Abstract
Cytokinesis is the last step of the cell cycle, producing two daughter cells inheriting equal genetic information. This process involves the assembly of an actomyosin ring during mitosis. In the fission yeast Schizosaccharomyces pombe, cytokinesis occurs at the geometric cell centre, a position which is defined by the interphase nucleus and the anilin-related Mid1 protein. The pom1Δ, tea1Δ and tea4Δ mutants are defective in restricting Mid1 as a band around the nucleus and misplace the division site. We previously reported that inhibition of the protein kinase Kin1 promoted failure of cytokinesis in pom1Δ and tea1Δ cells but the mechanism involving Kin1 remained elusive. Here we investigated the contribution of Kin1 in cytokinesis. We show that Kin1-GFP has a dynamic cell-cycle regulated distribution. Like pom1Δ and tea1Δ, tea4Δ exhibits a strong genetic interaction with kin1Δ. Using a conditional repressible kin1 allele that only alters interphase nuclear centering, we observed that Kin1 down-regulation severely compromised actomyosin ring formation and septum synthesis in tea4Δ cells. In addition, nuclear displacement induced either by overexpression of a putative catalytically inactive Kin1 mutant, by chemically mediated microtubule depolymerization or by mutation in the par1Δ gene impaired cytokinesis in tea4Δ but not tea4+ cells. We propose that nuclear mispositioning exacerbates the tea4Δ, pom1Δ and tea1Δ cell division phenotype. Our work reveal that nuclear centering becomes essential when Pom1/Tea1/Tea4 function is compromised and that Kin1 expression level is a key regulatory element in this situation. Our results suggest the existence of distinct overlapping control mechanisms to ensure efficient cell division.
Keywords: Actomyosin, metabolism, Alleles, Cytokinesis, genetics, physiology, Down-Regulation, genetics, physiology, Microtubule-Associated Proteins, metabolism, Protein Kinases, metabolism, Protein-Serine-Threonine Kinases, metabolism, Schizosaccharomyces, cytology, genetics, physiology, Schizosaccharomyces pombe Proteins, metabolism
Keywords: cytokinesis, mitosis, cell polarity, cell cycle, Schizosaccharomyces pombe, cytoskeleton, contractile actomyosin ring
INTRODUCTION
Cytokinesis is the last step of the cell division cycle and promotes the physical separation of daughter cells. In animals and fungi, partition of cells is brought about by constriction of an actomyosin-based contractile ring assembled during mitosis 1. Failure of cytokinesis may contribute to genome instability by producing aneuploid cell compartments after cell division.
Schizosaccharomyces pombe (S. pombe) has proven to be an excellent model organism to study eukaryotic cytokinesis. Like animal cells, S. pombe divides through the use of a contractile actomyosin ring (CAR). After spindle breakdown, cell division occurs by the constriction of the CAR at the geometric cell centre. A glucan-based septal material is concomitantly synthesized following the constricting ring in order to physically separate daughter cell cytosplasms. The septum is then digested to promote cell separation.
In contrast to animal cells, fission yeast CAR assembly starts early in mitosis. An important question is how the cell entering mitosis specifies the CAR assembly area and hence the division site. Early in mitosis, specification of the cell division site is defined by the position of the late interphase nucleus and the anilin-related protein Mid1. During interphase, a major pool of Mid1 is present in the nucleus and a sub-population moves out of the nucleus and associates with the neighboring cell cortex as a band. At mitotic commitment, nuclear Mid1 completely exits the nucleus and associates with the cell cortex prior to chromosome segregation 2–8. Recent studies 9, 10 have suggested a model with two parallel non overlapping mechanisms for CAR formation 11. First, Mid1 establishes a broad band of dots that dictates the site of ‘cortical nodes’ formation, which are protein complexes that include Rlc1-containing myosin II hexamers. Later, cortical nodes coalesce during the ‘lateral condensation’ process to form a ring structure 8, 12–14. Concomitantly, F-actin cables are formed in parallel in the cell centre and are packed as a ring. A homogenous ring structure is achieved by the incorporation of the FER/CIP4 homology (FCH) domain protein Cdc15 9.
The fission yeast cell is rod-shaped and interphase cell growth, including plasma membrane extension and cell wall remodeling, is restricted to the opposite ends. Early in G2, growth is activated only in the mother-inherited ‘old’ cell end. Growth activation in the ‘new’ end produced by cytokinesis, referred to as New End Take-Off (NETO), is trigerred in later in G2 15. Polarized growth is controlled by complex interactions between cortical polarity factors, such as Tea1 and Tea4, and dynamic organization of the cytoskeleton 16. Tea1 and Tea4 are also required for the cell end association of the Pom1 kinase that is involved in bipolar growth 17, 18 and Mid1 localization at the cell centre. In pom1Δ, tea4Δ and tea1Δ cells, Mid1 is not restricted as a band around the nucleus. As a consequence, the CAR is not centered in these cells, even though the nucleus remains in the middle of the cell 4, 6. In addition, Pom1, Tea1 and Tea4 have recently been shown to regulate Tip Occlusion, an inhibitory mechanism that prevents septum synthesis in the cell ends 19. The F-actin cytoskeleton is focused in the growing areas as motile patches and cables and contributes to growth during interphase. At mitotic commitment, growth ceases and F-actin structures are no longer present at the cell end but relocate to the division site 20.
In fission yeast, Kin1 is the sole member of the evolutionarily conserved family of KIN1/PAR-1/MARK serine/threonine kinases. These enzymes show diverse functions in eukaryotic cells such as embryonic polarity, asymmetry of cell division, cell differentiation, cell cycle progression and intracellular signalling 21. A fission yeast kin1Δ deletion mutant suggests a role for Kin1 in cell morphogenesis, F-actin polarity and cell separation 22–24. In kin1Δ interphase cells, the nucleus is eccentric, being closer to the new cell end. Thus, CAR and septum are asymmetrically synthesized in mitosis 22.
We have described a synthetic lethal interaction between kin1Δ and pom1Δ mutations. Simultaneous inhibition of these kinases leads to strong defects in CAR assembly and septation, preventing cytokinesis. In addition to Pom1, a deletion mutant of Tea1 showed extensive cytokinetic defects when combined with kin1Δ 25. However, the exact contribution of Kin1 in cytokinesis remained elusive.
Here we investigated the functional requirement of Kin1 in cytokinesis. We examined the dynamic cell-cycle regulated distribution of a Kin1-GFP fusion protein. Among a set of polarity mutants, we determined that only tea4Δ exhibited a genetic interaction with kin1Δ similar to pom1Δ and tea1Δ mutants. Next, we used a conditional repressible kin1 allele that showed nuclear and septum positioning defects but had no effect on cell shape, cell separation and F-actin polarity in wild type cells. In a tea4Δ strain, repression of this kin1 allele induces severe cytokinesis defects. We propose that Kin1 dependent nuclear centering is required for cytokinesis efficiency in tea4Δ cells. Consistent with this hypothesis, alternative chemically and genetically induced mispositioning of the nucleus in tea4Δ cells impaired cytokinesis. Thus, if Mid1-mediated cell division site specification is compromised, we show that maintenance of the nucleus in the middle of the cell becomes essential to prevent cytokinesis failure.
RESULTS
Kin1 has a dynamic cell-cycle regulated cortical distribution
To futher understand Kin1 function, we decided to reinvestigate its cellular localization in living cells. In early G2 cells, indirect immunofluorescence experiments by Drewes and Nurse showed that a C-terminally Kin1-13myc tagged protein was detected at the new cell end. Later in G2, the signal was detected on both ends and disappeared during mitosis without evidence for protein degradation 24. However, a global localization analysis of fission yeast proteins systematically tagged with the Yellow Fluorescent Protein (YFP) indicated that Kin1-YFP expressing cells could show a signal in the middle of the cell 26. To clarify Kin1 localization, we prepared a strain expressing a C-terminally GFP tagged Kin1 fusion protein, Kin1-GFP, integrated at its endogenous locus (Materials and Methods). The kin1-GFP strain exhibited a wild type phenotype. In living interphase cells, Kin1-GFP signal captured either in static images (Fig. 1A) or by time-lapse video microscopy (Fig. 1C) was localized at the cell tips (arrows, Fig. 1A). The signal appeared as dynamic dots close to the plasma membrane (supplementary movie S1). Kin1-GFP was detected on the new cell end in early G2 as previously reported 24 but also on the old end soon after growth resumption (arrow, Fig. 1C) conversely to the previous report 24. This result suggests that the Kin1-GFP signal may be more sensitive than Kin1-13myc.
Figure 1. Kin1-GFP has a dynamic cell-cycle regulated localization at the cell cortex that is partially dependent on F-actin polymerization.
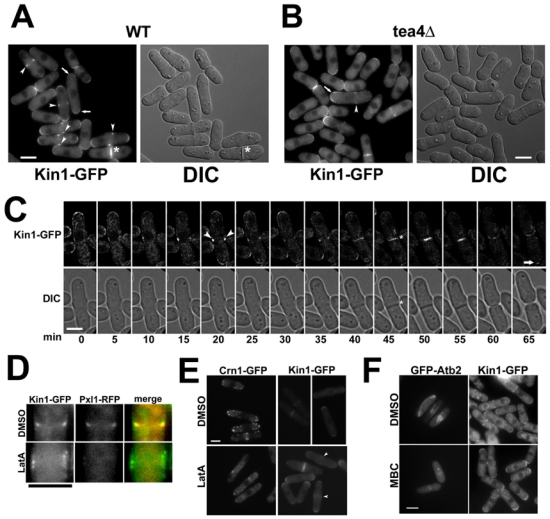
(A) kin1-GFP (WT) and (B) tea4Δ kin1-GFP (tea4Δ) cells were cultured to mid log phase. Representative images of GFP and DIC signals of live cells were taken. Arrows show cell end localization of Kin1-GFP and arrowheads indicate the signal in the cell middle. Asterisks show the presence of Kin1-GFP at the septum site. (C) Kin1-GFP and DIC images of kin1-GFP cells were captured by time-lapse video microscopy. A representative dividing cell is shown at 5 min intervals. Kin1-GFP appears as a ring in the central region of the cell (arrowheads), at the septum synthesis site (asterisk) and at the old (arrow) and the new cell ends at cell separation. (D) Kin1-GFP Pxl1-RFP were treated with 1% DMSO or 25 μM Latrunculin A, (LatA) for 10 min, medial signals of anaphase cells are shown. (E) Crn1-GFP and Kin1-GFP cells were treated with 1% DMSO or 25 μM LatA for 10 min. (F) GFP-Atb2 and Kin1-GFP cells were treated with 1% DMSO or 25 μg/ml MBC for 20 min. Bars: 5 μm.
Indeed, in mitotic cells, we detected a Kin1-GFP signal at the division site (arrowheads, Fig. 1A,C). Kin1-GFP appeared as a ring overlying the cell equator prior to nuclei separation at the onset of anaphase B (Fig. 1C). At cell ends, the signal was still present but progressively disappeared at mitotic exit. After mitosis, Kin1-GFP was detected at the septum synthesis site and/or plasma membrane invagination (asterisk, Fig. 1A,C). Then, Kin1-GFP was distributed on both sides of the septal structure, consistent with its detection at the new cell ends in early G2.
To confirm that Kin1-GFP was present in the CAR, we prepared a Kin1-GFP Pxl1-RFP expressing strain. The paxilin homolog Pxl1 is an actin polymerization dependent component of the CAR 27. Anaphase cells showed a colocalisation between Kin1-GFP and Pxl1-RFP as medial rings (DMSO, Fig. 1D), suggesting that Kin1-GFP is present in the CAR during anaphase. At the end of mitosis, Pxl1-RFP shrinked centripetally as the CAR constricted whereas Kin1-GFP associated with the synthesized septal material and/or invaginating plasma membranes (data not shown).
We examined the role of F-actin on Kin1-GFP localization using the actin depolymerizing drug latrunculin A (LatA). Depolymerization of F-actin was controlled by monitoring the localization of the GFP tagged F-actin patch component coronin Crn1-GFP 28. In DMSO treated cells, Crn1-GFP is present as dots in interphase cell ends or in the middle of the cell after mitosis. Crn1-GFP localization was completely abolished by LatA (Fig. 1E). In contrast, LatA had no effect on cell end or septum localizations for Kin1-GFP (Fig. 1E). However, we observed that the medial ring signal was enlarged in anaphase cells (arrowheads, LatA, Fig. 1E). To confirm this effect, we examined Kin1-GFP Pxl1-RFP anaphase cells treated with LatA. Pxl1-RFP signal was removed from the middle of the cell as previously published 27 whereas Kin1-GFP remained as a cortical band (LatA, Fig. 1D). Thus, F-actin polymerization is required for Kin1-GFP association as a tight ring in anaphase cells but is dispensable for other Kin1 localizations.
Kin1-GFP cells were also treated with the microtubule (MT) depolymerizing drug carbendazim (MBC) for 20 minutes. Treatment of alpha tubulin tagged GFP-Atb2 expressing cells in parallel showed no signal in contrast to DMSO treated cells. In the presence of MBC, Kin1-GFP was still localized at the cell cortex in cell ends, in the CAR and the septum synthesis site (Fig. 1F). Kin1-GFP remained at the cortex even after a prolonged absence of MTs (data not shown). Thus MTs are not required for Kin1 association with the cell cortex nor for its polarization.
Genetic interactions between kin1Δ and polarity mutants
Kin1-GFP localization and the pleiotropic defects of kin1Δ suggest distinct cellular functions for Kin1 during the cell cycle. Here, we focused on Kin1 function in cytokinesis. We previously reported that the kin1Δ mutation was synthetically lethal with the pom1Δ mutant. Pom1Δ kin1Δ cells were unable to complete cytokinesis preventing colony formation. In addition, we observed strong similar cytokinetic defects in a kin1Δ tea1Δ double mutant25.
This raised the question of whether other polarity mutants, when combined with kin1Δ, exhibit cell division defects similar to those observed in a kin1Δ tea1Δ mutant. For this purpose, we selected genes which, like pom1 and tea1, also function at the cell ends in polarized growth control 16. Double mutants were grown to mid log phase and cytokinesis efficiency was compared to kin1Δ tea1Δ cells by fluorescence microscopy (Fig. 2A). Kin1Δ cells synthesize normal septa (Fig. 2A) but exhibit a high septation index and a small percentage of multiseptate cells (Fig. 2B), indicating a cell separation defect as previously shown 22–24.
Figure 2. Kin1Δ tea1Δ and kin1Δ tea4Δ mutants show severe cytokinesis and growth defects.
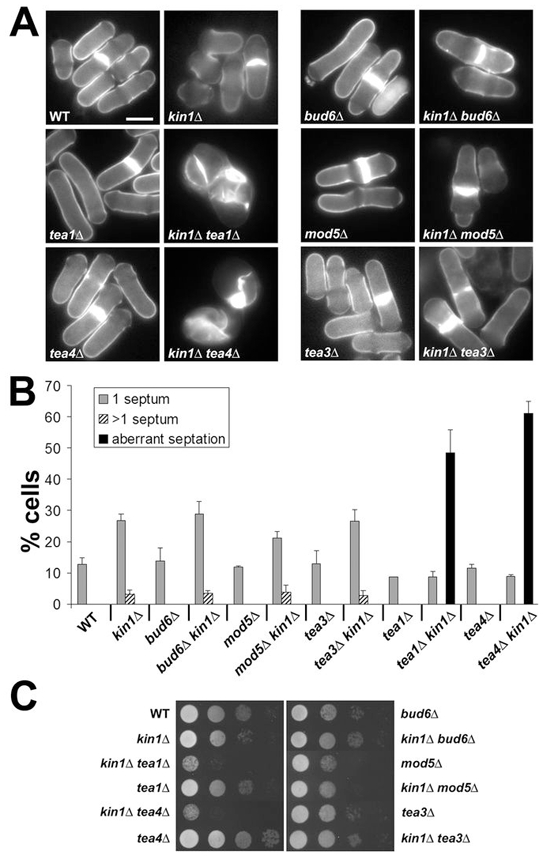
The indicated wild-type (WT), polarity and kin1Δ mutants were grown at 25°C to mid log phase. (A) Cells were stained with methyl blue. Bar: 5 μm. (B) Percentage of cells with 1 normal septum (1 septum), more than 1 normal septum (>1 septum) or aberrant septal deposits (aberrant septation) were scored in the indicated strains (n>200, in duplicate). (C) Growth of the indicated strains were tested by 10-fold serial dilutions on YES plates and incubated for 3 days at 25°C.
Tea4 binds to Tea1 and regulates NETO and symmetry of cell division 29, 30. Kin1Δ tea4Δ cells showed a severe cytokinetic phenotype (Fig. 2A), with aberrant septal material randomly deposited in a majority of cells (Fig. 2B). Moreover, double mutant cells completey lose their polarity and appeared almost round. This phenotype is strickingly similar to that of kin1Δ tea1Δ (Fig. 2A,B). This result was confirmed by comparing growth rates of mutants on agar plates, where kin1Δ tea1Δ and kin1Δ tea4Δ showed a significant growth reduction compared to single mutants (Fig. 2C).
We studied Kin1-GFP localization in a tea4Δ background. As shown on Fig. 1B, Kin1-GFP was present predominantly at one cell end throughout interphase, this may correspond to the actively growing cell end, since tea4Δ cells show a monopolar growth pattern in interphase 29, 30. During mitosis, the tip signal was detected on the most distant cell end relative to the asymmetric ring (arrow, Fig. 1B). Kin1-GFP was also detected at the division site in tea4Δ (arrowhead, Fig. 1B). We conclude that correct localization of Kin1-GFP does not require Tea4.
Three other monopolar growing mutants, bud6Δ, mod5Δ, and tea3Δ, corresponding to genes encoding Tea1 interacting proteins 31–34, have been studied. Kin1Δ bud6Δ, kin1Δ mod5Δ and kin1Δ tea3Δ cells were viable and showed septation (Fig. 2A,B) and growth (Fig. 2C) patterns similar to that of kin1Δ.
Hence, only pom1Δ, tea1Δ and tea4Δ, among the polarity mutants tested, show a strong synthetic cytokinetic phenotype when combined with kin1Δ, a majority of cells failing septation.
Kin1 down-regulation promoted widening of the division site area
Kin1Δ cells exhibit cell wall and cell shape defects, delocalized F-actin, asymmetry of cell division and inhibition of cell separation 22–24. To further clarify the role of Kin1 in cytokinesis, we studied a repressible kin1 allele (Fig. 3). We used either the nmt81GFP-kin1 25 or a non-tagged nmt81-kin1 versions (hereafter collectively referred to as nmt81-controlled kin1), in which the kin1 gene is under the control of the weak nmt81 thiamine-dependent repressible promoter. Both alleles exhibited identical phenotypes (data not shown). Promoter expression in the activated condition (ON) was close to that of the endogenous Kin1 whereas, in the repressed condition (OFF), Kin1 expression was strongly reduced to levels that were not detectable by the anti-Kin1 antibody (see Fig. 4A).
Figure 3. Kin1 down-regulation promotes mis-specification of the division site.
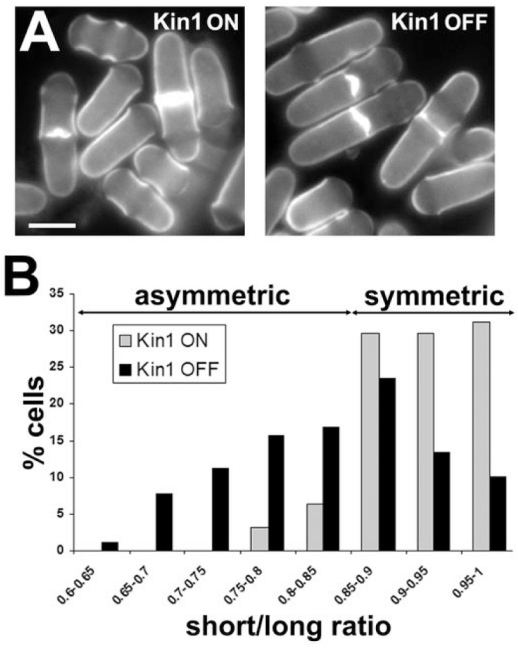
nmt81-controlled kin1 cells were cultured in the absence (ON) and in the presence (OFF) of thiamine for 16h at 30°C to repress Kin1 expression. (A) cells were stained with methyl blue in the Kin1 ON and Kin1 OFF conditions. (B) Percentage of cells showing a short/long cell length ratio of the indicated values. The short and long distances separating the septum to cell ends were measured in septated cells of the Kin1 ON and Kin1 OFF conditions and a short/long distance ratio was calculated. Ratios <0.85 and >0.85 refer to asymmetric and symmetric septa, respectively. Bars: 5 μm.
Figure 4. Kin1 down-regulation alters the dynamic assembly and disassembly of the CAR and septation in tea4Δ mitotic cells.
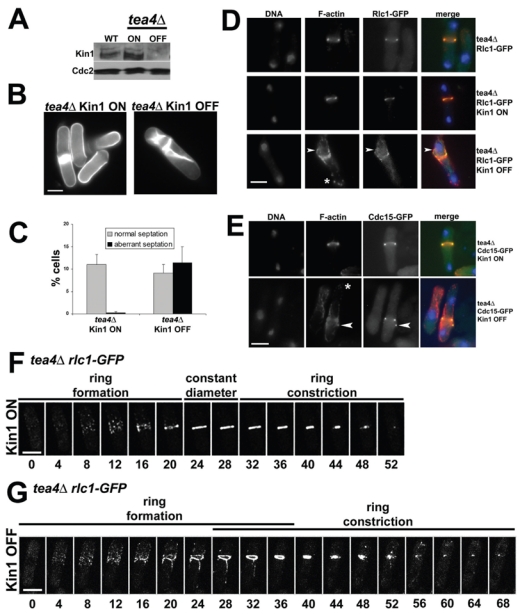
Tea4Δ nmt81-controlled kin1 cells were cultured in the absence (Kin1 ON) and in the presence (Kin1 OFF) of thiamine 16h at 30°C. (A) Western blot analysis of Kin1 expression levels in the ON and OFF conditions in tea4Δ compared to endogenous Kin1 level (WT) using an anti-Kin1 antibody. Western blot of Cdc2 was used as a loading control. (B) tea4Δ Kin1 ON and tea4Δ Kin1 OFF cells stained with methyl blue. (C) Percentage of tea4Δ Kin1 ON and Kin1 OFF or tea4Δ Rlc1-GFP Kin1 ON and Kin1 OFF total cells (n>200, in duplicate) exhibiting normal (normal septation) or aberrant (aberrant septation) septal structures. (D) tea4Δ rlc1-GFP and tea4Δ rlc1-GFP nmt81-controlled kin1 and (E) tea4Δ cdc15-GFP nmt81-controlled kin1 cells were cultured in EMM in the absence (Kin1 ON) and in the presence (Kin1 OFF) of thiamine for 16h at 30°C and were stained with DAPI (DNA, blue) and rhodamine-conjugated phalloidin (F-actin, red). Fluorescent signals and superposition of images are shown (merge). Asterisks indicate the presence of F-actin patches in the cell end and arrowheads show the presence of extra F-actin cables, Rlc1-GFP or Cdc15-GFP dots outside the CAR. (F, G) tea4Δ rlc1-GFP nmt81-controlled kin1 cells were cultured in the absence (Kin1 ON) and in the presence (Kin1 OFF) of thiamine for 16h at 30°C. Rlc1-GFP ring assembly during mitosis (ring formation), its presence as a packed ring (constant diameter) and its subsequent constriction (ring constriction) were recorded by time-lapse video microscopy. Maximal projections of Z stacks of representative cells (from n=22 in each condition) are shown at 4 minute intervals, t=0 was set as the image with first detected Rlc1-GFP signal. Bars: 5 μm.
The Kin1 ON cells showed a wild type phenotype. In Kin1 down-regulating cells, we observed that septa were orthogonal to the long cell axis but their positions seemed to be frequently eccentric (Kin1 OFF, Fig. 3A). Cell shape or interphase F-actin polarity defects were not observed (supplementary figure S1) conversely to the deletion strain.
We calculated the ratio between the short and the long daughter cell length in Kin1 ON and Kin1 OFF septated cells. In the Kin1 ON condition, 90.6% of cells showed a ratio > 0.85 (n=64, Fig. 3B) indicating that septa were almost at the geometric centre. After Kin1 repression, the septum position area relative to the cell ends was enlarged with more heterogeneous length ratios ranging from 0.65 to 1. Only 46% of cells showed a ratio > 0.85 (n=89, Fig. 3B). Thus, Kin1 repression promoted asymmetric positioning of the septum as well as the CAR (data not shown). Mispositioning of the division plane was not dependent on Mid1 since Mid1 was properly localized in Kin1 ON and Kin1 OFF cells (supplementary figure S2) and kin1Δ mid1Δ double mutant showed additive phenotypes (supplementary figure S3).
Since position of the CAR and the subsequent septum is specified in late G2 and early mitosis by the position of the nucleus 35–37, we monitored the position of the nucleus relative to the cell ends in late G2 cells (≥ 10 μm) and we observed that a higher proportion of nuclei were misplaced towards the new cell end (according to the birthscar) in the Kin1 OFF (28±1.1% with a short/long ratio < 0.85, n=206) compared to the Kin1 ON condition (16.4±1.8%, n=186). However, nuclear dynamics of interphase nuclei were not significantly different between Kin1 ON (supplementary movie S2) and Kin1 OFF (supplementary movie S3). The mean distance covered by the nuclei during interphase were very similar, 61.6±12.5 nm and 49.9±8.8 nm for Kin1 ON (n=23) and Kin1 OFF (n=19) respectively, suggesting that Kin1 down regulation does not increase nuclear oscillations. Thus, Kin1 repression impaired solely nuclear centering, consistent with frequent asymmetric CAR and septum position.
Kin1 down-regulation promoted abnormal septum synthesis in tea4Δ cells
Kin1Δ tea4Δ cells showed severe polarity defects, being unable to propagate as rods (Fig. 2A). Hence, septation failure may be a consequence of a lack of polarity controls rather than a cytokinesis defect per se. To clarify this issue, we out-crossed tea4Δ to the nmt81-controlled kin1 allele to conditionally repress Kin1 in this mutant (Fig. 4A).
Tea4Δ Kin1 ON behaved as the tea4Δ single mutant. Kin1 down-regulation promoted aberrant septum material synthesis in tea4Δ: a main asymmetric septal structure was observed together with aberrant septal deposits along the cortex or stretched structures that run along the main cell axis were detected (Fig. 4B). Half of cells with aberrant septal material were still rod-shaped. The septation index increased from 11.3% (Kin1 ON) to 20.4% (Kin1 OFF), including 11.4% of aberrant septations (Fig. 4C). Similar results were obtained with a tea1Δ nmt81-controlled kin1 strain: 5% of aberrant septated cells were observed (data not shown). Thus, Kin1 down-regulation in tea1Δ and tea4Δ mutants predominantly impaired cytokinesis rather than cell polarity control. The phenotype of tea4Δ Kin1 OFF was less severe than pom1Δ Kin1 OFF 25 and allowed us to carry out a detailed analysis of CAR formation during mitosis.
Kin1 down-regulation in tea4Δ mitotic cells altered the timing of both CAR assembly and disassembly
We examined how CARs formed in mitotic tea4Δ cells when Kin1 was down-regulated. Upon Kin1 repression, we observed a defect in F-actin incorporation into the CAR at mitosis compared to Kin1 ON. In 84% of anaphase cells, F-actin patches and/or cables were present outside the main F-actin ring structure (n=32, arrowhead and asterisk, Fig. 4D). In contrast, tea4Δ single mutant cells did not show F-actin mislocalization during anaphase (Fig. 4D).
To determine if other CAR components were also displaced, we used GFP-tagged Rlc1 and Cdc15. Rlc1 myosin light chain incorporates into the CAR early in mitosis 38 whereas Cdc15-GFP incorporation is a criterion for its maturation 9. Rlc1-GFP appeared as a ring in late anaphase that constricts with the F-actin ring after spindle disassembly 8. In tea4Δ Kin1 ON, the Rlc1-GFP signal (middle panel, Fig. 4D) behaved essentially as in the tea4Δ single mutant (upper panel, Fig. 4D). In tea4Δ Kin1 OFF, 62.5% of late anaphase cells showed additional Rlc1-GFP dots outside the main ring structure (arrowhead, lower panel, Fig. 4D; n=32). Similarly, Cdc15-GFP dots were detected outside the main CAR structure in 45.7% of late anaphase tea4Δ Kin1 OFF cells (arrowhead, Fig. 4E; n=35). We also examined Mid1 localization in these cells but Mid1 is mislocalized on the cortex in a tea4Δ-dependent manner as previously shown 6, whatever Kin1 expression level (supplementary figure S2).
Taken together, detection of F-actin, Rlc1-GFP and Cdc15-GFP in tea4Δ cells indicated that Kin1 down-regulation induces a defect in late anaphase CAR structure once assembly was initiated.
CAR assembly defects detected in tea4Δ Kin1 OFF late anaphase cells (Fig. 4D,E) can be attributed to improper ring assembly in early metaphase and/or precocious disassembly once formed. To clarify these CAR defects, Rlc1-GFP was recorded by time-lapse video microscopy in mitotic cells. The Rlc1-GFP signal is first detected as a broad band of dots referred to as ‘cortical nodes’ in early mitosis 13. This broad band exactly reflect the early mitotic Mid1 cortical band 8, 13. The Rlc1-GFP signal then coalesces as a packed ring by lateral condensation 9, followed by a constant diameter period and ring constriction.
In tea4Δ Kin1 ON cells, the mean duration of lateral condensation was 21.3±3.1 minutes (n=22, see t=20 minutes, Fig. 4F; see also supplementary movie S4). Following the ring formation period, a 4–8 minute interval of constant diameter for Rlc1-GFP ring was observed in 81% of n=22 cells (from t=24 to t=28 minutes, Fig. 4F). The mean duration of ring constriction was 24.3±3.7 minutes in n=22 cells (t=32 to t=52 minutes, Fig. 4F).
In tea4Δ Kin1 OFF cells, the broad band of Rlc1-GFP dots was larger than in the Kin1 ON condition and biased towards one cell end (compare t=12 minutes, Fig. 4F and 4G; see supplementary movie S5). The mean duration of lateral condensation was longer (26.7±6.3 minutes, n=22). Importantly, lateral condensation was not completed in 36.3% of cells at t=28 minutes, a time at which 100% of tea4Δ Kin1 ON cells showed a packed ring. This delay is due to the presence of abnormal stretched structures or very distant dots that were gradually incorporated into the ring (ring formation, Fig. 4G). Thus, Kin1 down-regulation enlarged the initial recruitment area of Rlc1-GFP dots and delayed their compaction into a packed ring. Regarding the constant diameter period, the tea4Δ Kin1 OFF cell population was heterogeneous: 1) 40.9% of cells rapidly triggered ring constriction in a 0–4 minute interval after a packed ring is detected (e.g. some cells initiated constriction prior than showing a packed ring as shown on Fig. 4G); 2) 45.4% have a stable ring for 4–8 minutes (81% in Kin1 ON), and 3) 13.7% showed an extended period with a packed ring (> 8 minutes). Finally, a subset of cells (45.4%) retained a short Rlc1-GFP medial dot over an extended period (see t=60 to t=68 minutes, Fig. 4G) or extra dots outside the division area at a time when all Kin1 ON cells showed no more Rlc1-GFP signal. Thus, Kin1 down-regulation in tea4Δ uncoupled packed ring formation and constriction but also altered ring disassembly in a subset of cells.
Kin1 repression phenotype in tea4Δ cells is exacerbated by cell elongation
It has been reported that Mid1 broad band is wider when cells are longer. As a consequence, cell elongation exacerbates the pom1Δ phenotype, increasing septation defects 6. We asked if cell elongation and widening of Mid1 localization would potentiate Kin1 repression phenotype in tea4Δ. We introduced the repressible nmt81-kin1 allele to cdc25-22 cells that are longer than wild type cells because the cdc25-22 cells are delayed in G2 even at the permissive temperature (25°C). Total cell length at division was increased from 15.1±1.1 to 25.2±2.9 μm independently of Kin1 expression level (Fig. 5A) and the percentage of asymmetric septa upon Kin1 repression was similar to cdc25+ cells (Fig. 5B). We introduced the tea4Δ mutation to cdc25-22 nmt81-kin1 cells. In the Kin1 ON condition, elongation of cells increased the percentage of aberrant septations from 2.6±0.6% to 16.9±1.4%, probably due to exacerbation of the tea4Δ phenotype. Furthermore, in the Kin1 OFF condition, elongation of cells strickingly enhanced the aberrant septation phenotype, from 23.3±1.6% to 68.8±4.5% (Fig. 5C,D). Therefore, Kin1 repression in tea4Δ cells was sensitive to cell length and Mid1 mislocalization.
Figure 5. The cytokinetic phenotype of Kin1 repression in tea4Δ cells is exacerbated by cell elongation.
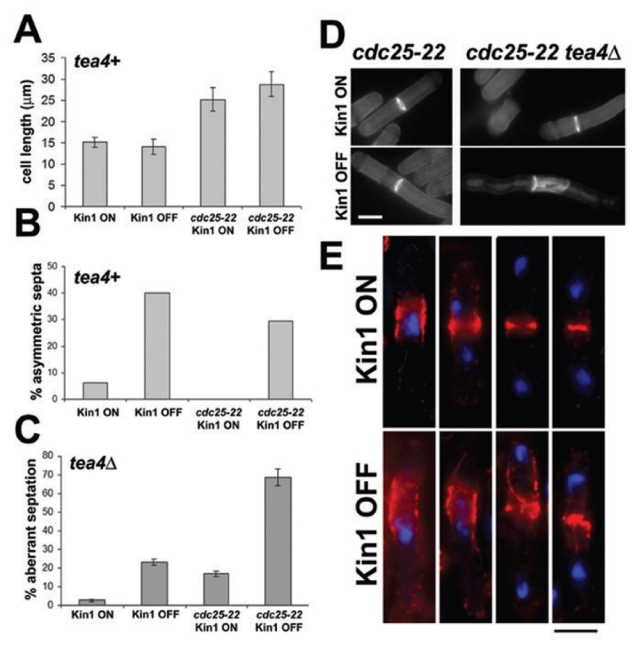
nmt81-controlled kin1 and cdc25-22 nmt81-controlled kin1 cells were cultured 16h at 25°C in the absence (Kin1 ON) and in the presence (Kin1 OFF) of thiamine: (A) total cell length of septated cells, (B) percentage of asymmetric septa (short/long ratio <0.85). Tea4Δ nmt81-controlled kin1 and tea4Δ cdc25-22 nmt81-controlled kin1 were cultured at 25°C in the Kin1 ON and Kin1 OFF conditions: (C) percentage of aberrant septations (n>30 in each sample, mean values of two independent experiments), (D) cdc25-22 nmt81-controlled kin1 and tea4Δ cdc25-22 nmt81-controlled kin1 were cultured at 25°C in the Kin1 ON and Kin1 OFF conditions and stained with methyl blue. (E) tea4Δ cdc25-22 nmt81-controlled kin1 cells were co-stained with DAPI (DNA, blue) and rhodamine-conjugated phalloidin (F-actin, red). Bars: 5 μm.
We examined CAR formation in these cells using F-actin and DNA co-staining (Fig. 5E). We observed a broad F-actin relocation signal beside the predivided nucleus in the Kin1 ON situation, consistent with a tea4Δ-dependent mislocalized Mid1 band that is extended by cell elongation 6. In the Kin1 OFF condition, this signal was dramatically expanded on the lateral cortex showing an enhanced widening of the recruitment area of F-actin. Later in mitosis, F-actin condensation as a packed ring was observed in Kin1 ON. In Kin1 OFF, F-actin condensation was defective or incomplete and F-actin structures emanating from the CAR at the end of anaphase were detected (Fig. 5E), probably corresponding to the extra septal material observed later (Fig. 5D).
We hypothesized that the nuclear centering deficiency in Kin1 OFF cells might be responsible for the CAR formation defects and aberrant cytokinesis in tea4Δ cells. To test this hypothesis, we used three different means for displacing the interphase nucleus prior to CAR assembly.
Overexpression of a putative catalytically inactive Kin1 mutant promotes cytokinesis failure in tea4Δ cells
When we overexpressed a putative catalytically inactive GFP-Kin1-K154R kinase (Material and methods) in wild type cells, we observed the displacement of the interphase nucleus towards one cell end where the mutant protein accumulated at the cortex. No effect on cell shape, F-actin polarity or cell separation were detected. The GFP signal gradually diminished towards the opposite cell end were F-actin was concentrated (arrow, tea4+, Fig. 6A). Hence, Kin1-K154R acts as a dominant negative mutant and its primary effect is to inhibit maintenance of the nucleus at the geometric cell centre. As a consequence, CARs were formed asymmetrically (arrowhead, tea4+, Fig. 6A).
Figure 6. Nuclear displacement induced defects in CAR formation and cytokinesis failure in tea4Δ andpom1Δ cells.
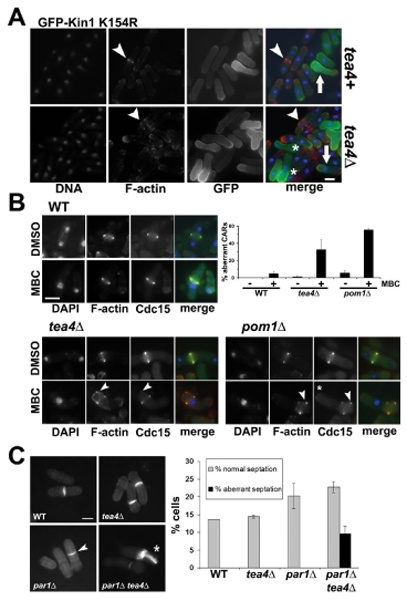
(A) Overexpression of kinase dead (GFP-Kin1 K154R) Kin1 versions in tea4+ or tea4Δ cells. After growth overnight at 30°C in the absence of thiamine (ON) in the culture medium, cells have been fixed and stained with DAPI (DNA, blue) and rhodamine-conjugated phalloidin (F-actin, red). Fluorescent signals and superposition of images are shown (merge). Arrowheads show normal (tea4+) or aberrant (tea4Δ) CARs and arrows indicate the displaced nuclei towards the cell end where GFP-Kin1 K154R accumulates. (B) Cdc15-GFP, tea4Δ cdc15-GFP and pom1Δ cdc15-GFP cells were treated for 2h with 50 μg/ml MBC or 1% DMSO as a control. Cells have been fixed and stained with DAPI (DNA, blue) and rhodamine-conjugated phalloidin (F-actin, red). Fluorescent signals and superposition of images are shown (merge). Arrowheads show abnormal F-actin structures or Cdc15-GFP dots outside the CARs and asterisks show cell end localized Cdc15-GFP. A histogram shows the percentage of aberrant CARs with both F-actin and Cdc15-GFP incorporation defects in the different strains with (+) or without (-) MBC. (C) Wild type (WT), tea4Δ, par1Δ and tea4Δ par1Δ cells have been grown at 30°C and stained with methyl blue. Arrowhead indicates a misplaced septum and asterisk shows an aberrant septation, histogram on the right shows the percentage of normal and aberrant septations in WT, tea4Δ, par1Δ and par1Δ tea4Δ cells (n>200 total cells, in duplicate). Bars: 5 μm.
We used this allele to conditionally move the nucleus within tea4Δ cells. When we overproduced Kin1-K154R in these cells, we observed nuclear displacement towards the cell end where GFP-Kin1-K154R concentrated (arrow, tea4Δ, Fig. 6A) as in tea4+. However, we also detected aberrant CARs (arrowhead, tea4Δ, Fig. 6A). We counted the percentage of binucleate post-mitotic cell compartments (4%, n>1000 cells) as a read out of ultimate cytokinesis abortion (asterisks, tea4Δ, Fig. 6A). This phenotype was not observed in tea4+ cells.
Depolymerization of interphase MTs in tea4Δ and pom1Δ mutants induced defective CAR formation
If nuclear mispositioning affects CAR formation efficiency in a tea4Δ background, we hypotesized that chemically induced nuclear displacement should affect CAR formation in tea4Δ but not tea4+ cells. MBC depolymerizes the MT network and, as a consequence, nuclei are no longer dynamically maintained in the cell centre in interphase but are heterogeneously displaced away from it 37. Cells are arrested with a CAR in metaphase-to-anaphase transition because of activation of the spindle assembly checkpoint. We monitored F-actin and Cdc15-GFP ring formation in MBC-treated cells. In wild-type cells, CAR formation was completed (see Cdc15-GFP incorporation in WT, Fig. 6B). Positions of CARs and nuclei were frequently uncoupled because nuclei moved after the CAR assembly site has been specified (WT, Fig. 6B). In tea4Δ cells, we observed that aberrant CARs were formed (arrowheads, bottom left panels, Fig. 6B; 32.6% of mitotically arrested cells with both F-actin and Cdc15-GFP incorporation defects, n=98). Consistently, we also observed aberrant CARs in MBC-treated pom1Δ cells (arrowhead and asterisk, bottom right panels, Fig. 6B; 55.7%, n=61). We conclude that MBC-mediated interphase nuclear displacement altered the efficiency of CAR formation in tea4Δ and pom1Δ but not in wild type cells.
The par1Δ tea4Δ double mutant exhibits cytokinesis defects
To support a role for nuclear positioning in CAR formation efficiency of tea4Δ cells, we used the par1Δ mutant. Par1 encodes a regulatory subunit of the phosphatase 2A (PP2A) heterotrimer. The par1Δ mutant shows a nuclear and septum (arrowhead, Fig. 6C; 25.6% of asymmetric septa versus 10.2% in wild type cells) mispositioning defect at 30°C as described 39. We out-crossed par1Δ to tea4Δ and examined septal structure at this temperature. Consistent with our hypothesis, par1Δ tea4Δ cells exhibited aberrant septations (asterisk, Fig. 6C) and polyploid cells (data not shown). The proportion of aberrant septation is 29.7% of septated cells (i.e. 9.6% of total cells) and is very similar to the proportion of asymmetric septa observed in the par1Δ single mutant (25.6%). Hence, the nuclear centering deficient par1Δ mutant interacted with tea4Δ with respect to cytokinesis, suggesting that par1Δ-mediated nuclear displacement alters cell division efficiency in tea4Δ cells.
DISCUSSION
Kin1 repression promotes nuclear mispositioning in wild type cells
In repressed nmt81-controlled kin1, nuclei were detected at offset positions closer to the new cell end in late G2 cells. As a consequence, the CAR and the subsequent septum formation were biased towards this cell end. Cell length of septated cells were not significantly different between the ON and OFF conditions, so asymmetry is not generated by a defect in arresting polarized growth in one cell end at mitotic onset. Thus, the primary defect of lowering Kin1 expression level in wild type cells is nuclear mispositioning. Nuclear dynamics between Kin1 ON and Kin1 OFF interphase cells are not significantly different, suggesting that nuclei acquired progressively an off-centered static position. Consistently, no microtubule network organization 22, 24 or dynamics (unpublished results) were detected in kin1Δ cells. Kin1 is present in the new cell end after cell separation until late G2 when cell division site is specified. This suggests that the nuclear centering function of Kin1 resides in this cell end. Consistently, overexpressed dominant negative GFP-Kin1-K154R accumulates with one cell end that correlates with nuclear proximity. Furthermore, this observation suggests a possible titration of a putative Kin1 binding partner at the cell ends while expressing Kin1-K154R.
Kin1 repression induces cytokinesis failure in a tea4Δ mutant
In addition to the previously described pom1Δ and tea1Δ, the present study revealed that the tea4Δ polarity mutant exhibits extensive septation defects when combined with kin1Δ. The absence of genetic interaction between tea3Δ, bud6Δ and mod5Δ mutants with kin1Δ indicates that monopolar growth per se is not responsible for promoting cytokinesis failure.
The conditional Kin1 OFF phenotype observed in tea1Δ and tea4Δ is strikingly similar to the Kin1 OFF effect in pom1Δ 25 albeit less severe, allowing a careful examination of the dynamic nature of CAR assembly defects. Our results strongly suggest that Tea1, Tea4 and Pom1 act in the same pathway. Furthermore, we observed that Kin1-GFP localization is not modified in a tea4Δ mutant nor in pom1Δ or tea1Δ (our unpublished results). Reciprocally, in kin1Δ cells, Tea1 24 and Pom1 (A. Paoletti, pers. comm.) cortical localizations are unaffected. Thus, Kin1 and Pom1/Tea1/Tea4 function in distinct control mechanisms to regulate cytokinesis.
Tip Occlusion is unlikely involved in synergistic defects between Kin1 OFF and tea4Δ
The report of Huang et al. 19 showed that Pom1, Tea1 and Tea4 are involved in an inhibitory mechanism that prevents septum synthesis in the cell ends (referred to as “Tip Occlusion”). The authors proposed that Tip Occlusion is essential for viability in the conditional mid1-18 mutant. A hypomorphic allele of cdc15, encoding a C-terminally GFP fused Cdc15, restored Tip Occlusion deficiency in mid1-18 although it did not restore cell viability. A proposed mechanism for Tip Occlusion is to delay incorporation of the septum synthesizing Cps1 protein until the CAR components are properly relocated from the cell ends to the central region 19. In our study, three lines of evidence rule out the possiblity that Tip Occlusion is involved in the tea4Δ Kin1 OFF phenotype. First, Cdc15-GFP was used to monitor CAR assembly and this allele did not rescue CAR or septation defects. Second, in cdc25-22 elongated cells where CAR formation occured significantly distant from the cell ends, aberrant septation was still observed. Third, kin1Δ cells did not influence the timing of Cps1 incorporation into the CAR (data not shown).
Regarding Kin1 localization in the cell ends (see below), we can not exclude the possibility that Kin1 also acts in a Tip Occlusion mechanism independent of the Tea4/Tea1/Pom1 system.
Mid1 mislocalization phenotype is exacerbated by Kin1 repression
It has been shown that the nucleus dynamically influences Mid1 and the cell division site positioning in fission yeast 35, 36. In kin1Δ cells, CAR formation 25 and septum synthesis 22 are only affected in their localization and Mid1 is properly localized around the nucleus (unpublished observations) as in Kin1 OFF cells (supplementary figure S2), suggesting that Kin1 is required for nuclear positioning but dispensable for Mid1 function and CAR assembly. Furthermore, a double deletion kin1Δ mid1Δ mutant showed additive phenotypes, indicating independent functions in CAR positioning (supplementary figure S3).
A common function of Pom1, Tea1 and Tea4 is to restrict Mid1 localization in the middle of the cell during interphase 4, 6. Consistently, pom1Δ cells show an extended Mid1 repartition around a half cell whereas the nucleus is still centered. At mitosis, the CAR formation area is eccentric but ultimately Rlc1-GFP (as well Mid1-GFP) coalesces into a packed ring. Interestingly, CAR position was still biased towards the nucleus, indicating that nuclear position is still influencing the site of CAR assembly in pom1Δ cells 6.
The tea1Δ mutant has a weaker phenotype on Mid1 localization than tea4Δ which is itself less proeminent than pom1Δ. These observations led to the conclusion that Pom1 is the major effector of Mid1 localization regulation and that contributions of Tea1 and Tea4 in this process are due to their function in anchoring Pom1 at the cortex 6. We observed that the Kin1 OFF phenotype was weaker in tea4Δ and even weaker in tea1Δ than in pom1Δ. In addition, the elongation of cells, that has been shown to exacerbate pom1Δ cytokinesis phenotype by expanding Mid1 medial band 6, increased cytokinesis defects in tea4Δ Kin1 OFF cells. Mid1 is mislocalized at the cell cortex in pom1Δ, tea1Δ and tea4Δ mutants and we were unable to detect an additional Mid1 mislocalization phenotype while repressing Kin1 (supplementary figure S2).
Nuclear displacement exacerbates Mid1 mispositioning phenotype
Upon pom1Δ cell centrifugation, Padte et al. showed that Mid1 still tracked the nucleus but the Mid1 band was broader6. However, CAR and septum formation were not studied in this situation. Kin1 repression, that solely mediates nuclear displacement, strongly impairs CAR formation in tea4Δ, tea1Δ and pom1Δ but not in wild type cells. Thus, our work reveals that maintenance of the nucleus in the middle of the cell is essential for CAR assembly in tea4Δ, tea1Δ and pom1Δ mutants.
Why Kin1 repression induced aberrant septation in tea4Δ cells? A possible explanation is that Kin1 OFF mediated extended CAR formation area delays CAR component incorporation in tea4Δ cells. As the CAR frequently constricts without being completed, it promotes septal material deposition in cells.
Consistent with our hypothesis, cytokinetic defects were observed in tea4Δ and/or pom1Δ cells when we used other means for nuclear displacements. Furthermore, when the nuclear centering deficient par1Δ mutant was combined with tea4Δ, we detected aberrant septations. Surprizingly, no interaction was previously reported between par1Δ and pom1Δ 39, maybe because aberrant septa were not discriminated from misoriented and misplaced septa resulting from the pom1Δ mutation. Par1Δ and kin1Δ mutations are synthetically lethal, indicating that Par1 and Kin1 have non-overlapping functions (unpublished results).
Kin1 shows a dynamic cell-cycle regulated association with the cell cortex
Using a C-terminally myc tagged Kin1 strain, Drewes and Nurse 24 detected Kin1 by immunofluorescence at the cell ends in interphase, consistent with its role in polarized growth. However, systematic gene tagging reports indicated a signal for Kin1 in the middle of a subset of cells 26, 40 but their exact cell cycle stage was not determined. When we reinvestigated Kin1 localization using a GFP tagged fusion protein, we efficiently detected Kin1-GFP at the cell ends and we also detected Kin1-GFP at the middle of living cells. The formaldehyde/glutaraldehyde fixation, used by Drewes and Nurse 24, prevented detection of Kin1-myc tagged protein by immunofluorescence at the middle of the cell whereas fixation by formaldehyde alone allowed observation of a Kin1-myc medial signal in mitotic cells (our unpublished results).
Using video-microscopy, we show that Kin1-GFP was recruited to the division site in two steps: first in the CAR in early mitosis and then as an invaginating signal when the septum is synthesized. Kin1 localization as a tight ring structure depends on an intact polymerized F-actin since LatA disrupted this signal. A kin1Δ strain did not show obvious CAR assembly defects but have cell separation defects 22, 23, 25. Thus, Kin1 may have no crucial function in CAR assembly or stability in a wild type background but rather may be recruited there to act in septum synthesis regulation.
Cortical anchoring of Kin1 is not dependent on F-actin but rather may be associated with membranes. This is consistent with the invaginating signal at septum synthesis that may correspond to daughter cell membrane closure. Accordingly, brefeldin A treatment that disrupts vesicular trafficking and plasma membrane organization abolished Kin1 polarization at the cell ends (data not shown).
Members of the KIN1/PAR-1/MARK serine/threonine kinase family have been detected at the cortex of cells in budding yeast, C. elegans, Drosophila and vertebrates 21. Hence, cortical localization of these kinases is a common theme, even though the underlying molecular mechanisms may be different. Furthermore, the MELK related kinase is specifically localized at the cortex from anaphase to telophase in Xenopus and human cultured cells, suggesting a potentially conserved recruitment event at the division site during mitosis 41.
In conclusion, our data show that mutations affecting nuclear centering have deleterious effects on CAR assembly and the subsequent septum synthesis in pom1Δ, tea4Δ and tea1Δ mutants. The slow diffusion between cortically-anchored Mid1 and the nucleus may be reponsible for exacerbating the pom1Δ, tea4Δ and tea1Δ phenotypes when the nucleus is not efficiently maintained in the cell centre. Our data show that the level of expression of Kin1 is a key regulatory element in this situation and reveal that two non-overlapping independent mechanisms (nuclear centering and Mid1 medial restriction) operate at mitotic onset to ensure faithful assembly of the division apparatus.
MATERIALS AND METHODS
S. pombe growth conditions and genetic manipulations
The fission yeast strains used in this study are listed in Table 1. Standard techniques and genetic approaches were used for the growth and manipulation of yeast cells as previously described 42. S. pombe strains were exponentially grown at 25°C and 30°C in YES rich medium or Edinburgh Minimal Medium (EMM) with appropriate supplements 42. EMM with 15 μM thiamine (Sigma) was used to repress expression from the nmt81 and nmt41 promoters. YES containing 100 mg of G418 (Gibco) per liter was used for selecting kanMX-expressing cells. Tetrad dissection was performed using a MSM Singer system and double mutants were selected on appropriate media. Methyl-2-benzimidazole-carbamate (MBC, Sigma) was diluted in DMSO at indicated final concentrations. Brefeldin A (Sigma) was diluted in ethanol and used at a final concentration of 100 μg/ml. Latrunculin A (Calbiochem) was diluted in DMSO and used at a final concentration of 25 μM.
TABLE 1.
S. pombe strains used in this study.
| strain | Genotype | source/reference |
|---|---|---|
| XLG029 | h- leu1-32 | Lab stock |
| XLG035 | h+ par1::ura4+ ura4-D18 ade6M216 | 39 |
| XLG053 | h- leu1-32 kin1::kanMX6 | 22 |
| XLG142 | h+ pom1::ura4+ ura4-D18 leu1-32 | 17 |
| FC486 | h- bud6::kanMX | 31 |
| XLG490 | h+ bud6::kanMX kin1::kanMX6 leu1-32 | This study |
| PN1687 | h- tea1::ura4+ ura4-D18 | 45 |
| XLG107 | h+ tea1::ura4+ kin1::kanMX6 ura4-D18 | 22 |
| XLG218 | nmt3GFPatb2 leu1-32 kanR | Lab stock |
| KS708 | h- mod5Δ::kanMX ura4-D18 ade6-M216 | 33 |
| XLG609 | h+ mod5Δ::kanMX kin1::kanMX6 leu1-32 | This study |
| CA2527 | h- wsh3/tea4::ura4+ leu1-32 ura4-D18 | 29 |
| XLG496 | h+ tea4::ura4+ kin1::kanMX6 leu1-32 ura4-D18 | This study |
| PN3231 | h+ tea3Δ::kanR leu1-32 | 34 |
| XLG441 | h- tea3::kanMX kin1::kanMX6 leu1-32 | This study |
| FC661 | h+ crn1-GFP-kanMX ade6- leu1-32 ura4-D18 | 28 |
| XLG288 | h+ kin1::kanMX-nmt81GFP-kin1 ura4-D18 leu1-32 | 25 |
| XLG646 | h- kin1::[kanR-nmt81-Kin1] leu1-32 | This study |
| XLG565 | h- tea1::ura4+ kin1::kanMX-nmt81GFP-kin1 ura4-D18 | This study |
| XLG511 | h+ tea4::ura4+ kin1::kanMX-nmt81GFP-kin1 ura4-D18 leu1-32 | This study |
| SP3890 | h- rlc1::GFP-kanMX leu1-32 ura4-D18 | 38 |
| XLG539 | h- tea4::ura4+ rlc1::GFP-kanMX leu1-32 ura4-D18 | This study |
| XLG547 | h+ tea4::ura4+ rlc1::GFP-kanMX kin1::kanMX-nmt81GFP-kin1 ura4-D18 leu1-32 | This study |
| XLG572 | h- kin1::GFP-kanMX6 ura4-D18 leu1-32 | This study |
| XLG578 | h- tea4::ura4+ kin1::GFP-kanMX6 leu1-32 ura4-D18 | This study |
| XLG624 | h- tea4::ura4+ par1::ura4+ ura4-D18 leu1-32 | This study |
| XLG648 | h- cdc25-22 kin1::[kanR-nmt81-Kin1] leu1-32 | This study |
| XLG649 | h+ cdc25-22 tea4::ura4+ kin1::[kanR-nmt81-Kin1] leu1-32 ura4-D18 | This study |
| XLG667 | h- kin1::GFP-kanMX6 cherryRFP-plx1+:leu+ura4-D18 leu1-32 | This study |
Chromosomally GFP tagged Kin1 strain
The C-terminal chromosomally GFP tagged Kin1 strain was obtained by oligonucleotide-mediated tagging 43. A five glycine linker was inserted between the end of kin1 and the start of GFP Open Reading Frames (ORFs). The following oligonucleotides were used: kin1-50s, 5′-GCG TTT CTT TTC ATC GAA TTA GTG GAA ACT CAT GGC AAT ATA AGA CAT TGG CTA GTC GTA TTC TTA ACG AAT TAA AAT TGG GCG GCG GCG GCG GCC GGA TCC CCG GGT TAA TTA ACA GTA AAG G-3′ and kin1-8as, 5′-GGT CTA GTT TCT CGT CTC TAA AAA GCT CAA CGT CTA ATA TAG TTC ACC TGA AGC GGT CTT ACT ATT AAA AAC CAT AAA CAG AAT TCG AGC TCG TTT AAA C-3′ with the pFA6a-GFP(S65T)-kanMX6 plasmid as a template. After transformation of h- leu1-32 cells with the PCR product, G418 resistant colonies were selected and analyzed by PCR and genomic sequencing to confirm integration at the kin1 locus. The selected kin1-GFP strain was back-crossed and we checked that cells exhibited a wild-type phenotype, indicating that the fusion protein is functional.
Overexpression of a Kin1 K154R mutant
Lysine to arginine mutation at position 154 was constructed by a two step PCR method. This residue was chosen after alignment with the Xenopus MELK protein kinase for which the corresponding K42R mutation led to a kinase dead enzyme 44. The mutagenized Kin1-K154R Open Reading Frame was inserted into pREP41GFP in NdeI cloning sites and sequenced. Kin1Δ cells transformed with pREP41GFP-Kin1-K154R were not rescued (data not shown), indicating that the kinase activity of Kin1 is required to fulfil its cellular functions. Transformation of pREP41GFP-Kin1-K154R to wild type and tea4Δ strains was performed as described 22. Overexpression was performed 16h in thiamine free medium at 30°C.
Microscopy techniques
For time-lapse video microscopy, cells were grown in EMM liquid medium with or without thiamine for nmt81-controlled Kin1 expression or YE5S liquid medium for Kin1-GFP at 25°C or 30°C. The nuclear dye Hoechst (Sigma) was added at 50 μg/ml to follow nuclear dynamics. Then 2 μl of cells were mounted on 2% EMM agar pads. GFP movies were captured using a spinning disc Nikon TE2000 microscope with a 100x 1.45 NA PlanApo and a HQ2 Roper camera. Nuclear movies were captured with a Leica DMIRE2 confocal microscope and a 100x PlanApo objective. Vizualization of Kin1-GFP in static cells was performed using a Leica DMRXA2 with a 100x 1.45 NA PlanApo objective and a HQ roper camera. DAPI/rhodamine-conjugated stained formaldehyde fixed cells were processed as previously described 22 and imaged using a DMRXA Leica microscope and a CoolSnap-ES camera. Fluorescent signals corresponding to the ON and OFF levels of the nmt81-controlled N-terminally GFP fused Kin1 protein were insufficient to be detected in our experimental microscopy conditions. Images were processed using Metamorph and ImageJ softwares.
Protein extracts and western blot analysis
Cells were harvested in ice-cold STOP buffer (10 mM EDTA, 150 mM NaCl, 50 mM NaF). Cell pellets were broken using glass beads (Sigma G8772). Protein extracts were prepared in RIPA buffer (10 mM Tris-HCl pH 7, 1% Triton X-100, 0.1% SDS, 2 mM EDTA, 150 mM NaCl, 50 mM NaF, 0.1 mM sodium vanadate, 10 μg/ml protease inhibitors (pepstatin, leupeptin, chymostatin, ICN), 1 mM PMSF). Protein concentration was determined by the DC Biorad method and 45 μg/lane of total proteins were separarated by SDS-PAGE and transferred onto polyvynilidene membranes Immobilon-P (Millipore). Kin1 was detected using a rabbit polyclonal antibody raised again a peptide located in the C-terminus of Kin1 at a 1:3000 dilution, and the Cdc2 protein was used as a loading control, detected by a monoclonal anti-PSTAIR antibody at a 1:5000 dilution (Sigma P7962). Immunodecorated proteins were revealed using the West Dura system (Pierce).
Acknowledgments
We thank J. Bähler, F. Chang, J. Hayles, P. Nurse, P. Perez, K. Sawin, K. Shiozaki and V. Simanis for the kind gift of strains. We thank C. Herbert and J.P. Tassan for critical reading of the manuscript and members of the laboratory for helpful discussions. We are very grateful to A. Paoletti for her invaluable expertise in live cell imaging at the Institut Curie (CNRS UMR144, Paris), for strains and critical comments on the manuscript. A. Cadou was supported by a PhD fellowship from the Région Bretagne (ARED2799). Fluorescence microscopy used the IFR140 microscopy platform and we thank S. Dutertre for her help in cell imaging processing.
Abbreviations
- CAR
contractile actomyosin ring
- DMSO
dimethyl sulfoxide
- LatA
latrunculin A
- MBC
methyl-2-benzimidazole-carbamate (carbendazim)
- MT
microtubule
References
- 1.Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–18. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Bähler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–16. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol Cell Biol. 2004;24:10621–35. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celton-Morizur S, Racine V, Sibarita JB, Paoletti A. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J Cell Sci. 2006;119:4710–8. doi: 10.1242/jcs.03261. [DOI] [PubMed] [Google Scholar]
- 5.Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–73. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padte NN, Martin SG, Howard M, Chang F. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr Biol. 2006;16:2480–7. doi: 10.1016/j.cub.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–19. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 8.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–34. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 9.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–16. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–88. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–8. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- 12.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 13.Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165:685–95. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin SG, Chang F. New end take off: regulating cell polarity during the fission yeast cell cycle. Cell Cycle. 2005;4:1046–9. [PubMed] [Google Scholar]
- 16.La Carbona S, Le Goff C, Le Goff X. Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex. Biol Cell. 2006;98:619–31. doi: 10.1042/BC20060048. [DOI] [PubMed] [Google Scholar]
- 17.Bahler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–70. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatebe H, Nakano K, Maximo R, Shiozaki K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr Biol. 2008;18:322–30. doi: 10.1016/j.cub.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Chew TG, Ge W, Balasubramanian MK. Polarity determinants Tea1p, Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell. 2007;12:987–96. doi: 10.1016/j.devcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Marks J, Hagan IM, Hyams JS. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci Suppl. 1986;5:229–41. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- 21.Tassan JP, Le Goff X. An overview of the KIN1/PAR-1/MARK kinase family. Biol Cell. 2004;96:193–9. doi: 10.1016/j.biolcel.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 22.La Carbona S, Allix C, Philippe M, Le Goff X. The protein kinase kin1 is required for cellular symmetry in fission yeast. Biol Cell. 2004;96:169–79. doi: 10.1016/j.biolcel.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Levin DE, Bishop JM. A putative protein kinase gene (kin1+) is important for growth polarity in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1990;87:8272–6. doi: 10.1073/pnas.87.21.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drewes G, Nurse P. The protein kinase kin1, the fission yeast orthologue of mammalian MARK/PAR-1, localises to new cell ends after mitosis and is important for bipolar growth. FEBS Lett. 2003;554:45–9. doi: 10.1016/s0014-5793(03)01080-9. [DOI] [PubMed] [Google Scholar]
- 25.La Carbona S, Le Goff X. Spatial regulation of cytokinesis by the Kin1 and Pom1 kinases in fission yeast. Curr Genet. 2006;50:377–91. doi: 10.1007/s00294-006-0099-5. [DOI] [PubMed] [Google Scholar]
- 26.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–7. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 27.Pinar M, Coll PM, Rincon SA, Perez P. Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol Biol Cell. 2008;19:1727–38. doi: 10.1091/mbc.E07-07-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelham RJ, Jr, Chang F. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat Cell Biol. 2001;3:235–44. doi: 10.1038/35060020. [DOI] [PubMed] [Google Scholar]
- 29.Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr Biol. 2005;15:1006–15. doi: 10.1016/j.cub.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 30.Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–91. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Glynn JM, Lustig RJ, Berlin A, Chang F. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr Biol. 2001;11:836–45. doi: 10.1016/s0960-9822(01)00235-4. [DOI] [PubMed] [Google Scholar]
- 32.Snaith HA, Samejima I, Sawin KE. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. Embo J. 2005;24:3690–9. doi: 10.1038/sj.emboj.7600838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snaith HA, Sawin KE. Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature. 2003;423:647–51. doi: 10.1038/nature01672. [DOI] [PubMed] [Google Scholar]
- 34.Arellano M, Niccoli T, Nurse P. Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr Biol. 2002;12:751–6. doi: 10.1016/s0960-9822(02)00821-7. [DOI] [PubMed] [Google Scholar]
- 35.Daga RR, Chang F. Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci U S A. 2005;102:8228–32. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolic-Norrelykke IM, Sacconi L, Stringari C, Raabe I, Pavone FS. Nuclear and division-plane positioning revealed by optical micromanipulation. Curr Biol. 2005;15:1212–6. doi: 10.1016/j.cub.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 37.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Goff X, Motegi F, Salimova E, Mabuchi I, Simanis V. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J Cell Sci. 2000;113(Pt 23):4157–63. doi: 10.1242/jcs.113.23.4157. [DOI] [PubMed] [Google Scholar]
- 39.Le Goff X, Buvelot S, Salimova E, Guerry F, Schmidt S, Cueille N, Cano E, Simanis V. The protein phosphatase 2A B′-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 2001;508:136–42. doi: 10.1016/s0014-5793(01)03047-2. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi A, Da-Qiao D, Tsutsumi C, Chikashige Y, Masuda H, Haraguchi T, Hiraoka Y. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14:217–25. doi: 10.1111/j.1365-2443.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 41.Chartrain I, Couturier A, Tassan JP. Cell-cycle-dependent cortical localization of pEg3 protein kinase in Xenopus and human cells. Biol Cell. 2006;98:253–63. doi: 10.1042/BC20050041. [DOI] [PubMed] [Google Scholar]
- 42.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 43.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–51. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Blot J, Chartrain I, Roghi C, Philippe M, Tassan JP. Cell cycle regulation of pEg3, a new Xenopus protein kinase of the KIN1/PAR-1/MARK family. Dev Biol. 2002;241:327–38. doi: 10.1006/dbio.2001.0525. [DOI] [PubMed] [Google Scholar]


