A crystal of nurse shark β2-microglobulin diffracted to 2.3 Å resolution and belonged to space group P3221, with unit-cell parameters a = b = 88.230, c = 67.146 Å and two molecules per asymmetric unit. The Matthews coefficient V M was calculated to be about 3.28 Å3 Da−1, corresponding to 62.5% solvent content.
Keywords: nurse shark, β2-microglobulin
Abstract
β2-Microglobulin (β2m) is an essential subunit of the major histocompatibility complex (MHC) class I molecule that helps to stabilize the structure of peptide–MHC I (pMHC I). It is also one of the typical immunoglobulin superfamily (IgSF) molecules in the adaptive immune system (AIS). Sharks belong to the cartilaginous fish, which are the oldest jawed vertebrate ancestors with an AIS to exist in the world. Thus, the study of cartilaginous fish β2m would help in understanding the evolution of IgSF molecules. In order to demonstrate this, β2m from a cartilaginous fish, nurse shark (Ginglymostoma cirratum), was expressed, refolded, purified and crystallized. Diffraction data were collected to a resolution of 2.3 Å. The crystal belonged to space group P3221, with unit-cell parameters a = b = 88.230, c = 67.146 Å. The crystal structure contained two molecules in the asymmetric unit. The results will provide structural information for study of the evolution of β2m and IgSF in the AIS.
1. Introduction
β2-Microglobulin (β2m) is part of the peptide–major histocompatibility complex (MHC) class I–β2m (pMHC I) ternary complex and plays an important role in T-cell antigen receptor (TCR) recognition and induction of the cytotoxic T lymphocyte (CTL) immune response. In the pMHC I complex, β2m is noncovalently related to the α1, α2 and α3 domains of the MHC I molecule to stabilize the MHC I heavy chain and epitope peptide and maintains the complex assembly (Chen, Kshirsagar, et al., 2010 ▶; Machold et al., 1995 ▶). Although it is commonly accepted that B2M genes are conserved across species such as fish, birds and mammals, they have several differences (Chen, Gao et al., 2010 ▶). Specifically, the extracellular domain of β2ms in teleosts, amphibians and mammals are 97, 97 and 99 residues in length, respectively (Hao et al., 2006 ▶), whereas shark β2m contains only 96 amino acids. An alignment of the amino-acid sequences of the extracellular domain of β2ms shows that nurse shark β2m (ns-β2m) has 36.5–42.1% identity to those from bony fish, chicken, mouse and human. More importantly, β2m crystal structures have been determined from mammals to bony fish and all β2ms have been classified into the family of immunoglobulin superfamily (IgSF) constant (C) molecules based on their number of strands and folding topology (Chen, Gao et al., 2010 ▶).
Cartilaginous fish, including sharks, are the most primitive extant jawed vertebrates. Fossil evidence reveals that they arose in the Carboniferous period (362–290 million years ago). About 45 families of sharks and their relatives are known to have lived during the late Palaeozoic and Mesozoic eras (Bird et al., 2002 ▶). Remarkably, sharks possess an adaptive immune system (AIS) including β2m, MHC I and II, immunoglobulin (Ig), T-cell receptors (TCRs) and so on (Stanfield et al., 2004 ▶). In order to determine and analyze the three-dimensional structure of β2m in cartilaginous fish, the nurse shark (Ginglymostoma cirratum) β2m gene was expressed and the protein was purified and crystallized. This is the first study of the the structure of β2m from a cartilaginous fish. The results will shed light on the evolution of IgSF C molecules in the AIS.
2. Materials and methods
2.1. Comparison of the sequence of ns-β2m with those of bony fish, chicken, mouse and human β2ms
The amino-acid sequence of ns-β2m was aligned with β2m sequences from grass carp, zebrafish, chicken, mouse and human. Both grass carp and zebrafish have two kinds of β2m. Sequence alignment was performed using DNAMAN v.5.5.2 (Raiola et al., 2004 ▶).
2.2. Expression and preparation of ns-β2m
The extracellular domain of the ns-β2m gene coding for 96 amino acids (GenBank accession No. GQ865623, nucleotides 8788–9060) was synthesized by Invitrogen Life Technologies (Shanghai). For the convenience of expressing ns-β2m, a unique NdeI restriction site was added to the 5′-terminus; a stop codon and a unique XhoI restriction site were added to the 3′-terminus in the order 5′-terminus to 3′-terminus. The gene was cloned without any tags and ligated into the cloning vector pMD18-T [Takara Biotechnology (Dalian) Co. Ltd]. The recombinant plasmid pMD18-T/ns-β2m was digested with NdeI and XhoI at 310 K for 10 h. Meanwhile, the prokaryotic expression vector pET21a(+) (Novagen, Merck KGaA, Darmstadt, Germany) was digested with NdeI and XhoI at 310 K for 10 h. The gene and the digested pET21a(+) were recovered and were ligated together by T4 DNA ligase for 10 h at 289 K. The recombinant plasmid pET21a/ns-β2m was transformed into Escherichia coli strain BL21 (DE3). The expression E. coli strain was grown in 2 l Luria–Bertani (LB) medium at 310 K. When the OD600 reached 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium to a final concentration of 1 mM. After 5 h, the bacteria were harvested by centrifugation at 6000g for 10 min at 277 K and resuspended in phosphate-buffered saline (PBS). After sonication, the sample was centrifuged at 16 000g for 10 min at 277 K and the pellet was collected as the protein was expressed as inclusion bodies. In principle, the process of extraction of inclusion bodies was carried out as described previously by Chen et al. (2008 ▶). The pellet was washed three times with a solution consisting of 0.5%(v/v) Triton X-100, 20 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM DTT and once with the same solution without Triton X-100. The inclusion bodies were dissolved overnight in urea buffer [8 M urea, 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM EDTA, 10%(v/v) glycerol, 10 mM DTT] with about 1 ml urea buffer per 30 mg protein.
2.3. Refolding and purification of the recombinant ns-β2m protein
Essentially, the refolding and purification of the ns-β2m protein was carried out as described previously by Garboczi et al. (1992 ▶). The ns-β2m inclusion bodies were gradually added to 500 ml refolding buffer (100 mM Tris–HCl pH 8.0, 2 mM EDTA, 400 mM l-arginine–HCl, 0.5 mM oxidized glutathione, 5 mM reduced glutathione) to 180 mg l−1. After incubation at 277 K for 12 h, the remaining soluble ns-β2m protein was concentrated and purified by chromatography on a Superdex 200 16/60 HiLoad (GE Healthcare) size-exclusion column with 20 mM Tris–HCl pH 8.0, 50 mM NaCl, which was followed by Resource-Q (GE Healthcare) anion-exchange chromatography with buffer A (10 mM Tris–HCl pH 8.0, 10 mM NaCl) and buffer B (10 mM Tris–HCl pH 8.0, 1 M NaCl) (Chen et al., 2008 ▶). Finally, about 12 mg protein was obtained after refolding from 90 mg ns-β2m inclusion bodies.
2.4. Crystallization of ns-β2m
The purified ns-β2m protein was buffer-exchanged into 20 mM Tris–HCl pH 8.0, 50 mM NaCl using Amicon Ultra-15 centrifugal filter devices (Millipore) with a molecular-weight cutoff of 10 000 and concentrated to 7 mg ml−1. Screening of crystallization conditions was set up manually using Index, Crystal Screen and Crystal Screen 2 (Hampton Research, Laguna Hills, California, USA) at 291 K using the sitting-drop vapour-diffusion method. Drops were prepared by mixing 1 µl protein solution with 1 µl reservoir solution and were equilibrated against 150 µl of the same reservoir solution in a VDX plate (Hampton Research). A crystal was obtained after 7 d in solution No. 39 [0.1 M HEPES pH 7.5, 2%(v/v) polyethylene glycol 400, 2.0 M ammonium sulfate] from Crystal Screen.
2.5. Data collection and processing
The crystal was first soaked in reservoir solution supplemented with 15%(v/v) glycerol as a cryoprotectant for several minutes and then flash-cooled directly in liquid nitrogen (77 K). The ns-β2m diffraction data were collected to 2.3 Å resolution on beamline NE3A at the High Energy Accelerator Research Organization (KEK) synchrotron facility (Tsukuba, Japan) at a wavelength of 1.0 Å using an ADSC Q270 imaging-plate detector. The data were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
A multiple amino-acid sequence alignment is shown in Fig. 1 ▶. The amino-acid sequence used in the alignment was the extracellular domain of β2m. The alignment shows that ns-β2m has 42.1, 41.1, 40.0, 38.9, 40.6, 37.5 and 36.5% identity to grass carp β2m type II, grass carp β2m type I, zebrafish β2m type I, zebrafish β2m type II, human β2m, mouse β2m and chicken β2m, respectively.
Figure 1.
Sequence alignment of ns-β2m with β2ms from other species. The alignment was performed using the DNAMAN program. The sequence identities between ns-β2m and β2m molecules from other species are listed after the sequences. The GenBank accession Nos. are as follows: grass carp type I, AB190815; grass carp type II, AB128864; zebrafish type I, NM_131163; zebrafish type II, NM_213126; chicken, M84767; mouse, M84364; human, NM_004048. Dashes represent identical residues to those in ns-β2m.
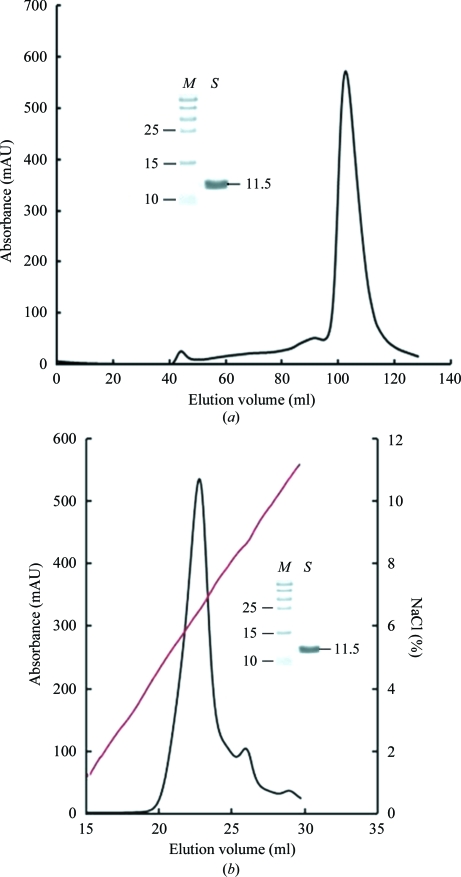
Refolding of inclusion bodies resulted in yields of 12–15% ns-β2m protein. The ns-β2m protein was first purified by Superdex 200 16/60 HiLoad size-exclusion chromatography (Fig. 2 ▶ a). The elution profile showed one peak corresponding to the expected monomeric ns-β2m protein, with a molecular mass of about 11.5 kDa. The collected protein was then purified by Resource-Q anion-exchange chromatography (Fig. 2 ▶ b). The first of the peaks was pooled and concentrated for crystallization.
Figure 2.
Purification of refolded ns-β2m by FPLC Superdex 200 16/60 HiLoad gel-filtration and Resouce-Q anion-exchange chromatography (GE Healthcare). (a) Gel-filtration profile of the refolded products. The load rate was 1 ml min−1 and elution was monitored at 280 nm. The inset shows a reduced SDS–PAGE gel (15%) of the corresponding purified protein. Lane M contains molecular-weight markers (labelled in kDa) and lane S contains the sample (ns-β2m protein). (b) The results of further purification of the refolded products by anion-exchange chromatography. The protein was eluted at an NaCl concentration of 5.6–7.0%. The inset shows a reduced SDS–PAGE gel (15%) of the corresponding purified protein. Lanes M and S contain molecular-weight markers (labelled in kDa) and sample, respectively.
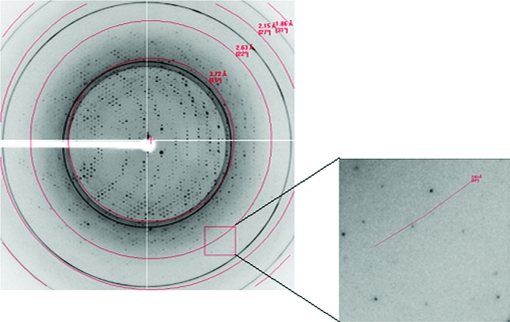
Crystallization trials were carried out using commercial crystallization kits. An ideal crystal (Fig. 3 ▶) appeared after 7 d using Crystal Screen condition No. 39 [0.1 M HEPES pH 7.5, 2%(v/v) polyethylene glycol 400, 2.0 M ammonium sulfate] without any need for further optimization. The ns-β2m crystal belonged to space group P3221, with unit-cell parameters a = b = 88.230, c = 67.146 Å, and diffracted to 2.3 Å resolution (Fig. 4 ▶). The Matthews coefficient V M was calculated to be about 3.28 Å3 Da−1, corresponding to 62.5% solvent content (Matthews, 1968 ▶). Selected data statistics are shown in Table 1 ▶.
Figure 3.
Picture of a well diffracting crystal of ns-β2m.
Figure 4.
Diffraction pattern of the ns-β2m molecule. High-resolution diffraction spots are highlighted by the box. The inset shows that the diffraction resolution is 2.3 Å.
Table 1. X-ray diffraction data-collection and processing statistics.
| Space group | P3221 |
| Unit-cell parameters (Å, °) | a = b = 88.230, c = 67.146, α = β = 90.000, γ = 120.000 |
| Volume of the unit cell (Å3) | 452673.4 |
| Resolution range (Å) | 50.00–2.30 (2.38–2.30) |
| Total No. of reflections | 153809 |
| No. of unique reflections | 12603 |
| Completeness (%) | 94.0 (100.0) |
| Average I/σ(I) | 33.7 (14.6) |
| Rmerge† (%) | 8.2 (20.3) |
| Average multiplicity | 11.9 (12.2) |
R
merge = 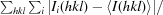
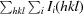 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity from multiple measurements.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity from multiple measurements.
As the nurse shark is a member of the cartilaginous fish, which are the oldest jawed vertebrates, the structural results might pave the way for further study of the evolution of IgSF C molecules. Structure determination and refinement is currently under way by molecular replacement using PDB entry 1kjv (Rudolph et al., 2002 ▶) as the starting model.
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (973 Program, 2007CB815805) and the Key National Natural Science Foundation of China (U0631009). We thank Professor George F. Gao and Dr Jianxun Qi (Institute of Microbiology, Chinese Academy of Sciences) for helpful suggestions. The authors declare no competing financial interests.
References
- Bird, S., Wang, T., Zou, J., Cunningham, C. & Secombes, C. J. (2002). J. Immunol. 168, 3329–3340. [DOI] [PubMed]
- Chen, H., Kshirsagar, S., Jensen, I., Lau, K., Simonson, C. & Schluter, S. F. (2010). Dev. Comp. Immunol. 34, 189–195. [DOI] [PubMed]
- Chen, W., Chu, F., Peng, H., Zhang, J., Qi, J., Jiang, F., Xia, C. & Gao, F. (2008). Acta Cryst. F64, 200–202. [DOI] [PMC free article] [PubMed]
- Chen, W., Gao, F., Chu, F., Zhang, J., Gao, G. F. & Xia, C. (2010). J. Biol. Chem. 285, 22505–22512. [DOI] [PMC free article] [PubMed]
- Garboczi, D. N., Hung, D. T. & Wiley, D. C. (1992). Proc. Natl Acad. Sci. USA, 89, 3429–3433. [DOI] [PMC free article] [PubMed]
- Hao, H.-F., Yang, T.-Y., Yan, R.-Q., Gao, F.-S. & Xia, C. (2006). Fish Shellfish Immunol. 20, 118–123. [DOI] [PubMed]
- Machold, R. P., Andrée, S., Van Kaer, L., Ljunggren, H. G. & Ploegh, H. L. (1995). J. Exp. Med. 181, 1111–1122. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Raiola, A., Camardella, L., Giovane, A., Mattei, B., De Lorenzo, G., Cervone, F. & Bellincampi, D. (2004). FEBS Lett. 557, 199–203. [DOI] [PubMed]
- Rudolph, M. G., Stevens, J., Speir, J. A., Trowsdale, J., Butcher, G. W., Joly, E. & Wilson, I. A. (2002). J. Mol. Biol. 324, 975–990. [DOI] [PubMed]
- Stanfield, R. L., Dooley, H., Flajnik, M. F. & Wilson, I. A. (2004). Science, 305, 1770–1773. [DOI] [PubMed]