Abstract
Sapovirus is a genus of caliciviruses that are known to cause enteric disease in humans and animals. There is considerable genetic diversity among the sapoviruses, which are classified into different genogroups based on phylogenetic analysis of the full-length capsid protein sequence. While several mammalian species, including humans, pigs, minks, and dogs, have been identified as animal hosts for sapoviruses, there were no reports of sapoviruses in bats in spite of their biological diversity. In this report, we present the results of a targeted surveillance study in different bat species in Hong Kong. Five of the 321 specimens from the bat species, Hipposideros pomona, were found to be positive for sapoviruses by RT-PCR. Complete or nearly full-length genome sequences of approximately 7.7 kb in length were obtained for three strains, which showed similar organization of the genome compared to other sapoviruses. Interestingly, they possess many genomic features atypical of most sapoviruses, like high G+C content and minimal CpG suppression. Phylogenetic analysis of the viral proteins suggested that the bat sapovirus descended from an ancestral sapovirus lineage and is most closely related to the porcine sapoviruses. Codon usage analysis showed that the bat sapovirus genome has greater codon usage bias relative to other sapovirus genomes. In summary, we report the discovery and genomic characterization of the first bat calicivirus, which appears to have evolved under different conditions after early divergence from other sapovirus lineages.
Introduction
The caliciviruses are a family of small non-enveloped viruses, and can be classified into five genera: Vesivirus, Lagovirus, Norovirus, Sapovirus and Nebovirus. They possess a non-segmented, polyadenylated, positive-sense ssRNA genome of about 7.5 to 8.5 kb in length, enclosed in an icosahedral capsid of 27 to 40 nm in diameter. Among them, noroviruses and sapoviruses (SaVs) are well known to cause enteric disease in a range of mammals, including humans, while vesiviruses and lagoviruses cause systemic diseases in specific animal hosts. Nebovirus is the most recently established genus in the family Caliciviridae [1], and its members are associated with enteric diseases in cattle [2], [3]. A putative sixth genus, Recovirus, has been proposed for a novel calicivirus detected in stool specimens from rhesus monkeys [4], [5]. Another new genus, Valovirus, has been proposed for a novel group of swine caliciviruses known as the St-Valérien-like viruses [6]. In addition, there exist other unclassified caliciviruses, such as the recently described chicken calicivirus [7].
The genus Sapovirus currently contains only one recognized species, the Sapporo virus, which was discovered in 1977 in Sapporo, Japan [8]. The SaV genome is approximately 7.1 to 7.5 kb in length, and may have two or three ORFs. ORF1 encodes a polyprotein that undergoes proteolytic cleavage to form the non-structural proteins and the major capsid protein VP1. ORF2 encodes the minor structural protein VP2. ORF3 encodes a small basic protein of unknown function [9], [10]. Interestingly, it is located in an overlapping reading frame within ORF1, and is present only in SaVs from selected genogroups. At present, SaVs are classified formally into 5 genogroups based on phylogenetic analysis of the full-length VP1 sequence, though additional genogroups have been proposed to accommodate some novel SaVs discovered in recent years. Further classification of SaVs into genotypes has also been undertaken, though taxonomic assignment at the genotype level appears to be less well-defined than at the genogroup level [11].
As mentioned above, both noroviruses and SaVs generally cause mild to asymptomatic enteric infections in human and animal hosts [12]. Human SaV infections are reported to be similar to or milder than human norovirus infections, but SaV infections have a shorter duration of viral shedding and are less associated with projectile vomiting [13]–[16]. Incidence of SaV-associated gastroenteritis infections remains less than norovirus-associated infections for both sporadic and outbreak settings, though various studies have reported increasing rates of SaV infections around the world [17]–[21]. The genetic diversity of SaVs is comparable to that of noroviruses, and the diversity of reported animal hosts is also similar. Noroviruses have been discovered in specimens from humans, pigs, cattle, dogs, sea lions, African lion, and mice [22]–[25]. In comparison, SaVs have been found in specimens from humans, pigs, dogs, minks and California sea lions [23], [25]–[27].
Bats (order Chiroptera of class Mammalia) constitute a significant portion of biological diversity in many ecosystems and have a wide geographical distribution [28]. We have previously discovered novel viruses in several local bat species [29]–[35], and there were many similar discoveries of novel bat viruses by researchers in other parts of the world. In particular, important human viral pathogens like the SARS virus, Nipah virus and Ebola virus were found to have originated from bats and contributed to substantial human morbidity and mortality in recent outbreaks. Taken together, these discoveries hint that these small mammals are important reservoirs of diverse and undiscovered animal viruses, with significant risk of zoonotic transmission to humans [36].
In the present study, we investigated the presence of unknown calicivirus diversity in bats by targeted RT-PCR screening. Novel SaV sequences were amplified from several faecal samples of the bat species Hipposideros pomona, and genome sequences were obtained for three strains of the bat SaV. Sequence analysis indicated that the novel virus possesses several genomic features atypical of SaVs, and phylogenetic analysis revealed that it descended from a lineage that had diverged early from other SaV.
Results
Surveillance and detection of novel SaVs in bats
A total of 728 anal swabs from different bat species in Hong Kong were obtained. No obvious signs of enteric disease, like anorexia and diarrhoea, were observed in the bats during the brief period of captivity needed for sampling.
RT-PCR using broadly reactive degenerate primers for a 185 nt fragment in the 3D-like RNA-dependent RNA polymerase (RdRp) region of the calicivirus ORF1 gene was positive in two specimens. Repeated screening using more sensitive specific primers revealed three additional positive specimens. Further information on the species and RT-PCR screening results are presented in Table 1, Table S1 and Figure S1. Sequence similarity search using BLASTN against the NCBI non-redundant nucleotide database did not reveal significant similarity to known SaV sequences. Another search using BLASTX against the NCBI non-redundant protein database produced hits to SaV sequences, with the most closely related sequence being the RdRp sequence of porcine SaV (GenBank accession number ACT98315) at 43% aa identity. A phylogenetic tree was constructed from the nucleotide alignment based on the length of the partial RdRp sequence obtained from bat SaV/TLC72 (GenBank accession number JQ267527) (Figure S2).
Table 1. Species distribution of specimens and RT-PCR surveillance results in the present study.
| Bat species | Number of animals screened by RT-PCR | Number (%) of animals with positive detection of SaV |
| Hipposideros armiger | 14 | - |
| Myotis ricketti | 103 | - |
| Miniopterus pusillus | 78 | - |
| Myotis chinensis | 18 | - |
| Rhinolophus sinicus | 65 | - |
| Tylonycteris pachypus | 14 | - |
| Hipposideros pomona | 321 | 5 (1.56%) |
| Pipistrellus abramus | 9 | - |
| Miniopterus schreibersii | 84 | - |
| Nyctalus noctula | 1 | - |
| Pipistrellus sp. | 2 | - |
| Scotophilus kuhlii | 1 | - |
| Rhinolophus affinis | 9 | - |
| Rhinolophus pusillus | 9 | - |
Genome sequencing and analysis of novel bat SaVs
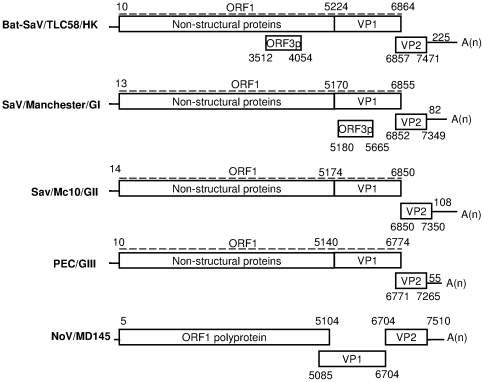
Complete or nearly full-length genome sequences (with incomplete 5′ ends) were obtained for three positive samples using the sequencing strategy as described in the Methods section. For two of the samples that were positive only for RT-PCR screening with specific primers, only sequences for short segments of the viral genome were obtained. Additional viral genome sequencing on these samples was unsuccessful due to limited clinical materials available and possibly low viral titres. The complete genome of bat SaV strain TLC58 (Genbank accession number JN899075) is 7696 nt in length and has a genomic G+C content of 60.2 mol%. Both the length and G+C content of the bat SaV genome are significantly higher than that of other known SaVs (Table 2). Each genome is predicted to contain 3 overlapping ORFs, comparable to the genome organization of SaVs in GI, GIV and GV (Figure 1). The 5′-UTR and the 3′-UTR are 9 nt and 225 nt in length, respectively. The length of the 3′-UTR is considerably longer than other SaVs (Table 2). The two other nearly full-length bat SaV genomes were found to be highly similar to that of the complete bat SaV/TLC58 genome in nucleotide sequence and genome organization, and were not analysed separately.
Table 2. Comparison of genomic features among selected SaV.
| Virus | Genogroup* | Genome length (nt) | Genomic G+C content (mol%) | ORF1 length (nt) | ORF2 length (nt) | 3′-UTR length (nt) | ORF2 reading frame relative to ORF1 | Presence of putative ORF3 |
| Bat SaV/TLC58/HK | N/A | 7696 | 60.2 | 6855 | 615 | 225 | +1 | Y |
| SaV/Manchester | I | 7431 | 50.7 | 6843 | 498 | 82 | −1 | Y |
| SaV/Mc10 | II | 7548 | 51.5 | 6837 | 501 | 108 | −1 | N |
| Porcine enteric calicivirus | III | 7320 | 53.5 | 6765 | 495 | 55 | −1 | N |
| SaV/Hu/Chiba/000671/1999/JP | IV | 7420 | 53.5 | 6816 | 504 | 91 | −1 | Y |
| SaV/Hu/Ehime475/2004/JP | V | 7500 | 49.8 | 6906 | 501 | 83 | −1 | Y |
| Canine SaV† | N/A | Incomplete | Incomplete | Incomplete | 501 | 141 | +1 | N |
| California sea lion 1 SaV† | N/A | Incomplete | Incomplete | Incomplete | 504 | Incomplete | −1 | N |
Only the formally recognized SaV genogroups are included.
Complete genome sequence is not available for canine SaV and California sea lion 1 SaV.
Sequence accession numbers are as follows: Bat SaV/TLC58/HK (JN899075), SaV/Manchester (X86560), SaV/Mc10 (NC_010624), porcine enteric calicivirus (PEC) (AF182760), SaV/Hu/Chiba/000671/1999/JP (AJ786349), SaV/Hu/Ehime475/2004/JP (DQ366344), canine SaV (JN387134), California sea lion 1 SaV (JN420370).
Figure 1. Genome organization of the bat SaV TLC58/HK.
The genome organization of the bat SaV TLC58/HK in comparison with the genome organization of human SaV GI strain Mancheseter, human SaV GII strain Mc10, porcine enteric calicivirus, and norovirus GII strain MD145.
The complete ORF1 is 6855 nt long, and encodes a large precursor polyprotein with an estimated molecular mass of 246.8 kDa. The polyprotein contains characteristic amino acid motifs conserved in caliciviruses: 2C-like NTPase at residue 482 (GPPGIGKT), VPg at residues 958 (KGKTK) and 972 (SEYEE), 3C-like protease at residue 1183 (GDCG), 3D-like RNA-dependent RNA polymerase at residues 1520 (GLPSG) and 1568 (YGDD), and VP1 at residue 1867 (PPG). It undergoes proteolytic processing to produce the nonstructural viral proteins and the major capsid protein VP1. Based on comparison with the ORF1 cleavage map of SaV/Mc10 [37], [38], a human SaV GII strain, we predicted the cleavage site that generates the major capsid protein to be located between residues 1740 (E) and 1741 (G). An in-frame AUG start codon is located in a favourable context for translation initiation (GUGUUUGUGAUGGA) just upstream to the cleavage site, which has also been reported in other caliciviruses [6], [39]. This sequence is noted to be similar to the 5′ UTR of the genome, and it was postulated that the site might permit internal translation initiation from subgenomic RNA [39]. The sequence identities of the bat SaV/TLC58 with other SaVs in the complete ORF1 protein sequence vary between 36.0% and 37.4% (Table 3). While comparison with caliciviruses of other genera, the ORF1 sequence identities are overall lower (15.6%–22.8%) than those between different SaVs (Table S2). For individual alignment of protease – polymerase, sequence identities with other SaVs (45.3%–48.4%) are overall higher than those with other genera (22.7%–32.1%) (Table 3 and Table S2). The VP1 is predicted to be 546 aa long, and has a molecular mass of 56.6 kDa. It shares 36.1% to 39.2% amino acid identities with VP1 of other SaVs (Table 3). Likewise analysis with caliciviruses of other genera reveals lower similarities with 14.9% to 23.4% sequence identities (Table S2).
Table 3. Comparison of genome identities and amino acid identities between the predicted polyproteins of bat SaV and the selected SaV.
| Viruses | Genogroup | Bat SaV/TLC34/HK | Bat SaV/TLC39/HK | Bat SaV/TLC58/HK | ||||||||||||
| Pairwise nt identity (%) | Pairwise amino acid identity (%) | Pairwise nt identity (%) | Pairwise amino acid identity (%) | Pairwise nt identity (%) | Pairwise amino acid identity (%) | |||||||||||
| Complete genome | ORF1 | Pro-Pol | VP1 | VP2 | Complete genome | ORF1 | Pro-Pol | VP1 | VP2 | Complete genome | ORF1 | Pro-Pol | VP1 | VP2 | ||
| SaV/Manchester | GI | 44.7 | 36.3 | 47.2 | 36.3 | 16.9 | 44.8 | 36.4 | 47.5 | 35.8 | 16.9 | 44.8 | 36.4 | 47.5 | 36.1 | 16.9 |
| SaV/Mc10 | GII | 45.2 | 37.1 | 48.1 | 37.5 | 17.4 | 45.2 | 37.1 | 48.2 | 37.5 | 17.4 | 45.2 | 37.1 | 48.2 | 37.3 | 17.4 |
| PEC | GIII | 45.3 | 36.5 | 45.4 | 36.3 | 15.5 | 45.3 | 36.7 | 45.7 | 36.6 | 15.5 | 45.3 | 36.6 | 45.7 | 36.3 | 15.5 |
| SaV/Chiba/000671 | GIV | 46.1 | 37.3 | 48.2 | 38.5 | 19.9 | 46.0 | 37.4 | 48.4 | 38.2 | 19.9 | 46.0 | 37.4 | 48.4 | 38.3 | 19.9 |
| SaV/Ehime475 | GV | 44.2 | 35.9 | 45.9 | 36.9 | 19.4 | 44.2 | 36.0 | 46.1 | 37.1 | 19.4 | 44.2 | 36.0 | 46.1 | 36.9 | 19.4 |
| Canine SaV | N/A | N/A | N/A | 46.7 | 39.3 | 16.5 | N/A | N/A | 47.0 | 39.2 | 16.5 | N/A | N/A | 47.0 | 39.2 | 16.5 |
| California sea lion 1 SaV | N/A | N/A | N/A | 45.2 | 38.9 | 17.9 | N/A | N/A | 45.3 | 38.9 | 17.9 | N/A | N/A | 45.3 | 38.9 | 17.9 |
| Bat SaV/TLC34/HK | N/A | - | - | - | - | - | 99.5 | 99.3 | 99.7 | 98.5 | 100 | 99.6 | 99.6 | 99.7 | 99.2 | 100 |
| Bat SaV/TLC39/HK | N/A | 99.5 | 99.3 | 99.7 | 98.5 | 100 | - | - | - | - | - | 99.8 | 99.6 | 100 | 99.0 | 100 |
| Bat SaV/TLC58/HK | N/A | 99.6 | 99.6 | 99.7 | 99.2 | 100 | 99.8 | 99.6 | 100 | 99.0 | 100 | - | - | - | - | - |
Pro-Pol: Protease - Polymerase.
The complete ORF2 is 615 nt long, with an overlapping region of 8 nt with the 3′ terminus of ORF1. Its reading frame is +1 relative to that of ORF1, unlike most other SaVs (Table 2). ORF2 encodes the minor structural protein VP2, which has an estimated molecular mass of 21 kDa. The mechanism of translation initiation in ORF2 of SaVs has not been fully elucidated. In the present case, a translational upstream ribosome binding site (TURBS) motif (CAUGGGACC; underline indicates region complementary to 18 S ribosomal rRNA sequence) could be identified at 24 nt upstream of the ORF2 start codon. Sequence identities for VP2 with other SaVs vary from 15.5% to 19.9% (Table 3). By comparison with caliciviruses of other genera, sequence identities are generally lower than those between SaVs. (4.8%–12.3%) (Table S2).
Phylogenetic and recombination analysis
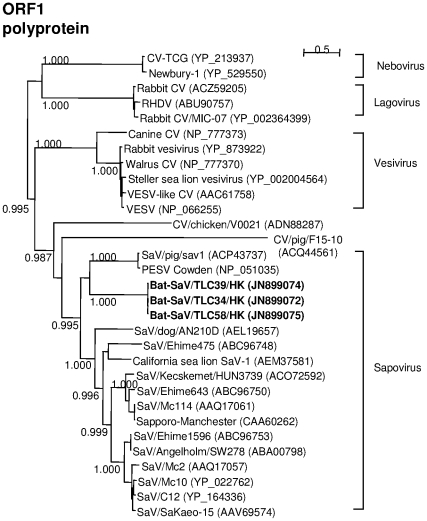
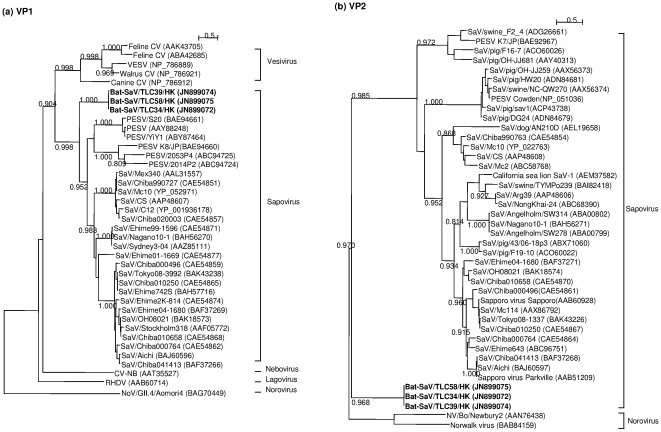
Phylogenetic trees were constructed using the predicted amino acid sequences of the ORF1 precursor polyprotein (Figure 2), VP1 and VP2 (Figure 3). The LG+G+F model was found to be the best-fit substitution model in all cases. Phylogenetic analysis was not performed for the putative ORF3 product as no homologous sequences were available. Sequence analysis with the Recombination Analysis Tool did not reveal any potential recombination breakpoints in the bat SaV sequences.
Figure 2. Unrooted maximum-likelihood tree based on full-length amino acid sequences of ORF1 precusor polyprotein.
SH-like aLRT branch support values of greater than 0.70 are shown besides major branches. Scale bar indicates the number of inferred substitutions per site.
Figure 3. Unrooted maximum-likelihood trees of VP1 and VP2.
The trees were constructed based on the full-length amino acid sequences of (a) VP1 major capsid protein, and (b) VP2 minor structural protein. SH-like aLRT branch support values of greater than 0.70 are shown besides major branches. Scale bar indicates the number of inferred substitutions per site.
There are subtle but important differences in the phylogenetic position of the bat SaV in the three phylogenetic trees. In the tree based on the full-length amino acid sequences of the ORF1 polyprotein, the bat SaVs are clustered tightly with the porcine SaVs in a monophyletic clade constituting the SaVs. However, in the VP1 tree, the bat SaVs are positioned just outside the clade of other SaVs. In the VP2 tree, the bat SaVs are located approximately equidistant from the GII noroviruses and porcine SaVs. The phylogenetic positions of the bat SaVs are supported by high Shimodaira-Hasegawa-like approximate likelihood ratio test (SH-like aLRT) branch support values as calculated by PhyML.
Although the phylogenetic positions of the novel bat virus are slightly divergent in the three trees, they generally show the bat SaV as being most closely related to the SaVs. In our opinion, there is insufficient ground for proposing a new genus for the novel virus under the current framework of taxonomic classification. The ORF1 polyprotein and VP1 capsid protein sequences of the novel bat virus showed obvious phylogenetic clustering with other SaV sequences. It should also be noted that the VP2 protein sequences are shorter and more divergent, and therefore are considered to be less useful in the phylogenetic classification of caliciviruses [4]. Lastly, the genome organization of the bat SaV is highly similar to that of the SaVs as shown above. Hence, together with relatively high sequence identities with other SaVs rather than with calicivirues in other genera (Table S2), we propose that the novel bat virus be classified as a new member of the genus Sapovirus in the family Caliciviridae.
Codon usage and compositional bias analysis
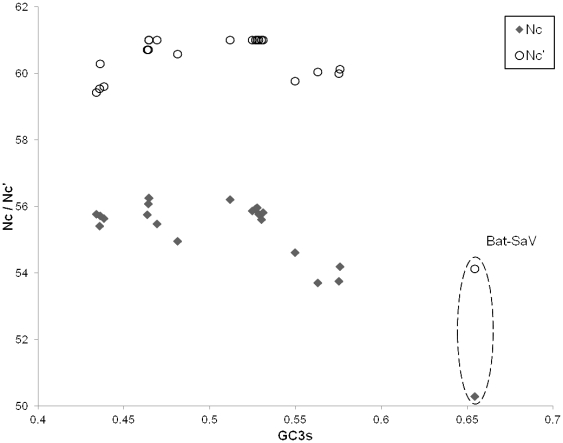
As genomic nucleotide composition is strongly associated with codon usage bias in viruses, we examined the codon usage in the genomes of the novel bat SaV and other SaVs given their different nucleotide composition. The bat SaV genome was found to have significantly greater codon usage bias than the other SaV genomes, as measured by their effective number of codons (Nc) (Figure 4). Adjusting Nc for background nucleotide composition (Nc′) did not significantly affect the observed difference in codon usage bias.
Figure 4. Scatterplot of codon usage.
The scratterplot of codon usage summary statistics Nc and Nc′ against the proportion of G or C nucleotides at the 3rd position of synonymous codons (GC3s), showing greater codon usage bias in the bat SaV genome relative to other SaV genomes. Unlike the porcine enteric caliciviruses, the observed difference in codon usage bias persists with adjustment of background nucleotide composition (Nc′).
Next, we examined CpG dinucleotide bias in the SaV genomes, as studies on other animal RNA viruses suggest that CpG suppression is a major factor in their genome evolution [40], [41]. Odds ratio of CpG and GpC dinucleotides (ρCG and ρGC) and the CpG/GpC ratio were calculated to assess the degree of CpG suppression. Results confirm the presence of significant CpG suppression (ρCG≤0.78) in examined SaV genomes, with the only exception being the bat SaV genome (Table 4). ρGC values are similar across examined SaV genomes, suggesting that the difference in CpG suppression is specific. All SaV genomes are found to have a slightly negative GC skew, and there is no major difference between the degree of GC skew in bat SaV and the other SaVs (Table 4). This suggests that the degree of cytosine deamination is not a major factor in the altered GC composition and CpG suppression in the bat SaV genome.
Table 4. CpG dinucleotide bias in selected SaV genomes, as assessed by the odds ratio of CpG (ρCG) and other measures.
| Virus | SaV genogroup | ρCG | ρGC | CpG/GpC ratio | GC skew |
| Bat SaV/TLC58/HK | N/A | 0.792 | 1.017 | 0.778 | −0.011 |
| SaV/Manchester | I | 0.530 | 0.954 | 0.556 | −0.017 |
| SaV/Mc10 | II | 0.587 | 0.938 | 0.626 | −0.032 |
| Porcine enteric calicivirus | III | 0.520 | 1.016 | 0.511 | −0.016 |
| SaV/Hu/Chiba/000671/1999/JP | IV | 0.665 | 0.976 | 0.681 | −0.045 |
| SaV/Hu/Ehime475/2004/JP | V | 0.490 | 0.968 | 0.507 | −0.036 |
Significantly less CpG suppression was found in the bat SaV genome, while a similar degree of negative GC skew was observed in all SaV genomes.
Discussion
Although the taxonomic classification of caliciviruses has improved with the availability of full-length gene sequences and robust phylogenetic methods [42], the increase in genetic diversity introduced by novel caliciviruses would necessitate further taxonomic revisions within the family. The International Committee on Taxonomy of Viruses has adopted a systematic polythetic approach towards virus taxonomy, but classification at or below the genus level may be complicated by the specific biology of diverse viruses. As a case in point, the proposed assignment of the novel bat virus to the genus Sapovirus might be opposed on the basis of an increased genomic G+C content, the different reading frame of ORF2, and the increased length of the 3′ UTR. On the other hand, the polythetic criteria for inclusion in the genus are not fully clear, and phylogenetic distances between viral gene sequences have assumed overriding importance in previous and current classifications. It should be noted that even phylogenetic analysis may be confounded by other factors such as the cleavage pattern of ORF1 polyprotein, which has not been determined experimentally for many caliciviruses.
Among the various notable genomic features and properties in the novel bat SaV, we were most intrigued by its remarkably high G+C genomic content. Most caliciviruses have a genomic G+C content of 44.2–57.4 mol%. Among them, the genomic G+C content of the SaVs lie within the relatively narrow range of 49.0–53.6 mol% in spite of their genetic diversity. Hence, the presently observed G+C genomic content of 60.2% is significantly higher than that for other SaVs or caliciviruses, and indeed would rank amongst mammalian RNA viruses with the highest G+C genomic content [43]. Relatively little is known about the evolution of genome composition in caliciviruses. A number of factors have been postulated to exert selectional pressure on the G+C content of viral genomes, including host body temperature, immune pressure, codon and nucleotide usage patterns [44]–[47]. Our results suggested that the increased G+C content is associated with a decrease in CpG suppression, but does not have a direct correlation with codon usage bias. We are unaware of any previous findings indicating that genomes of bat viruses are under less CpG suppression, thus the observed reduction in CpG suppresion is unlikely to result from host-related factors. The greater codon usage bias in the bat SaV genome is another interesting genome feature, which could be associated with altered dinucleotide frequencies. The association could be tested by Markov modelling of the dinucleotide and codon frequencies in the SaV genomes, although the small genome sizes and the presently small number of complete genomes would limit the usefulness of this approach [48].
The novel SaV described presently is the first known member of the Caliciviridae in bats. The approach to its discovery is based on the established strategy of targeted genetic screening informed by conserved sequences of related viruses. Although this “homology-based” strategy has been successful in the discovery of numerous viruses, the advent of affordable high-density microarrays and high-thoughput sequencing has given rise to virus discovery through metagenomics. Indeed, the first canine SaVs were discovered recently by metagenome sequencing of canine diarrhoea samples on a high-throughput pyrosequencer [23]. Important advantages of the new method include detection of novel viruses not closely related to known viruses, and the capacity to detect multiple divergent viruses in cases of co-infection. However, metagenomics sequencing can suffer from possible bias during sample preparation [49], and it is unlikely to detect very low titres of viruses in a specimen, such as the three bat faecal samples that were positive upon repeat screening with specific PCR primers in the present study. While we anticipate the increasing utilization of the metagenomics approach, existing methods such as viral culture, electron microscopy and targeted nucleic acid amplification would continue to serve important roles in virus discovery.
As Hong Kong is a highly urbanized city, the local roosting sites of bats are mainly man-made structures, such as water tunnels and abandoned mines. Hipposideros pomona is very common and widespread throughout Hong Kong countryside areas. It is a small-sized leaf-nosed bat with body weight ranged from 6–8 g. It possesses a small nose leaf which is simple, small, and lacking of lateral leaflets (Figures S3 and S4). This species may aggregate in small chambers or enclosures where the air flow is relatively limited. The 5 SaV-infected specimens were all captured in a place called Tai Lam – Shek Kong located next to a major country park of Hong Kong, and this roosting site shares similar ecological characteristic with other sampled roosting sites. Due to the extremely high human population density in Hong Kong, direct contact between humans and bats is relatively frequent. Fortunately, no local case of bat zoonosis has ever been reported [36]. The relatively large genetic distance between the present bat SaV and other mammalian SaVs suggests that the zoonotic risk posed by this virus is likely to be low, though this should be confirmed with further in vitro and in vivo studies.
There are two main limitations in the current study. First and foremost, clinical information on the sampled bats is limited to the brief period of captivity needed for sample collection, which is unlikely to reflect the disease association of the virus accurately. In other words, the scope of the study is limited to surveillance of viral diversity and possible discovery of new viruses. Secondly, the number of samples for the novel virus is quite small, despite the use of specific PCR primers for screening and the relatively large number of samples collected. Thus, we were unable to draw conclusions on the seasonality of its detection or its host specificity. To address these limitations, long-term follow-up studies would be required to identify sufficient positive samples with associated clinical data. Increasing the scale of surveillance would also help, though there are practical geographical and logistic constraints in our locality.
In conclusion, we identified a novel bat SaV with several genomic features and properties that set it apart from other members of the genus Sapovirus. Phylogenetic analysis suggests that its ancestral lineage had diverged early from the other SaVs and evolved under different conditions. Further discovery and characterization of additional strains would enhance our understanding of the evolutionary history of the SaVs and other caliciviruses.
Materials and Methods
Surveillance and sample collection
The study was approved by the Department of Agriculture, Fisheries and Conservation, HKSAR; and Committee on the Use of Live Animals in Teaching and Research, The University of Hong Kong. Bats from 14 different locations in rural areas of Hong Kong, including water tunnels, closed mines, sea caves and forested areas, were captured over a 36-month period. Anal swabs were collected by an experienced veterinary surgeon, and kept in viral transport medium at 4°C before processing.
RNA extraction
Viral RNA was extracted from the anal swabs using a QIAamp Viral RNA mini kit (Qiagen). The RNA was eluted into 50 µl RNase-free water and was used as the template for RT-PCR.
RT-PCR of the RdRp region using conserved primers, and sequencing
Screening was performed by amplifying a 185 nt fragment in the RdRp region of the ORF1 gene of caliciviruses. Conserved degenerate primers (5′-GAYTAYTCNMRRTGGGAYTC-3′ and 5′- GGCATNCCNGAKGGNAYNCC -3′) were designed from the multiple sequence alignment of the available calicivirus gene sequences in NCBI GenBank. First-strand cDNA synthesis was performed using SuperScript III kit (Invitrogen) according to manufacturer's instructions. The PCR mixture (25 µl) contained cDNA, PCR buffer (10 mM Tris/HCl pH 8.3, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin), 200 µM of each dNTP and 1.0 U AmpliTaq Gold polymerase (Applied Biosystems). PCR cycling conditions were as follows: hot start at 94°C for 7 min, followed by 50 cycles of 94°C for 1 min, 50°C for 1 min and 72°C for 1 min with a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystems). Standard precautions were taken to avoid PCR contamination and no false-positive signal was observed in the negative controls. The PCR products were gel-purified using a QIAquick gel extraction kit (Qiagen). Both strands of the PCR products were sequenced twice with an ABI Prism 3730xl DNA Analyser (Applied Biosystems), using the two PCR primers.
RT-PCR screening of bat sapovirus using specific primers
Additional RT-PCR screening was performed on the same samples using specific primers designed from the RdRp nucleotide sequences of bat SaVs obtained from previous rounds of RT-PCR and sequencing, as RT-PCR screening with specific primers usually offers higher sensitivity than a comparable screening with consensus degenerate primers. Sequences of the specific primers are as follows: forward primer 5′- CACAATGCAGCCAGCCA-3′ and reverse primer 5′- GGTGCGCGTGGTGAACAC-3′. PCR cycling conditions were as follows: hot start at 94°C for 7 min, followed by 50 cycles of 94°C for 1 min, 52°C for 1 min and 72°C for 1 min with a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystems). Standard precautions were taken to avoid PCR contamination and no false-positive signal was observed in the negative controls. PCR product purification and sequencing were performed as above.
Cloning of PCR product and sequencing
Purified PCR products were cloned into a pCR2.1-TOPO vector (Invitrogen) according to manufacturer's instructions. The vector was then used to transform the competent Escherichia coli strain DH5α by electroporation. Positive transformants were identified by blue–white screening, and eight colonies were selected for DNA sequencing of the construct using the M13 forward and reverse primers according to the manufacturer's instructions. Sequencing reactions were performed as described above.
Genome sequencing
Viral genome sequences were obtained using strategies we had previously used for other RNA viruses [50]–[52]. RNA extraction and cDNA generation were performed as described above. PCR primers were designed by targeting conserved regions, which were identified from the multiple alignment of genomes of related SaVs, as primer-binding sites. Additional primers for subsequent rounds of PCR were designed based on the results of earlier rounds of genome sequencing. The complete set of primer sequences is available from the authors upon request. The 5′ and 3′ ends of the viral genomes were sequenced following amplification of the segments by rapid amplification of cDNA ends, which was performed using the SMARTer RACE cDNA Amplification kit (Clontech) according to the manufacturer's instructions.
Phylogenetic and genome analysis
ORFs were located using the ORF Finder tool at NCBI (http://www.ncbi.nlm.nih.gov/projects/gorf/). Annotation of the predicted proteins was performed by BLAST sequence similarity search against annotations in the NCBI RefSeq database. Multiple sequence alignments were constructed using MUSCLE version 3.8.31 [53], and phylogenetic informative regions were extracted using BMGE [54]. Maximum-likelihood phylogenetic trees were constructed using PhyML version 3 [55], under the best-fit protein evolution model as selected by ProtTest 3 [56]. Branch support values were estimated by calculation of SH-like aLRT values [57]. Recombination detection was performed by analysing the translated sequences of ORF1 and ORF2 separately using the Recombination Analysis Tool [58].
Codon usage and compositional bias analysis
The full-length ORF1 and ORF2 coding sequences were extracted from selected SaV genomes and concatenated for codon usage analysis (see Table 4 for the list of included genome sequences). Codon usage and summary statistic of codon usage bias (Nc and Nc′) were calculated using the INCA package version 2.1 [59], where Nc is the effective number of codons in the coding regions of the genome [60], and Nc′ is the effective number of codons adjusted for background nucleotide composition [61]. For CpG dinucleotide bias analysis, odds ratio of CpG and GpC dinucleotides and the CpG/GpC ratio were calculated as described in previous studies [40], [41]. Odds ratio of ≤0.78 indicates significant suppression of the dinucleotide, same as the interpretation criteria of previous studies. Symmetrized nucleotide frequencies and dinucleotide odds ratio were not considered in the present study, as SaV genomes consist of positive-sense ssRNA only. To investigate the possible effects of cytosine deamination, genomic GC skew, which is the ratio (G-C)/(G+C), was calculated for the SaV genomes. The strength of the GC skew had been suggested to correlate with the degree of cytosine deamination [41], [44], [62].
Supporting Information
Geographical distribution of the bat specimens in the present study.
(TIF)
Neighbor-joining tree of partial RdRp nucleotide sequences. The tree was constructed based on the length of the nucleotide sequence in the RdRp region obtained from bat SaV/TLC72.
(TIF)
Photo showing Hipposideros pomona is in the drainage at Tai Lam – Shek Kong.
(TIF)
Photo showing Hipposideros pomona possesses a small nose leaf.
(TIF)
Epidemiology of the tested bat specimens.
(DOC)
Amino acid identity of the Bat SaV/TLC58/HK with representative caliciviruses of other genera.
(DOC)
Acknowledgments
We thank Director Alan Chi-Kong Wong, Siu-Fai Leung, Chik-Chuen Lay, Ping-Man So and K. F. Chan [HKSAR Department of Agriculture, Fisheries, and Conservation (AFCD)] and Hong Kong Police Force for facilitation and support; Chung-Tong Shek, Cynthia S. M. Chan and Joseph W. K. So from AFCD for their excellent technical assistance and collection of animal specimens. Photos and ecological information of the bats roosting sites are reproduced with kind permission from AFCD. Views expressed in this paper are those of the authors only, and may not represent the opinion of the AFCD or the Government of the HKSAR. We are grateful for the generous support of Mr Hui Hoy and Mr Hui Ming in the genomic sequencing platform.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is partly supported by the Research Grant Council; University Development Fund, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; and the Shaw Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smiley JR, Chang KO, Hayes J, Vinje J, Saif LJ. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. Journal of virology. 2002;76:10089–10098. doi: 10.1128/JVI.76.20.10089-10098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Martino B, Di Profio F, Martella V, Ceci C, Marsilio F. Evidence for recombination in neboviruses. Veterinary microbiology. 2011;153:367–372. doi: 10.1016/j.vetmic.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 3.Kaplon J, Guenau E, Asdrubal P, Pothier P, Ambert-Balay K. Possible novel nebovirus genotype in cattle, France. Emerging infectious diseases. 2011;17:1120–1123. doi: 10.3201/eid1706.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farkas T, Sestak K, Wei C, Jiang X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. Journal of virology. 2008;82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farkas T, Dufour J, Jiang X, Sestak K. Detection of norovirus-, sapovirus- and rhesus enteric calicivirus-specific antibodies in captive juvenile macaques. The Journal of general virology. 2010;91:734–738. doi: 10.1099/vir.0.015263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.L'Homme Y, Sansregret R, Plante-Fortier E, Lamontagne AM, Ouardani M, et al. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus genes. 2009;39:66–75. doi: 10.1007/s11262-009-0360-3. [DOI] [PubMed] [Google Scholar]
- 7.Wolf S, Reetz J, Otto P. Genetic characterization of a novel calicivirus from a chicken. Archives of virology. 2011;156:1143–1150. doi: 10.1007/s00705-011-0964-5. [DOI] [PubMed] [Google Scholar]
- 8.Chiba S, Sakuma Y, Kogasaka R, Akihara M, Horino K, et al. An outbreak of gastroenteritis associated with calicivirus in an infant home. Journal of medical virology. 1979;4:249–254. doi: 10.1002/jmv.1890040402. [DOI] [PubMed] [Google Scholar]
- 9.Clarke IN, Lambden PR. Organization and expression of calicivirus genes. The Journal of infectious diseases. 2000;181(Suppl 2):S309–316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- 10.Atmar RL, Estes MK. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clinical microbiology reviews. 2001;14:15–37. doi: 10.1128/CMR.14.1.15-37.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L'Homme Y, Brassard J, Ouardani M, Gagne MJ. Characterization of novel porcine sapoviruses. Archives of virology. 2010;155:839–846. doi: 10.1007/s00705-010-0651-y. [DOI] [PubMed] [Google Scholar]
- 12.Bank-Wolf BR, Konig M, Thiel HJ. Zoonotic aspects of infections with noroviruses and sapoviruses. Veterinary microbiology. 2010;140:204–212. doi: 10.1016/j.vetmic.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Rockx B, De Wit M, Vennema H, Vinje J, De Bruin E, et al. Natural history of human calicivirus infection: a prospective cohort study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2002;35:246–253. doi: 10.1086/341408. [DOI] [PubMed] [Google Scholar]
- 14.Chiba S, Nakata S, Numata-Kinoshita K, Honma S. Sapporo virus: history and recent findings. The Journal of infectious diseases. 2000;181(Suppl 2):S303–308. doi: 10.1086/315574. [DOI] [PubMed] [Google Scholar]
- 15.Percival S, Chalmers R, Embrey M, Hunter P, Sellwood J, et al. Norovirus and sapovirus. Microbiology of Waterborne Diseases. London: Academic Press; 2004. pp. 433–444. [Google Scholar]
- 16.Logan C, O'Sullivan N. Detection of viral agents of gastroenteritis: Norovirus, Sapovirus and Astrovirus. Future Virology. 2007;3:61–70. [Google Scholar]
- 17.Svraka S, Vennema H, van der Veer B, Hedlund KO, Thorhagen M, et al. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. Journal of clinical microbiology. 2010;48:2191–2198. doi: 10.1128/JCM.02427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang XL, Lee BE, Tyrrell GJ, Preiksaitis JK. Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004–2007. The Journal of infectious diseases. 2009;199:547–551. doi: 10.1086/596210. [DOI] [PubMed] [Google Scholar]
- 19.Tam CC, Rodrigues LC, Viviani L, Dodds JP, Evans MR, et al. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut. 2011 doi: 10.1136/gut.2011.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khamrin P, Maneekarn N, Peerakome S, Tonusin S, Malasao R, et al. Genetic diversity of noroviruses and sapoviruses in children hospitalized with acute gastroenteritis in Chiang Mai, Thailand. Journal of medical virology. 2007;79:1921–1926. doi: 10.1002/jmv.21004. [DOI] [PubMed] [Google Scholar]
- 21.Monica B, Ramani S, Banerjee I, Primrose B, Iturriza-Gomara M, et al. Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. Journal of medical virology. 2007;79:544–551. doi: 10.1002/jmv.20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martella V, Lorusso E, Decaro N, Elia G, Radogna A, et al. Detection and molecular characterization of a canine norovirus. Emerging infectious diseases. 2008;14:1306–1308. doi: 10.3201/eid1408.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, et al. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. The Journal of general virology. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martella V, Campolo M, Lorusso E, Cavicchio P, Camero M, et al. Norovirus in captive lion cub (Panthera leo). Emerging infectious diseases. 2007;13:1071–1073. doi: 10.3201/eid1307.070268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scipioni A, Mauroy A, Vinje J, Thiry E. Animal noroviruses. Veterinary journal. 2008;178:32–45. doi: 10.1016/j.tvjl.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Guo M, Evermann JF, Saif LJ. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Archives of virology. 2001;146:479–493. doi: 10.1007/s007050170157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Shan T, Wang C, Cote C, Kolman J, et al. The fecal viral flora of California sea lions. Journal of virology. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson DE, Reeder DM. Mammal species of the world : a taxonomic and geographic reference. Baltimore: Johns Hopkins University Press; 2005. [Google Scholar]
- 29.Lau SK, Woo PC, Lai KK, Huang Y, Yip CC, et al. Complete genome analysis of three novel picornaviruses from diverse bat species. Journal of virology. 2011;85:8819–8828. doi: 10.1128/JVI.02364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau SK, Poon RW, Wong BH, Wang M, Huang Y, et al. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. Journal of virology. 2010;84:11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Lau SK, Woo PC, Wong BH, Wong AY, Tsoi HW, et al. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology. 2010;404:106–116. doi: 10.1016/j.virol.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau SK, Woo PC, Li KS, Huang Y, Wang M, et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo PC, Lau SK, Li KS, Poon RW, Wong BH, et al. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau SK, Li KS, Huang Y, Shek CT, Tse H, et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. Journal of virology. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong S, Lau S, Woo P, Yuen KY. Bats as a continuing source of emerging infections in humans. Reviews in medical virology. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka T, Katayama K, Ogawa S, Hansman GS, Kageyama T, et al. Proteolytic processing of sapovirus ORF1 polyprotein. Journal of virology. 2005;79:7283–7290. doi: 10.1128/JVI.79.12.7283-7290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oka T, Yamamoto M, Katayama K, Hansman GS, Ogawa S, et al. Identification of the cleavage sites of sapovirus open reading frame 1 polyprotein. The Journal of general virology. 2006;87:3329–3338. doi: 10.1099/vir.0.81799-0. [DOI] [PubMed] [Google Scholar]
- 39.Hansman GS, Oka T, Takeda N. Sapovirus-like particles derived from polyprotein. Virus research. 2008;137:261–265. doi: 10.1016/j.virusres.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Rima BK, McFerran NV. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. The Journal of general virology. 1997;78(Pt 11):2859–2870. doi: 10.1099/0022-1317-78-11-2859. [DOI] [PubMed] [Google Scholar]
- 41.Karlin S, Doerfler W, Cardon LR. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? Journal of virology. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berke T, Matson DO. Reclassification of the Caliciviridae into distinct genera and exclusion of hepatitis E virus from the family on the basis of comparative phylogenetic analysis. Archives of virology. 2000;145:1421–1436. doi: 10.1007/s007050070099. [DOI] [PubMed] [Google Scholar]
- 43.Kapoor A, Simmonds P, Lipkin WI, Zaidi S, Delwart E. Use of nucleotide composition analysis to infer hosts for three novel picorna-like viruses. Journal of virology. 2010;84:10322–10328. doi: 10.1128/JVI.00601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenbaum BD, Rabadan R, Levine AJ. Patterns of oligonucleotide sequences in viral and host cell RNA identify mediators of the host innate immune system. PloS one. 2009;4:e5969. doi: 10.1371/journal.pone.0005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ElHefnawi M, Alaidi O, Mohamed N, Kamar M, El-Azab I, et al. Identification of novel conserved functional motifs across most Influenza A viral strains. Virology journal. 2011;8:44. doi: 10.1186/1743-422X-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobo FP, Mota BE, Pena SD, Azevedo V, Macedo AM, et al. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PloS one. 2009;4:e6282. doi: 10.1371/journal.pone.0006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. Journal of molecular evolution. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 48.Tse H, Cai JJ, Tsoi HW, Lam EP, Yuen KY. Natural selection retains overrepresented out-of-frame stop codons against frameshift peptides in prokaryotes. BMC genomics. 2010;11:491. doi: 10.1186/1471-2164-11-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim KH, Bae JW. Amplification Methods Bias Metagenomic Libraries of Uncultured Single-Stranded and Double-Stranded DNA Viruses. Applied and environmental microbiology. 2011;77:7763–7768. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo PC, Lau SK, Lam CS, Lai KK, Huang Y, et al. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. Journal of virology. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo PC, Lau SK, Huang Y, Lam CS, Poon RW, et al. Comparative analysis of six genome sequences of three novel picornaviruses, turdiviruses 1, 2 and 3, in dead wild birds, and proposal of two novel genera, Orthoturdivirus and Paraturdivirus, in the family Picornaviridae. The Journal of general virology. 2010;91:2433–2448. doi: 10.1099/vir.0.021717-0. [DOI] [PubMed] [Google Scholar]
- 52.Tse H, Chan WM, Tsoi HW, Fan RY, Lau CC, et al. Rediscovery and genomic characterization of bovine astroviruses. The Journal of general virology. 2011;92:1888–1898. doi: 10.1099/vir.0.030817-0. [DOI] [PubMed] [Google Scholar]
- 53.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC evolutionary biology. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 56.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Systematic biology. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 58.Etherington GJ, Dicks J, Roberts IN. Recombination Analysis Tool (RAT): a program for the high-throughput detection of recombination. Bioinformatics. 2005;21:278–281. doi: 10.1093/bioinformatics/bth500. [DOI] [PubMed] [Google Scholar]
- 59.Supek F, Vlahovicek K. INCA: synonymous codon usage analysis and clustering by means of self-organizing map. Bioinformatics. 2004;20:2329–2330. doi: 10.1093/bioinformatics/bth238. [DOI] [PubMed] [Google Scholar]
- 60.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 61.Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Molecular biology and evolution. 2002;19:1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- 62.Cardon LR, Burge C, Clayton DA, Karlin S. Pervasive CpG suppression in animal mitochondrial genomes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3799–3803. doi: 10.1073/pnas.91.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographical distribution of the bat specimens in the present study.
(TIF)
Neighbor-joining tree of partial RdRp nucleotide sequences. The tree was constructed based on the length of the nucleotide sequence in the RdRp region obtained from bat SaV/TLC72.
(TIF)
Photo showing Hipposideros pomona is in the drainage at Tai Lam – Shek Kong.
(TIF)
Photo showing Hipposideros pomona possesses a small nose leaf.
(TIF)
Epidemiology of the tested bat specimens.
(DOC)
Amino acid identity of the Bat SaV/TLC58/HK with representative caliciviruses of other genera.
(DOC)