript.
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Chai, J., Wu, J. W., Yan, N., Massagué, J., Pavletich, N. P. & Shi, Y. (2003). J. Biol. Chem. 278, 20327–20331. [DOI] [PubMed]
- Chayen, N. E. (1997). J. Appl. Cryst. 30, 198–202.
- Edelstein, L. C. & Collins, T. (2005). Gene, 359, 1–17. [DOI] [PubMed]
- Ivanov, D., Stone, J. R., Maki, J. L., Collins, T. & Wagner, G. (2005). Mol. Cell, 17, 137–143. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Nallamsetty, S., Austin, B. P., Penrose, K. J. & Waugh, D. S. (2005). Protein Sci. 14, 2964–2971. [DOI] [PMC free article] [PubMed]
- Pengue, G., Calabrò, V., Bartoli, P. C., Pagliuca, A. & Lania, L. (1994). Nucleic Acids Res. 22, 2908–2914. [DOI] [PMC free article] [PubMed]
- Peterson, F. C., Hayes, P. L., Waltner, J. K., Heisner, A. K., Jensen, D. R., Sander, T. L. & Volkman, B. F. (2006). J. Mol. Biol. 363, 137–147. [DOI] [PMC free article] [PubMed]
- Schumacher, C., Wang, H., Honer, C., Ding, W., Koehn, J., Lawrence, Q., Coulis, C. M., Wang, L. L., Ballinger, D., Bowen, B. R. & Wagner, S. (2000). J. Biol. Chem. 275, 17173–17179. [DOI] [PubMed]
- Stone, J. R., Maki, J. L., Blacklow, S. C. & Collins, T. (2002). J. Biol. Chem. 277, 5448–5452. [DOI] [PMC free article] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Wang, Z.-X., Kueh, J. L. L., Teh, C. H.-L., Rossbach, M., Lim, L., Li, P., Wong, K.-Y., Lufkin, T., Robson, P. & Stanton, L. W. (2007). Stem Cells, 25, 2173–2182. [DOI] [PubMed]
- Wang, Z.-X., Teh, C. H.-L., Kueh, J. L. L., Lufkin, T., Robson, P. & Stanton, L. W. (2007). J. Biol. Chem. 282, 12822–12830. [DOI] [PubMed]
- Williams, A. J., Blacklow, S. C. & Collins, T. (1999). Mol. Cell. Biol. 19, 8526–8535. [DOI] [PMC free article] [PubMed]
- Williams, A. J., Khachigian, L. M., Shows, T. & Collins, T. (1995). J. Biol. Chem. 270, 22143–22152. [DOI] [PubMed]
- Yu, H., Kunarso, G., Hong, F. H. & Stanton, L. W. (2009). J. Biol. Chem. 284, 31327–31335. [DOI] [PMC free article] [PubMed]
- Zhang, W., Walker, E., Tamplin, O. J., Rossant, J., Stanford, W. L. & Hughes, T. R. (2006). Nucleic Acids Res. 34, 4780–4790. [DOI] [PMC free article] [PubMed]