Abstract
Background
The Bone Morphogenetic Protein 4 gene (BMP4) is located in chromosome 14q22-q23 which has shown evidence of linkage for isolated nonsyndromic cleft lip with or without cleft palate (NSCL/P) in a genome wide linkage analysis of human multiplex families. BMP4 has been shown to play crucial roles in lip and palatal development in animal models. Several candidate gene association analyses also supported its potential risk for NSCL/P, however, results across these association studies have been inconsistent. The aim of the current study was to test for possible association between markers in and around the BMP4 gene and NSCL/P in Asian and Maryland trios.
Methodology/Principal Findings
Family Based Association Test was used to test for deviation from Mendelian assortment for 12 SNPs in and around BMP4. Nominal significant evidence of linkage and association was seen for three SNPs (rs10130587, rs2738265 and rs2761887) in 221 Asian trios and for one SNP (rs762642) in 76 Maryland trios. Statistical significance still held for rs10130587 after Bonferroni correction (corrected p = 0.019) among the Asian group. Estimated odds ratio for carrying the apparent high risk allele at this SNP was 1.61 (95%CI = 1.20, 2.18).
Conclusions
Our results provided further evidence of association between BMP4 and NSCL/P.
Introduction
Cleft lip with or without cleft palate (CL/P) is the most common human craniofacial malformation with a prevalence in newborns between 1/500–1/2000 worldwide [1], [2], accounting for approximately one third of all congenital abnormalities [3]. In etiologic studies, CL/P is usually classified as syndromic or nonsyndromic based on the concurrent existence of other congenital malformations or developmental abnormalities. Almost 70% of the children born with a CL/P phenotype are nonsyndromic cases [4]–[7]. As the lip and primary palate have distinct developmental origins from the secondary palate, clefts of these areas can be further subdivided into cleft lip with or without cleft palate and cleft palate alone to maximize homogeneity of cleft phenotype [8]. Although a large number of candidate gene studies have been conducted in this field, and several genome-wide association studies (GWAS) [9]–[12] have recently provided more clues, specific causal variants and biological mechanisms responsible for occurrence of nonsyndromic cleft lip with or without cleft palate (NSCL/P) remain unclear.
The bone morphogenetic protein 4 gene (BMP4) on chromosome 14q22-q23 is one of the promising candidate genes for NSCL/P. Animal models show it is predominately expressed in epithelia of the maxillary and mandibular primordia [13], [14], as well as the mesenchymal tissue around the frontonasal process and lateral maxillary prominence in the C578L/6J mice [15]. When the exogenous antagonist Noggin for BMP2 and BMP4 was administered to the region of beak fusion, visible clefts of the upper beak were shown in 10 of 13 chick embryos [14]. Further, bilateral fusion delay of the lip was reported in all nine mice with BMP4 conditional null allele at 12 embryonic days [16]. In humans, the chromosomal region around 14q22-q23 gave evidence of linkage (significant at a genome-wide level) to a gene controlling NSCL/P in multiplex NSCL/P families recruited from various populations [17], [18]. However, results from published association studies have been inconsistent [9]–[12], [19]–[22] suggesting a need for further research.
Here we tested for linkage and association between markers in and around BMP4 gene and NSCL/P using 297 case parent trios from Maryland and three Asian populations to further clarify the potential role BMP4 may play in the etiology of this common and complex disorder. Our results provided further evidence of association between BMP4 and NSCL/P.
Materials and Methods
Ethics statement
Study protocols were reviewed and approved by the institutional review boards in all participating institutions. All adult participants (including adult probands and probands' parents) and parents or guardians of pediatric participants provided written informed consent.
Sample description
A total of 297 NSCL/P probands and their parents were recruited from four cleft lip and palate treatment centers: Maryland (76 trios from Johns Hopkins and University of Maryland hospitals), Taiwan (146 trios from Chang Gung Memorial Hospital), Singapore (35 trios from KK Women's and Children's Hospital), and Korea (40 trios from Yonsei Medical Center) (Table 1). All probands had received clinical genetic assessment to check for other birth defects or developmental delays and were all diagnosed as NSCL/P cases.
Table 1. Gender of NSCL/P probands.
| Site | Male | Female | Total |
| Maryland | 44 | 32 | 76 |
| Korea | 22 | 18 | 40 |
| Singapore | 24 | 11 | 35 |
| Taiwan | 95 | 51 | 146 |
| Total | 185 | 112 | 297 |
SNP selection, DNA, and genotyping
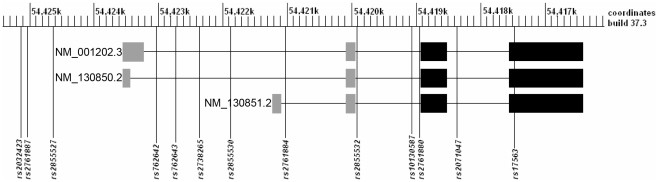
A total of 13 SNPs (Table 2 and 3, and Fig. 1) were genotyped in and around BMP4 with a goal of identifying an average of one SNP per 5 kilobase pairs (kb) of physical distance where one SNP locates in exon 4 (rs17563), another nine SNPs locate in introns and the additional 3 SNPs reside in the 5′ untranslated region of the gene basing on variant NM_130850.2 or NM_001202.3 (http://www.ncbi.nlm.nih.gov/gene/652).
Table 2. TDT analysis for 12 SNPs in and around BMP4 in Asian trios.
| SNP name | Position | Minor | MAF | FAM* | T | NT | OR (95%CI) | P Value | Bonferroni corrected P Value |
| allele | (%) | ||||||||
| rs17563 | 54417522 | 2 | 25.4 | 122 | 77 | 72 | 1.07 (0.78,1.48) | 0.6877 | 1.0000 |
| rs2071047 | 54418411 | 1 | 45.9 | 136 | 81 | 98 | 0.83 (0.62,1.11) | 0.2045 | 0.9358 |
| rs2761880 | 54418986 | 1 | 26.7 | 119 | 79 | 66 | 1.20 (0.86,1.66) | 0.2842 | 0.9819 |
| rs10130587 | 54419110 | 2 | 48.7 | 139 | 113 | 70 | 1.61 (1.20,2.18) | 0.0016 | 0.0190 |
| rs2855532 | 54419965 | 1 | 43.0 | 145 | 104 | 82 | 1.27 (0.95,1.69) | 0.1082 | 0.7469 |
| rs2761884 | 54421052 | 1 | 42.4 | 153 | 113 | 88 | 1.28 (0.97,1.70) | 0.0769 | 0.6172 |
| rs2855530 | 54421917 | 1 | 49.6 | 148 | 110 | 92 | 1.20 (0.91,1.58) | 0.2083 | 0.9394 |
| rs2738265 | 54422399 | 2 | 42.3 | 148 | 112 | 84 | 1.33 (1.01,1.77) | 0.0489 | 0.4521 |
| rs762643 | 54422767 | 1 | 43.1 | 108 | 75 | 67 | 1.12 (0.81,1.56) | 0.5020 | 0.9998 |
| rs762642 | 54423053 | 2 | 43.3 | 141 | 84 | 104 | 0.81 (0.61,1.08) | 0.1449 | 0.8472 |
| rs2761887 | 54425052 | 2 | 43.1 | 150 | 115 | 84 | 1.37 (1.03,1.81) | 0.0283 | 0.2914 |
| rs2032423 | 54425147 | 1 | 43.0 | 151 | 113 | 87 | 1.30 (0.98,1.72) | 0.0683 | 0.5721 |
FAM*: number of informative families, T: number of transmitted alleles, NT: number of un-transmitted alleles.
Table 3. TDT analysis for 12 SNPs in and around BMP4 in Maryland trios.
| SNP name | Position | Minor allele | MAF (%) | FAM* | T | NT | OR (95%CI) | P Value | relative difference ratio |
| rs17563 | 54417522 | 1 | 47.7 | 36 | 26 | 24 | 1.08 (0.62, 1.89) | 0.7789 | - |
| s2071047 | 54418411 | 1 | 43.5 | 37 | 25 | 24 | 1.04 (0.60, 1.82) | 0.8817 | 5.23 |
| rs2761880 | 54418986 | 1 | 3.9 | 5 | 4 | 2 | 2.00 (0.37, 10.92) | 0.4178 | 85.39 |
| rs10130587 | 54419110 | 1 | 40.8 | 33 | 23 | 22 | 1.05 (0.58, 1.88) | 0.8837 | - |
| rs2855532 | 54419965 | 1 | 46.1 | 38 | 19 | 33 | 0.58 (0.33, 1.01) | 0.0515 | −7.21 |
| rs2761884 | 54421052 | 1 | 43.7 | 38 | 22 | 31 | 0.71 (0.41, 1.23) | 0.2188 | −3.07 |
| rs2855530 | 54421917 | 2 | 48.1 | 41 | 35 | 24 | 1.46 (0.87, 2.45) | 0.1530 | - |
| rs2738265 | 54422399 | 2 | 45.1 | 35 | 20 | 29 | 0.69 (0.39, 1.22) | 0.1954 | −6.62 |
| rs762643 | 54422767 | 1 | 41.0 | 35 | 18 | 30 | 0.60 (0.33, 1.08) | 0.0833 | −1.41 |
| rs762642 | 54423053 | 2 | 39.3 | 39 | 36 | 21 | 1.71 (1.00, 2.94) | 0.0469 | 9.24 |
| rs2761887 | 54425052 | 2 | 44.7 | 33 | 18 | 28 | 0.64 (0.36, 1.16) | 0.1442 | −3.71 |
| rs2032423 | 54425147 | 1 | 45.1 | 37 | 22 | 28 | 0.79 (0.45, 1.37) | 0.3994 | −4.88 |
FAM*: number of informative families.
T: number of transmitted alleles, NT: number of un-transmitted alleles.
Relative difference ratio: Relative difference ratio for MAFs between Asian and Maryland parents.
Figure 1. Schematic genomic structure of BMP4 and coordinates of 13 genotyped SNPs.
Three variants (NM_001202.3, NM_130850.2 and NM_130851.2) of the human BMP4 gene are shown in the figure with 4 exons each aligned from left to right. Filled gray and black boxes represent the untranslated and translated exons respectively. All three variants encode an identical protein through translation of exons 3 and 4. Scale on top of the figure shows position of the gene and coordinates for the 13 genotyped SNPs basing on GRCh build 37.3. Minor allele frequency for one SNP (rs2855527) was less than 1% in both Asian and Maryland founders and was excluded from association analysis.
SNPs with “SNP scores" >0.6 (an assessment of design quality of the Illumina assay based on a proprietary algorithm), high validation levels in dbSNP (including validation on multiple platforms), and high heterozygosity levels were given priority when selecting SNPs. All 3 SNPs released by the HapMap project (HapMap Data Rel 24/phaseII Nov08, on NCBI B36 assembly, dbSNP b126) were included in our marker panel. The genomic DNA was extracted from peripheral blood using the protein precipitation method and stored at −20°C. Aliquots (4 µg) of each genomic DNA sample were dispensed into a bar-coded 96-well microtiter plate at a concentration of 100 ng/µl and genotyped by Illumina's GoldenGate chemistry [23] at the Genetic Resources Core Facility (GRCF) of Johns Hopkins University. Two duplicates and four controls from the Centre d'Etude du Polymorphisme Humain (CEPH) collection were included on each plate to evaluate genotyping consistency within and between plates.
Statistical analysis
(1) SNP screening and preliminary analysis
We first screened SNPs for minor allele frequency (MAF), pairwise linkage disequilibrium (LD) measured as r2, and Hardy-Weinberg equilibrium (HWE) for parents within each population (and in three Asian populations combined) using Haploview (v4.2, http://www.broadinstitute.org/haploview/haploview) [24]. SNPs with MAF>1%, genotyping call rate >80% in each group, and showing adherence to HWE at p>0.01 were eligible for analysis. The relative difference ratio was calculated as the difference in MAFs for parents in Maryland and the combined Asian samples, divided by the MAF in Asian parents for each SNP.
(2) Association analysis
We performed transmission disequilibrium test (TDT) analysis, originally proposed by Spielman et al. [25] on each eligible marker, for haplotypes of SNPs in each LD block, as well as for haplotypes of 2–5 SNPs in sliding windows using Family Based Association Test (FBAT; http://www.biostat.harvard.edu/~fbat/fbat.htm) [26] and PLINK (v1.07; http://pngu.mgh.harvard.edu/purcell/plink/) [27]. Bonferroni correction (http://www.quantitativeskills.com/sisa/calculations/bonfer.htm) was used to correct multiple comparisons. The overall empiric P value for the gene was generated from 10 000 permutations using Haploview as well (v4.2, http://www.broadinstitute.org/haploview/haploview) [24].
Results
Preliminary analysis
Among the 13 SNPs genotyped in and around BMP4, rs2855527 was the only one with MAF<1% in Maryland (0.5%) and in Asian parents, and was excluded from further analysis. The remaining 12 SNPs were eligible because all were compatible with HWE (at p>0.01) and their genotyping call rates were >80% (83.6% to 100%).
The minor allele differed for three SNPs (rs17563, rs10130587 and rs2855530) between Asian and Maryland parents among these 12 SNPs. MAF for rs2761880 was low (3.9%) in Maryland parents compared to Asian parents (26.7%). MAFs for the other 8 SNPs were similar in Asian and Maryland parents, ranging from 42.3%–45.9% and 39.3%–46.1%, respectively (Table 2 and 3). The relative difference ratios for MAFs were less than 10% for these 8 SNPs (Table 3). LD patterns among these 12 SNPs were similar in Asian and Maryland parents (Fig S1). In Asian parents, the first 3 and last 7 SNPs formed LD blocks respectively; in Maryland parents, the first 2 and last 8 SNPs formed separate LD blocks. SNP rs10130587 was an independent SNP in both Maryland and Asian parents.
Association analysis
When deviation from strict Mendelian inheritance of individual SNPs was analyzed in Asian trios, three SNPs (rs10130587, rs2738265 and rs2761887) showed nominally significant evidence of linkage and association (p<0.05) with NSCL/P. Significance held for rs10130587 after strict Bonferroni correction (corrected p = 0.019). The overall empiric P value for rs10130587 by Haploview was 0.018. Estimated odds ratio (OR) for carrying the apparent high-risk allele (MAF = 48.7%) at rs10130587 was 1.61(95%CI = 1.20, 2.18). No significant evidence of linkage and association with NSCL/P was particularly evident for rs10130587 in any of the three Asian groups or in a specific cleft type (cleft lip and palate or cleft lip only) in combined Asian population after Bonferroni correction (Table S2).
Sliding window haplotype analysis of these 12 SNPs in and around BMP4 generated a total of 38 tests. All haplotypes including rs10130587 were significantly associated with NSCL/P in combined Asian group. The Bonferroni corrected P value for the most significant haplotype (rs2761880-rs10130587) was not significant (0.0626) (Table S1). No significant linkage and association was seen in haplotype analysis of SNPs in each LD block (data not shown).
When 76 Maryland trios were analyzed, SNP rs762642 in the second LD block gave nominally significant evidence of linkage and association (p = 0.0469), however, this result was not significant after Bonferroni correction (Table S1). In sliding window haplotype analysis, majority of the haplotypes including rs762642 was nominally significant, but the most significant haplotype (rs762643- rs762642-rs2761887) was no longer significant after Bonferroni correction (Table S1). No significant linkage and association was seen in haplotype analysis of SNPs in each LD block (data not shown).
Discussion
BMP4 is located in 14q22-q23, and has 4 exons and 3 introns among which exons 3 and 4 encode protein (http://genome.ucsc.edu/cgi-bin). The protein encoded by this gene is a member of the bone morphogenetic protein family, part of the transforming growth factor-beta superfamily [28]. Animal experiments and genome wide linkage analysis in multiplex NSCL/P families repeatedly suggested BMP4 play a potential role in risk for NSCL/P [13]–[18]. Several candidate gene association analyses [19]–[22] have also contributed evidence that BMP4 is an important candidate gene for NSCL/P, although there has been no consistency across studies.
SNP rs762642 is the most frequently reported SNP in literature on BMP4 and its influence on NSCL/P [9]–[12], [19], [22]. One case-parent trio study from Chile tested the haplotype of rs1957860-rs762642 and found significant evidence for linkage and association [19]. For individual SNP analysis, as with all previously published reports, no evidence was shown for rs762642 either in our Asian or Maryland trios (after Bonferroni correction). And we did not find evidence in our sliding window haplotype analysis or in haplotype analysis for SNPs in identified LD block that included rs762642 after Bonferroni correction.
SNP rs17563 lies in exon 4 of BMP4, but evidence from association analysis with this marker has not been consistent. Two case-control studies conducted mainly in Han Chinese population [20], [21] and one resequencing study [29] using samples from six different populations showed evidence of association between rs17563 and NSCL/P. However, evidence of linkage and association was not seen for rs17563 in a case-parent trio study conducted in two Scandinavia populations using log-linear models [22]. Our negative results are consistent with Jugessur's analysis [22] although our sample size was quite limited.
Although no evidence of linkage and association was seen for rs762642 and rs17563 with NSCL/P, intriguing result was found for another SNP (rs10130587) in our Asian trios (which was significant after Bonferroni correction). SNP rs10130587 was recently identified by the 1000 genomes program (http://www.ncbi.nlm.nih.gov/projects, 2011.12.8) with frequencies for allele C as 48.4% and 38.5% in combined Asian (Han Chinese in Beijing, Han Chinese South, and Japanese individuals) and CEPH samples separately (Table S3) [30]. Because MAF for this intronic SNP was high (48.7%) in our Asian parents, this marker should be informative and may be in LD with an unknown causal variant. The significant linkage and association observed in Asian NSCL/P trios was not replicated in our smaller sample of Maryland trios which also had a slightly lower MAF (40.8%) compared to our Asian founders. Because rs10130587 was not genotyped in any previous candidate gene and GWAS studies [9]–[12], [19]–[22], our result can be considered an independent analysis although 52.9% (157 trios) of our sample [31] went into one GWAS [10]. It is possible that association for NSCL/P could have been identified with this SNP were it included in marker panels in previous association studies, including those failing to detect any evidence of linkage and association between BMP4 and NSCL/P [9]–[12], [17], [22]. The arrays are always changing and it is likely that the version used for GWAS was optimized for Caucasians because most of the early HapMap sequencing was done with CEPH samples. Our results supported the published evidence for some association between BMP4 and NSCL/P, and builds upon both experimental animal and linkage studies in humans.
Supporting Information
Linkage disequilibrium as measured by r2 in BMP4 among parents of NSCL/P probands from Asian and Maryland trios. White: r2 = 0. Shades of gray: 0<r2<1. Black: r2 = 1. BMP4, Bone Morphogenetic Protein 4; NSCL/P, nonsyndromic cleft lip with or without cleft palate.
(DOC)
Nominally significant tests from FBAT analysis for sliding window haplotypes (size 2–5 SNPs each) for 12 markers in BMP4 in Asian and Maryland trios.
(DOC)
TDT analysis for rs10130587 in BMP4 in trios from different Asian sites and by cleft type in combined Asian population.
(DOC)
Frequency for allele C in rs10130587 in BMP4 by race group using data from 1000genomes.
(DOC)
Acknowledgments
We thank all participants who donated samples for this multi-center study of oral clefts, as well as the staff at each participating site and institution.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by grants from the National Institute of Dental & Craniofacial Research (No. R21-DE-013707 and R01-DE-014581) and Fogarty Institution (No. D43-TW006176). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- 2.Hashmi SS, Waller DK, Langlois P, Canfield M, Hecht JT. Prevalence of nonsyndromic oral clefts in Texas: 1995–1999. Am J Med Genet A. 2005;134:368–372. doi: 10.1002/ajmg.a.30618. [DOI] [PubMed] [Google Scholar]
- 3.Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin Cell Dev Biol. 2005;16:683–693. doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell LE. Twin studies in oral cleft research. In: Wyszynski DF, editor. Cleft lip and palate. Oxford: Oxford University Press; 2002. pp. 214–221. [Google Scholar]
- 5.Mossey PA, Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski DF, editor. Cleft lip and palate. Oxford: Oxford University Press; 2002. pp. 127–158. [Google Scholar]
- 6.Lidral AC, Murray JC. Genetic approaches to identify disease genes for birth defects with cleft lip/palate as a model. Birth Defects Res (Part A) 2004;70:893–901. doi: 10.1002/bdra.20096. [DOI] [PubMed] [Google Scholar]
- 7.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374:1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 9.Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, et al. Key susceptibility locus for nonsyndrominc cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 10.Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 13.Barlow AJ, Francis-West PH. Ectopic application of recombinant BMP-2 and BMP-4 can change patterning of developing chick facial primordia. Development. 1997;124:391–398. doi: 10.1242/dev.124.2.391. [DOI] [PubMed] [Google Scholar]
- 14.Ashique AM, Fu K, Richman JM. Endogenous bone morphogenetic proteins regulate outgrowth and epithelial survival during avian lip fusion. Development. 2002;129:4647–4660. doi: 10.1242/dev.129.19.4647. [DOI] [PubMed] [Google Scholar]
- 15.Paiva KB, Silva-Valenzuela MG, Massironi SM, Ko GM, Siqueira FM, et al. Differential Shh, Bmp and Wnt gene expressions during craniofacial development in mice. Acta histochemica. 2010;112:508–517. doi: 10.1016/j.acthis.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Sun X, Braut A, Mishina Y, Behringer RR, et al. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- 17.Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, et al. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals genotype-specific differences in linkage and association results. Hum Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suazo J, Santos JL, Jara L, Blanco R. Association between bone morphogenetic protein 4 gene polymorphisms with nonsyndromic cleft lip with or without cleft palate in a Chilean population. DNA Cell Biol. 2010;29:59–64. doi: 10.1089/dna.2009.0944. [DOI] [PubMed] [Google Scholar]
- 20.Lin JY, Chen YJ, Huang YL, Tang GP, Zhang L, et al. Association of bone morphogenetic protein 4 gene polymorphisms with nonsyndromic cleft lip with or without cleft palate in Chinese children. DNA Cell Biol. 2008;27:601–605. doi: 10.1089/dna.2008.0777. [DOI] [PubMed] [Google Scholar]
- 21.Jianyan L, Zeqiang G, Yongjuan C, Kaihong D, Bing D, et al. Analysis of interactions between genetic variants of BMP4 and environmental factors with nonsyndromic cleft lip with or without cleft palate susceptibility. Int J Oral Maxillofac Surg. 2010;39:50–56. doi: 10.1016/j.ijom.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, et al. Genetic determinants of facial clefting:analysis of 357 candidate genes using two national cleft studies from Scandinavia. PLoS ONE. 2009;4:e5385. doi: 10.1371/journal.pone.0005385. doi: 10.1371/journal.pone.0005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliphant A, Barker DL, Stuenlpnagel JR, Chee MS. BeadArray™ Technology: Enabling an accurate, cost efficient approach to high-throughput genotyping. Biotechniques. 2002;32:S56–S61. [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Spielman RS, McGinnis RE, Ewenst WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 26.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, et al. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–411. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Zhang TX, Wu T, Hetmanski JB, Ruczinski I, et al. The FGF & FGFR gene family and risk of cleft lip with/without cleft palate. Cleft Palate-Cran J. 2011 doi: 10.1597/11-132. In-Press. doi: http://dx.doi.org/10.1597/11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage disequilibrium as measured by r2 in BMP4 among parents of NSCL/P probands from Asian and Maryland trios. White: r2 = 0. Shades of gray: 0<r2<1. Black: r2 = 1. BMP4, Bone Morphogenetic Protein 4; NSCL/P, nonsyndromic cleft lip with or without cleft palate.
(DOC)
Nominally significant tests from FBAT analysis for sliding window haplotypes (size 2–5 SNPs each) for 12 markers in BMP4 in Asian and Maryland trios.
(DOC)
TDT analysis for rs10130587 in BMP4 in trios from different Asian sites and by cleft type in combined Asian population.
(DOC)
Frequency for allele C in rs10130587 in BMP4 by race group using data from 1000genomes.
(DOC)