The chromosomal passenger complex (CPC) is a key mitotic regulator composed of Aurora B kinase, Survivin, Borealin and INCENP.[1]The centromeric complex is involved in the correction of mis-attached microtubules by sensing tension on kinetochores.
Aurora kinase phosphorylates microtubule binding factors, decreases their affinity for microtubules, and in turn, spindle microtubules are destabilized around kinetochores. The main Aurora B substrates involved in this dynamic are Ndc80/Hec1, a subunit of the Kinetochore-Microtubule-Network (KMN), and the microtubule-destabilizing factors MCAK and Stathmin/OP18. In the past, the main question was to elucidate the mechanism by which tension is sensed on centromere/kinetochore and how it may regulate Aurora B kinase activity.[2]Recently, Liu et al. have elegantly solved this question.[3]By using FRET-based bio-sensors and playing with the targeting of Aurora B, these authors have established that the spatial separation of Aurora B kinase from kinetochore substrates senses chromosome biorientation. In the absence of tension, kinetochore substrates in the vicinity of the kinase are phosphorylated and their affinity for microtubules is lowered. Kinetochore-microtubules attachments are thus destabilized. Conversely when tension is exerted, kinetochore substrates are pulled away from the kinase, their phosphorylation decreases and therefore microtubules are stabilized around kinetochores (Figure 1 A).[3, 4]
Figure 1. Aurora B kinase, an immobile centromeric passenger.
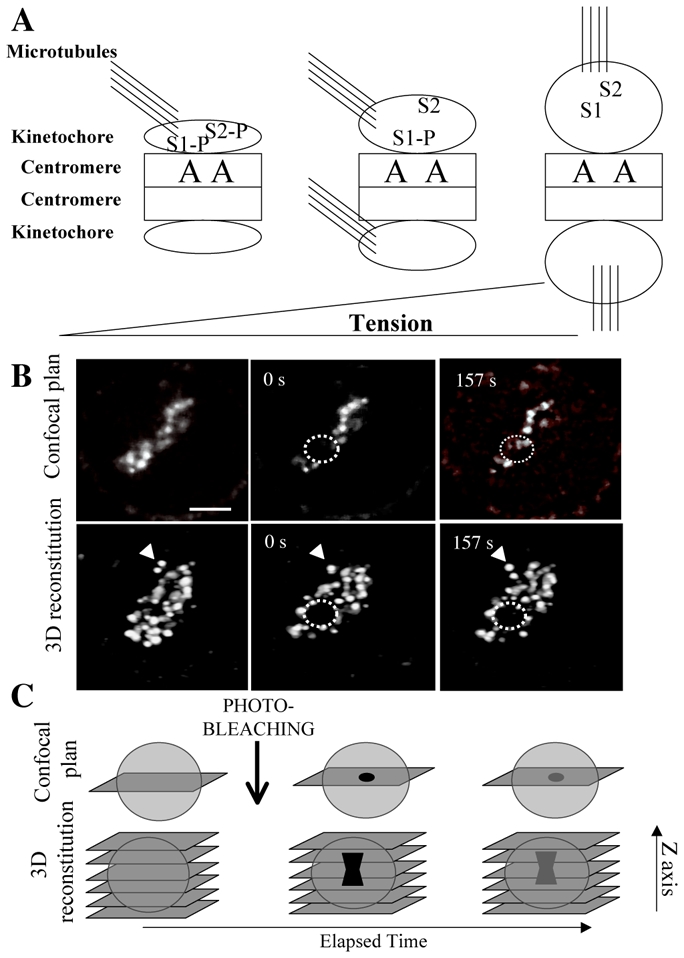
Part A: Aurora B kinase (A) present on the centromeres phosphorylates substrates (S1, S2) on kinetochores. Upon tension kinetochores are stretched and Aurora B substrates moves away from the enzyme.[3,4]In the presence of phosphatases, they are dephosphorylated. Centromeres are represented by squares, microtubules by lines and kinetochores by ellipses.
Part B: Analysis of Aurora B-GFP recovery after photobleaching on centromeres.
Both a confocal plan and a 3D reconstitution image are recorded at the indicated times (0s: just post bleaching and 157s: last imaged time). The dotted lines show the photo-bleached area and the arrowhead was placed to illustrate how misinterpretation may occur.
Although on the confocal plan, some fluorescence is noted in the bleached area (157 s) the fluorescence has not recovered the bleached region as visualized on the reconstituted images (157 s post-bleaching). Aurora B-GFP is thus mostly immobile on centromere.6
Part C: Description of the 3D-FRAP technique.[6]First image the whole cell (Z-sectioning), then choose a confocal plan and realize the bleaching of few centromeres. Perform Z-stack imaging just after bleaching and at the end of the experiment. All experiences in which too many movements are noted in the reconstituted images should not be analyzed. This allows the discrimination between cell movement and recovery of fluorescence on bleached centromeres.
Which implication of this model for Aurora kinase and its partners?
Aurora kinase phosphorylates close substrates and the cascade of phosphorylation is regulated by the tension across kinetochores (Figure 1A). Therefore, Aurora kinase activity should not be modulated during mitosis. As a consequence of this model, the kinase must be mostly immobile on the inner centromere. Actually, the C-terminus of Borealin is necessary for targeting the complex to the centromere and, Aurora B is incorporated into the CPC via binding to the IN-box of INCENP. One may thus expect that the three subunits are poorly dynamic on centromeres. Conflicting data were published on the dynamic of Aurora B and INCENP[5–9]but we have reported that Aurora B, INCENP and Borealin are fully immobile during prometaphase and metaphase.[6, 8]
All this gives light to the 3D-FRAP technique we have developed.[6]Indeed bleached centromeres are tiny, very dynamic structures and we have noted that mobility of the structures themselves may be a source of misinterpretation of FRAP data (Figure 1 B). The movement of fluorescent objects could be wrongly interpreted as a recovery of fluorescence. We have suggested performing a 3D reconstitution of the structure at different times during the experiment and especially at the end of recovery (Figure 1B, C). In such experiments, another source of artefact may also be introduced by the use of drugs that prevents anaphase onset, like MG132. Passenger protein level is tightly controlled by proteolysis and preventing such regulation induces an artificial pool of free proteins that may interfere with and destabilize the complex.
The 3D-FRAP is thus a convenient technique for studying the dynamic of centromeric proteins. It has revealed that Survivin was mobile on centromere and then sticky to its partners on microtubules. Further experiments are needed to ascertain the role of Survivin but it may compete with other Aurora B substrates or have a direct role on microtubule nucleation around kinetochores.[10]The distinct behaviour of Survivin on centromere and on microtubules may suggest the existence of at least two CPC complexes.
More data are necessary for elucidating the role of the CPC, its organisation within the centromere and for analysing dynamically Aurora B kinase substrates. In the near future, the definition of the interfaces between inner and outer centromeres and centromere/kinetochore will progress with the high-resolution fluorescent microscopies.
Acknowledgments
We would like to thank Dr Stéfan Dimitrov for support and discussion. This work was supported by ANR (Project R05075CC). The team is sponsored by “La Ligue Nationale contre le Cancer (Equipe labelisée La Ligue)”.
References
- 1.Ruchaud S, et al. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. Review. [DOI] [PubMed] [Google Scholar]
- 2.Kelly AE, et al. Curr Opin Cell Biol. 2009;21:51–8. doi: 10.1016/j.ceb.2009.01.004. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, et al. Science. 2009;323:1350–3. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santaguida S, et al. EMBO J. 2009 doi: 10.1038/emboj.2009.173. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murata-Hori M, et al. J Cell Biol. 2002;159:45–53. doi: 10.1083/jcb.200207014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delacour-Larose M, et al. Cell Cycle. 2004;3:1418–26. doi: 10.4161/cc.3.11.1203. [DOI] [PubMed] [Google Scholar]
- 7.Beardmore VA, et al. J Cell Sci. 2004;117:4033–42. doi: 10.1242/jcs.01242. [DOI] [PubMed] [Google Scholar]
- 8.Delacour-Larose M, et al. Cell Cycle. 2007;6:1878–85. doi: 10.4161/cc.6.15.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahonen LJ, et al. Chromosoma. 2009;118:71–84. doi: 10.1007/s00412-008-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosa J, et al. Mol Biol Cell. 2006;17:1483–93. doi: 10.1091/mbc.E05-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]


