OsAREB8 from rice (O. sativa), a member of the AREB/ABF family of bZIP transcription factors, was expressed, purified and crystallized using the sitting-drop vapour-diffusion method. A crystal of OsAREB8 in complex with its cognate DNA diffracted X-rays to 3.65 Å resolution.
Keywords: AREB/ABF family, stress response, Oryza sativa
Abstract
The AREB/ABF family of bZIP transcription factors play a key role in drought stress response and tolerance during the vegetative stage in plants. To reveal the DNA-recognition mechanism of the AREB/ABF family of proteins, the bZIP domain of OsAREB8, an AREB/ABF-family protein from Oryza sativa, was expressed in Escherichia coli, purified and crystallized with its cognate DNA. Crystals of the OsAREB8–DNA complex were obtained by the sitting-drop vapour-diffusion method at 277 K with a reservoir solution consisting of 50 mM MES pH 6.4, 29% MPD, 2 mM spermidine, 20 mM magnesium acetate and 100 mM sodium chloride. A crystal diffracted X-rays to 3.65 Å resolution and belonged to space group C222, with unit-cell parameters a = 155.1, b = 206.7, c = 38.5 Å. The crystal contained one OsAREB8–DNA complex in the asymmetric unit.
1. Introduction
The plant hormone abscisic acid (ABA) controls drought stress response and tolerance in plants. ABA induces many drought-responsive genes in plants (Yamaguchi-Shinozaki & Shinozaki, 2006 ▶; Nakashima et al., 2009 ▶; Fujita et al., 2011 ▶). In the ABA signalling pathway ABA is recognized by its receptors, PYL/PYR/RCAR proteins (Ma et al., 2009 ▶; Park et al., 2009 ▶), and the ABA–receptor complex inhibits the phosphatase activity of the group A protein phosphatases 2C (PP2Cs) by direct interaction (Miyazono et al., 2009 ▶; Melcher et al., 2009 ▶). The inhibition of PP2Cs results in the activation of subclass III protein kinases of the SNF1-related protein kinases 2 (SnRK2s). The activated SnRK2s phosphorylate downstream substrates such as the AREB/ABF family of bZIP transcription factors to activate ABA-responsive gene expression.
AREB1 is one of the AREB/ABF-family transcription factors in Arabidopsis thaliana, which are positive regulators of ABA signalling involved in drought stress tolerance (Uno et al., 2000 ▶; Fujita et al., 2005 ▶; Furihata et al., 2006 ▶; Yoshida et al., 2010 ▶). AREB1 possesses four conserved phosphorylation sites (C1–C4), which are the targets of SnRK2s, and a bZIP-type DNA-binding domain between the C3 and C4 phosphorylation sites. AREB1 from Arabidopsis belongs to a group A subfamily of bZIP (basic region leucine zipper) groups (Jakoby et al., 2002 ▶). In this group, the amino-acid sequences of the four conserved phosphorylation sites and the bZIP-type DNA-binding domain are highly conserved. AREB1 is activated by ABA-dependent multisite phosphorylation of the conserved domain (Furihata et al., 2006 ▶) in order to recognize a conserved cis-element (designated the ABA-responsive element; ABRE; PyACGTGG/TC) which exists in the promoters of ABA-inducible genes (Giraudat et al., 1994 ▶; Busk & Pagès, 1998 ▶).
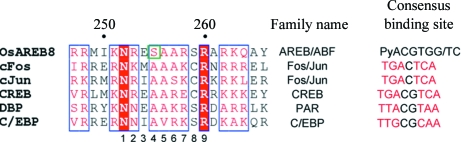
The bZIP domain of the AREB/ABF family possesses three or four leucine repeats in its C-terminal leucine-zipper region and a highly conserved N-terminal basic region for the recognition of the ABRE sequence. In general, bZIP domains recognize divergent DNA sequences using five conserved signature sequences in the basic region (Fujii et al., 2000 ▶; Fig. 1 ▶). For example, the CREB and Fos/Jun families have an N1 XXA4A5 XXC8R9 sequence (where X represents a variable amino-acid residue) in these basic regions for the recognition of CREB and AP-1 sites, respectively. In contrast, the AREB/ABF family has an N1 XXS4A5 XXS8R9 sequence in its basic region for the recognition of the ABRE sequence. Although the Ser residue at position 8 is also observed in other bZIP families, such as the PAR and C/EBP families, the Ser residue at position 4 is unique to the AREB/ABF family. Although the structures of CREB, Fos/Jun and C/EBP in complex with these cognate DNAs have been reported (Glover & Harrison, 1995 ▶; Schumacher et al., 2000 ▶; Miller et al., 2003 ▶), the structure of the AREB/ABF family is still unknown. The rice AREB/ABF-family transcription factor OsAREB8 (Os06g0211200), which is also known as OsbZIP46, OsABF2 and ABL1, regulates ABRE-dependent gene expression in response to ABA, auxin and several abiotic stress responses such as drought and oxidative stresses (Nijhawan et al., 2008 ▶; Hossain et al., 2010 ▶; Yoshida et al., 2010 ▶; Yang et al., 2011 ▶). To elucidate the structural basis of the DNA-binding mechanism of the AREB/ABF family of bZIP proteins, the bZIP domain of OsAREB8 was expressed in Escherichia coli, purified and crystallized with a double-stranded DNA containing the ABRE sequence.
Figure 1.
Amino-acid sequence comparison of the basic regions of bZIP transcription factors and their consensus sequences. The characteristic serine residue of OsAREB8 is indicated by a green square. The alignment figure was prepared using ESPript (Gouet et al., 1999 ▶). The most conserved palindromic bases in the consensus binding sites are shown in red.
2. Materials and methods
2.1. Overexpression, purification and crystallization
The gene fragment for the OsAREB8 bZIP domain (residues 235–308 of OsAREB8) was amplified by PCR and cloned into the NdeI/HindIII site of pET-26b plasmid (Novagen). The constructed plasmid (pET26b-OsAREB8) was transformed into E. coli strain Rosetta (DE3) to express OsAREB8. The transformants were cultured in LB medium supplemented with 20 µg ml−1 kanamycin at 310 K. OsAREB8 expression was induced by the addition of 1 mM (final concentration) isopropyl β-d-1-thiogalactopyranoside (IPTG) when the optical density of the medium at 600 nm reached 0.6. The cells were cultured for a further 14 h at 298 K. After cultivation, the cells were harvested by centrifugation at 5000g for 10 min. The harvested cells were resuspended in 10 mM Tris–HCl pH 7.5 and disrupted by sonication. To remove contaminant DNA derived from E. coli, 0.01%(v/v) (final concentration) polyethyleneimine was added to the disrupted solution. After centrifugation at 40 000g for 30 min, the supernatant was supplemented with 2 mM (final concentration) MgCl2 and treated with Benzonase nuclease (Novagen). The treated solution was applied onto a TOYOPEARL AF-Heparin HC-650 (Tosoh) column. The column was washed with 10 mM Tris–HCl pH 7.5, 250 mM NaCl, and the bound OsAREB8 was eluted with 10 mM Tris–HCl pH 7.5, 1 M NaCl. The eluted protein was further purified using a 6 ml Resource S (GE Healthcare) column pre-equilibrated with 10 mM Tris–HCl pH 7.5 and was eluted with a linear gradient of 0–1 M NaCl. The purified protein was dialyzed against 10 mM Tris–HCl pH 7.5, 50 mM NaCl for crystallization. All purification experiments were performed at 277 K.
For crystallization of the OsAREB8–DNA complex, 14 bp blunt-end double-stranded DNAs containing the ABRE sequence (5′-CTGCCACGTGGCAG-3′, 5′-CTATCACGTGGCAG-3′ and 5′-TGCCACGTGGCAGG-3′; the ABRE sequence is indicated in bold) were added to the OsAREB8 solution. The OPC-purified oligonucleotides were purchased from Operon Biotechnology (Tokyo, Japan) and were annealed by heating to 368 K for 1 min and slow cooling to 277 K in 10 mM Tris–HCl pH 7.5, 50 mM NaCl. Because bZIP domains form a homodimer to recognize the DNA sequence, the purified OsAREB8 and the double-stranded DNA were mixed in a 2:1 molar ratio. The OsAREB8–DNA complex was concentrated to 500 µM for crystallization. All crystallization experiments were performed at 277 K using the sitting-drop vapour-diffusion method. Each drop was prepared by mixing 1 µl OsAREB8–DNA solution and 1 µl reservoir solution.
2.2. Data collection and processing
The crystals were mounted on cryoloops and flash-cooled in a nitrogen stream at 95 K for data collection. An X-ray diffraction data set was collected on the AR-NW12A beamline at the Photon Factory (PF; Tsukuba, Japan) using an ADSC Quantum 210r detector and an X-ray wavelength of 1.0000 Å. The diffraction data were indexed and integrated with the program XDS (Kabsch, 2010 ▶) and scaled with the program SCALA from the CCP4 suite (Winn et al., 2011 ▶). The anisotropy of the data was analyzed by the Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/; Strong et al., 2006 ▶). A self-rotation function was calculated using the program MOLREP (Vagin & Teplyakov, 2010 ▶). The initial structure of the OsAREB8–DNA complex was determined by the molecular-replacement method using the program MOLREP with the coordinates of the MafG–DNA complex (PDB entry 3a5t; the amino-acid sequence identity of the MafG bZIP domain to OsAREB8 is 37%; Kurokawa et al., 2009 ▶). A total of 5% of the reflections were randomly selected to provide a test set for R free calculations. The initial model was refined using the program REFMAC5 (Murshudov et al., 2011 ▶).
3. Results and discussion
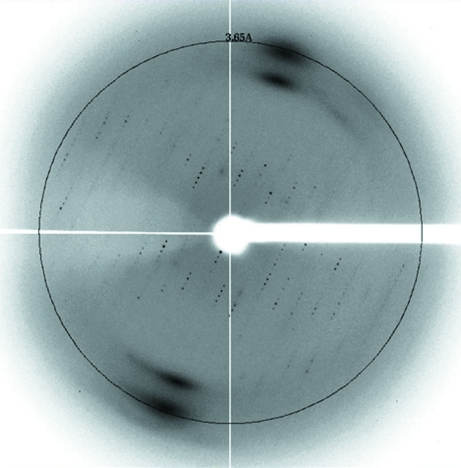
Recombinant OsAREB8 was expressed in E. coli and purified by two column-chromatographic steps. We used DNase to remove contaminant DNA from E. coli. This treatment increased the yield of OsAREB8. DNase and digested DNA fragments should be removed by the two column-chromatographic steps. SDS–PAGE analysis revealed that the OsAREB8 was more than 95% pure (Fig. 2 ▶). We only obtained diffraction-quality crystals when we used double-stranded DNA with sequence 5′-CTGCCACGTGGCAG-3′ for cocrystallization. The best crystal of the OsAREB8–DNA complex was obtained using a reservoir solution consisting of 50 mM MES pH 6.4, 29% MPD, 2 mM spermidine, 20 mM magnesium acetate, 100 mM NaCl (Fig. 3 ▶). Because the crystallization conditions contained a high concentration of MPD, we did not use a cryoprotectant when we flash-cooled the crystals. The crystal of the OsAREB8–DNA complex diffracted X-rays to a resolution of 3.65 Å (Fig. 4 ▶) and belonged to space group C222, with unit-cell parameters a = 155.1, b = 206.7, c = 38.5 Å. Data-collection statistics are summarized in Table 1 ▶. The data set was severely anisotropic. Analysis using the Diffraction Anisotropy Server showed that the suggested diffraction limits (F/σ < 3) along the a* and b* directions were 4.6 and 4.1 Å, respectively, although the suggested diffraction limit along the c* direction was 3.6 Å. In this study, the R merge in the highest resolution shell was relatively high. This could be related to the severe anisotropy of the data.
Figure 2.
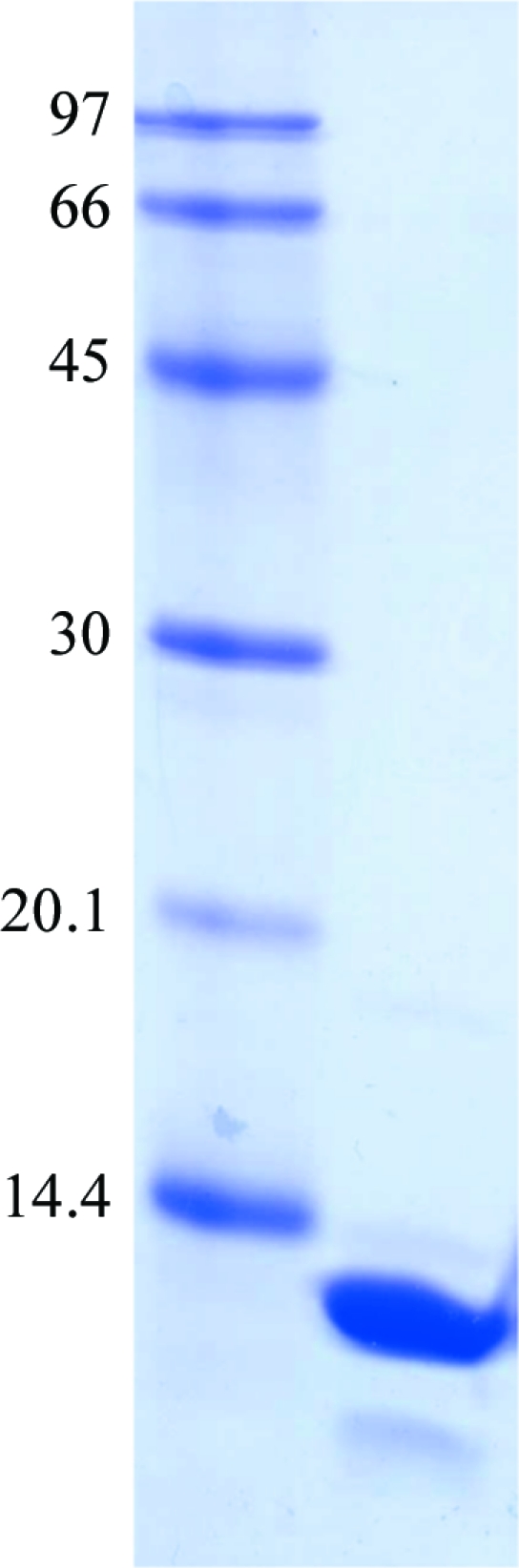
SDS–PAGE (15%) of OsAREB8. The approximate molecular weight of OsAREB8 (residues 235–308) is 8800.
Figure 3.
Crystals of OsAREB8 in complex with its cognate DNA.
Figure 4.
X-ray diffraction image of OsAREB8 in complex with its cognate DNA. The circle indicates a resolution of 3.65 Å.
Table 1. Summary of data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Beamline | PF AR-NW12A |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 95 |
| Crystal-to-detector distance (mm) | 301.4 |
| Oscillation width (°) | 1.0 |
| Total No. of frames | 180 |
| Space group | C222 |
| Unit-cell parameters | |
| a (Å) | 155.1 |
| b (Å) | 206.7 |
| c (Å) | 38.5 |
| Resolution range (Å) | 20.0–3.65 (3.85–3.65) |
| Total No. of observations | 51535 (7819) |
| No. of unique observations | 7285 (1067) |
| Mutliplicity | 7.1 (7.3) |
| Completeness (%) | 99.4 (100) |
| Rmerge† (%) | 11.0 (78.4) |
| 〈I/σ(I)〉 | 13.4 (2.9) |
R
merge = 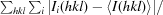
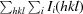 , where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is its average.
, where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is its average.
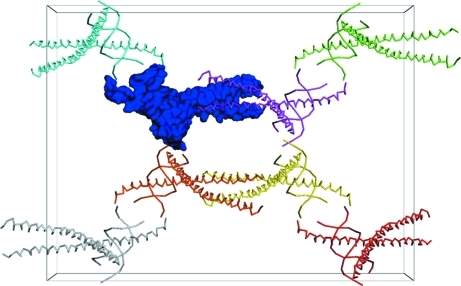
According to Matthews coefficient analysis (Matthews, 1968 ▶), the crystal appeared to contain two or three OsAREB8–DNA complexes in the asymmetric unit. Assuming the presence of two or three complexes per asymmetric unit, the Matthews coefficient of the crystal would be 2.96 or 1.97 Å3 Da−1, respectively. However, analysis of the self-rotation function did not reveal any significant peaks that indicate the presence of two or three complexes per asymmetric unit. In addition, the results of molecular replacement suggested that the crystal contained only one OsAREB8–DNA complex per asymmetric unit (Fig. 5 ▶). This means that the solvent content of the crystal of the OsAREB8–DNA complex is very high (the Matthews coefficient and solvent content of the crystal are 5.92 Å3 Da−1 and 79.25%, respectively). The R factor and MOLREP score after MOLREP were 0.626 and 0.545, respectively. The R factor, R free and FOM values after 30 cycles of refinement by REFMAC5 were 0.284, 0.377 and 0.596, respectively. Further model building and refinement are in progress.
Figure 5.
Crystal packing of the OsAREB8–DNA complex in the centred orthorhombic cell. The molecules in the asymmetric unit are shown as a darker blue surface, while symmetry-related molecules in the unit cell are shown as Cα traces. This image was created with PyMOL (DeLano, 2002 ▶).
Acknowledgments
The synchrotron-radiation experiments were performed on beamline AR-NW12A at the Photon Factory (Proposal No. 2008S2-001). This work was supported by the Targeted Proteins Research Program (TPRP) of the Ministry of Education, Culture, Sports, Science and Technology, Japan, by the Program for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN) and by the Science and Technology Research Partnership for Sustainable Development (SATREPS) of the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA).
References
- Busk, P. K. & Pagès, M. (1998). Plant Mol. Biol. 37, 425–435. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Fujii, Y., Shimizu, T., Toda, T., Yanagida, M. & Hakoshima, T. (2000). Nature Struct. Biol. 7, 889–893. [DOI] [PubMed]
- Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., Hiratsu, K., Ohme-Takagi, M., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2005). Plant Cell, 17, 3470–3488. [DOI] [PMC free article] [PubMed]
- Fujita, Y., Fujita, M., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2011). J. Plant Res. 124, 509–525. [DOI] [PubMed]
- Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2006). Proc. Natl Acad. Sci. USA, 103, 1988–1993. [DOI] [PMC free article] [PubMed]
- Giraudat, J., Parcy, F., Bertauche, N., Gosti, F., Leung, J., Morris, P. C., Bouvier-Durand, M. & Vartanian, N. (1994). Plant Mol. Biol. 26, 1557–1577. [DOI] [PubMed]
- Glover, J. N. & Harrison, S. C. (1995). Nature (London), 373, 257–261. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Hossain, M. A., Cho, J.-I., Han, M., Ahn, C.-H., Jeon, J.-S., An, G. & Park, P. B. (2010). J. Plant Physiol. 167, 1512–1520. [DOI] [PubMed]
- Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T. & Parcy, F. (2002). Trends Plant Sci. 7, 106–111. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kurokawa, H., Motohashi, H., Sueno, S., Kimura, M., Takagawa, H., Kanno, Y., Yamamoto, M. & Tanaka, T. (2009). Mol. Cell. Biol. 29, 6232–6244. [DOI] [PMC free article] [PubMed]
- Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A. & Grill, E. (2009). Science, 324, 1064–1068. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Melcher, K. et al. (2009). Nature (London), 462, 602–608.
- Miller, M., Shuman, J. D., Sebastian, T., Dauter, Z. & Johnson, P. F. (2003). J. Biol. Chem. 278, 15178–15184. [DOI] [PubMed]
- Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H.-J., Asano, A., Miyauchi, Y., Takahashi, M., Zhi, Y., Fujita, Y., Yoshida, T., Kodaira, K.-S., Yamaguchi-Shinozaki, K. & Tanokura, M. (2009). Nature (London), 462, 609–614. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nakashima, K., Ito, Y. & Yamaguchi-Shinozaki, K. (2009). Plant Physiol. 149, 88–95. [DOI] [PMC free article] [PubMed]
- Nijhawan, A., Jain, M., Tyagi, A. K. & Khurana, J. P. (2008). Plant Physiol. 146, 333–350. [DOI] [PMC free article] [PubMed]
- Park, S.-Y. et al. (2009). Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed]
- Schumacher, M. A., Goodman, R. H. & Brennan, R. G. (2000). J. Biol. Chem. 275, 35242–35247. [DOI] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2000). Proc. Natl Acad. Sci. USA, 97, 11632–11637. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yamaguchi-Shinozaki, K. & Shinozaki, K. (2006). Annu. Rev. Plant Biol. 57, 781–803. [DOI] [PubMed]
- Yang, X., Yang, Y.-N., Xue, L.-J., Zou, M.-J., Liu, J.-Y., Chen, F. & Xue, H.-W. (2011). Plant Physiol. 156, 1397–1409. [DOI] [PMC free article] [PubMed]
- Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2010). Plant J. 61, 672–685. [DOI] [PubMed]