Abstract
In this work we extended the study of genes controlling the formation of specific differentiation structures called “domes” formed by the rat mammary adenocarcinoma cell line LA7 under the influence of DMSO. We have reported previously that an interferon-inducible gene, rat-8, and the β-subunit of the epithelial sodium channel (ENaC) play a fundamental role in this process. Now, we used a proteomic approach to identify proteins differentially expressed either in DMSO-induced LA7 or in 106A10 cells. Two differentially expressed proteins were investigated. The first, tropomyosin-5b, strongly expressed in DMSO-induced LA7 cells, is needed for dome formation because its synthesis inhibition by the antisense RNA technology abolished domes. The second protein, maspin, strongly expressed in the uninduced 106A10 cell line, inhibits dome formation because 106A10 cells, transfected with rat8 cDNA (the function of which is required for the organization of these structures), acquired the ability to develop domes when cultured in presence of an antimaspin antibody. Dome formation in these cultures are accompanied by ENaC β-subunit expression in the absence of DMSO. Therefore, dome formation requires the expression of tropomyosin-5b, in addition to the ENaC β-subunit and the rat8 proteins, and is under the negative control of maspin.
A well defined cellular modification resulting in the formation of hemispheric structures called “domes” is an in vitro model for studying differentiation of the mammary gland (1). This model is based on two cellular clones, LA7 and 106, both derived from a rat adenocarcinoma cell line, RAMA25 (2). After exposure to differentiating agents such as DMSO, LA7 but not 106A10 cells are able to form domes (1). The identification of the genes involved in the process of dome formation has been carried out by using a transcript-based differential expression detection technique (cDNA library subtraction) (3, 4). This work has shown that the formation of domes requires the expression of two genes: rat8, a homologue of the human gene 9–27, encoding for the protein Leu-13, which forms a complex with other proteins at the surface of human B lymphocytes (5–7), and the amiloride-sensitive epithelial sodium channel (ENaC) β-subunit (4). The expression of the β-subunit of this channel, which is essential for its induction in other systems (8), normally is repressed in LA7 cells by the activity of gene 133. Induction of dome formation by DMSO is a result of the repression of the expression of this gene (4). Gene 133 is the rat orthologous of the human epithelial membrane protein 3 (EMP3) (9).
In this work we used the proteomic approach to identify differentially expressed proteins in the LA7 cells under the influence of DMSO and in 106A10 cells. This approach was suggested by the possibility of an incomplete correlation between mRNA and protein levels expressed by a particular gene (10, 11), owing to the existence of posttranscriptional mechanisms that control the rate of synthesis and the half-life of proteins (12). Many differentially expressed proteins have been revealed in our system by using this approach, and we have concentrated on the study of two of these proteins, tropomyosin-5b (Tm-5b) and maspin. We show that both play an important role in dome formation.
Materials and Methods
Cells, Media, and Differentiation Inducers.
The cell lines LA7 and 106A10, both clonal derivatives from the Rama-25 line and isolated from a 7,12-dimethylbenz[a]anthracene-induced adenocarcinoma in Sprague–Dawley mammary rat tissue (2), were cultured as described (1). For proteomic analysis, cells were cultured in triplicate, each of them derived from a different frozen culture. The LA7 cells, after having grown to confluence, were exposed to 1.5% DMSO as inducer of cell differentiation (13). Cells were harvested by scraping to avoid using trypsin to prevent extracellular protein digestion.
Sample Preparation for Proteomic Analysis.
LA7 and 106A10 cell pellets were resuspended in lysis buffer (8 M urea/4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/40 mM Tris base/65 mM dithioerythritol). Total protein estimation was carried out by standard procedures (14). For analytical or preparative runs, 45 μg or 2 mg of total proteins was loaded, respectively, on a two-dimensional (2D) polyacrylamide gel.
High-Resolution 2D Gel Electrophoresis.
2D electrophoresis was performed as described previously (15–17), with the following minor modifications. Isoelectric focusing (first dimension) was carried out on nonlinear, wide-range immobilized pH gradients (pH 3–10, 18-cm long) by using the IPGphor system (Amersham Pharmacia). After the first-dimensional separation, the IPG strips were loaded on the top of a vertical 9–16% SDS-polyacrylamide linear gradient gel. For analytical purposes, the gels were silver-stained as described (18, 19). Preparative gels were stained for 1 h with 0.2% Coomassie blue R-250 in 50% methanol/10% acetic acid and destained in 50% methanol/2% acetic acid.
Image Acquisition and Data Analysis.
Electrophoretogram images were obtained with a computing densitometer (Molecular Dynamics 300S; Amersham Pharmacia) and processed with the Melanie 3 computer system (Geneva Bioinformatics, Geneva). Gels were calibrated by comigration of LA7 and 106A10 cells with human serum proteins as references. The electrophoretic coordinates used for serum proteins were as described (20). Quantitative variations of proteins were expressed as volumes of spots. To correct for variability from silver staining, relative volumes to the sum of the volume of all spots in each gel were calculated with the Melanie 3 (Medical Electrophoresis Analysis Interactive Expert) system.
Matrix-Assisted Laser Desorption Ionization/Time of Flight Mass Spectrometry.
Protein identification by matrix-assisted laser desorption ionization/time of flight mass fingerprinting was carried out according to the procedure described (21). Excised spots from preparative 2D gels were digested with trypsin, and the corresponding peptides were extracted from gel, dried, and resuspended with 0.1% trifluoroacetic acid in 50% acetonitrile. A portion (0.5 μl) of this solution was loaded together with 0.5 μl of matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid) on the sample probe of a Voyager Elite mass spectrometer (PerSeptive Biosystems). Analysis was performed in the delay extraction reflection mode, and peptide masses were obtained by using the ms-fit search program.
mRNA Isolation and Northern Analysis.
Total RNA was isolated by the single-step acid-guanidinium isothiocyanate-phenol-chloroform extraction method (22). For Northern blot analysis, total RNA (5–10 μg) from each cell line was fractionated by electrophoresis as described (23). Probes were prepared by using [32-P]α-dCTP and the random priming labeling kit Ready Prime (Amersham Pharmacia). Ribosomal cDNA or 36B4 cDNA, the expression level of which is independent from the action of inducers (24), was used as a loading control.
Reverse Transcription–PCR (RT-PCR) Assay.
Uninduced LA7 and 106A10 cells and LA7 and 106A10 cultured in the presence of DMSO were used for RNA isolation. One microgram of RNA was treated with 1 unit of RNase-free DNase (GIBCO/BRL) for 15 min at room temperature to remove any possible DNA contamination. Complementary DNA-strand synthesis was performed essentially as reported already (25). PCR analysis was performed by using 10% of the first-strand reaction. RT-PCR fragments were detected after 35 amplification cycles. Concurrently, additional samples containing LA7 and 106A10 RNAs after DNase treatment and before retrotranscription were subjected to PCR in identical conditions to exclude the presence of genomic DNA. For PCR amplification of the ENaC β-subunit, we used primers and PCR conditions as described (4).
Antisense Oligonucleotide Methodology.
For the inhibition study of Tm-5b mRNA expression, three oligonucleotides of 20 bases were synthesized: the antisense oligomer (5′-CTTTCTCCGCACCGCCTCCA-3′), designed as the complementary sequence to the Tm-5b mRNA, between nucleotides 118 and 137 of the mRNA sequence (accession no. M60669); the sense oligomer (5′-TGGAGGCGGTGCGGAGAAAG-3′) from the same region and the scrambled oligomer (5′-TCCCACCTTATCCTCCGCCG-3′), a scrambled sequence with the same nucleotides used for the antisense oligomer. The experiment was performed as already described (3). Cells were maintained in culture for 60–72 h, inspected for dome formation, photographed, and harvested for RNA extraction.
Antibody Antimaspin.
Polyclonal antimaspin antibody (Santa Cruz Biotechnology) was used in Western blot analysis according to the manufacturer's recommendation. When used as a blocking antibody (26), it was added to the 106A10 cell culture 16 h after the cells were plated and diluted 1:50 in the culture medium. The cells were inspected for dome formation 24 h after the antibody addition, examined microscopically, photographed by using ×4 and ×20 objectives, and harvested for RNA preparation. Immunofluorescent detection of maspin was performed on 106A10 cells grown for 24–36 h as described already (3). Cells were methanol-fixed for 10 min and then incubated with antimaspin polyclonal antibody according to the manufacturer's protocol (Santa Cruz Biotechnology). The secondary antibody used was rhodamine-conjugated (Vector Laboratories).
Results
Proteomic Analysis of DMSO-Induced LA7 and Uninduced 106A10 Cells.
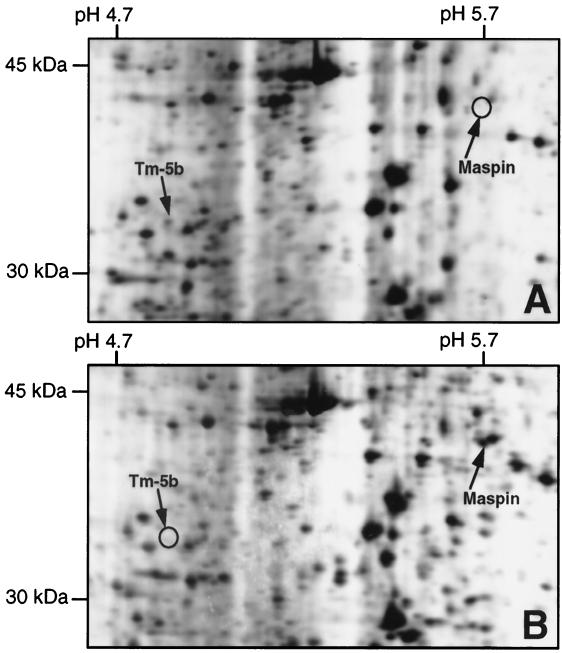
Total cell extracts obtained from DMSO-induced LA7 and uninduced 106A10 cells were used to run standard 2D-protein gel electrophoresis within an isoelectric focusing range of pH 3–10. 2D gel electrophoresis allowed the separation of about 2,000 different proteins per gel and allowed us to obtain high-resolution polypeptide maps of LA7 and 106A10 cells. For each of the two cell types, three different gels were run. The proteins were visualized from each gel after silver staining. The gels then were digitized by a laser densitometer and analyzed by specific software programs. Computer-assisted analysis of the results, performed by using the informatic program melanie 3, allowed us to obtain a semiquantitative analysis of the electrophoretic spots (27) and to identify more than 200 differentially expressed proteins between the two reported conditions (see Fig. 1). Fifty of these spots were removed from the gels and analyzed by mass spectrometry. The results obtained by using matrix-assisted laser desorption ionization/time of flight were compared with protein databases; 14 matches to known proteins in swiss-prot and trembl databases were identified.
Figure 1.
Silver-stained details of 2D gel electrophoresis polypeptide maps of DMSO-induced LA7 cells (A) and uninduced 106A10 cells (B). Spots were named by protein identification by mass spectrometry analysis. Differential expression of Tm-5b and maspin proteins is indicated with ○ in B and A, respectively.
We focused our attention on two of these newly identified proteins, Tm-5b and maspin, because of their strong asymmetric expressions in opposite ways, as determined by densitometric analysis. Tm-5b protein was expressed at least 50–60 times more abundantly in DMSO-induced LA7 cells (see Fig. 1A). Maspin was expressed 70–80 times more abundantly in untreated 106A10 cells but expressed only minimally in the DMSO-induced LA7 cell line (see Fig. 1B).
Northern Confirmation of Tm-5b and Maspin Differential Expression.
To investigate whether the Tm-5b and maspin differential expression found at the protein level was also present at the mRNA level, we performed Northern blot analysis on LA7 and 106A10 cells that were both induced by DMSO and uninduced.
As shown in Fig. 2A, Tm-5b mRNA was highly expressed in DMSO-induced LA7 cells (lane 2), but at a very low level in untreated 106A10 cells (lane 3); after DMSO induction, Tm-5b expression in 106A10 cells was higher (lane 4), almost as in uninduced LA7 cells (lane 1), but still lower then in DMSO-induced LA7 cells.
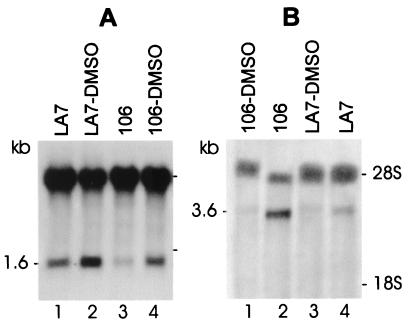
Figure 2.
Northern blot analysis of Tm-5b (A) and maspin (B). Cells were cultured as described in the text and exposed to DMSO. Northern blots were hybridized with radioactive Tm-5b- and maspin-specific probes. (A) Uninduced LA7 (lane 1), DMSO-induced LA7 (lane 2), uninduced 106A10 (lane 3), and DMSO-induced 106A10 (lane 4). (B) DMSO-induced 106A10 (lane 1), uninduced 106A10 (lane 2), DMSO-induced LA7 (lane 3), and uninduced LA7 (lane 4). A cDNA probe for 28S ribosomal RNA was used as a loading control in both A and B.
In contrast, maspin mRNA was highly expressed in untreated 106A10 cells (Fig. 2B, lane 2) and expressed at a very low level in the LA7 cells (lane 4), but minimally expressed in LA7 and 106A10 cell lines, both of which were DMSO-induced (lanes 1 and 3). Therefore, the mRNA expression pattern of Tm-5b and maspin respectively parallels that detected at the protein level by 2D gel electrophoresis analysis.
Role of Tm-5b in Dome Formation.
To investigate the role of Tm-5b in the process of dome formation, LA7 cultures were exposed to antisense-specific oligonucleotides directed to the 5′ end of the Tm-5b mRNA (GenBank accession no. M60669) in the presence of the inducer DMSO. The antisense treatment prevented the formation of domes in the presence of DMSO (Fig. 3B), whereas domes were formed, as usual, in LA7 cells after DMSO induction with no addition of antisense oligonucleotides (Fig. 3A); sense and scrambled oligonucleotides had no effect on dome formation (data not shown).
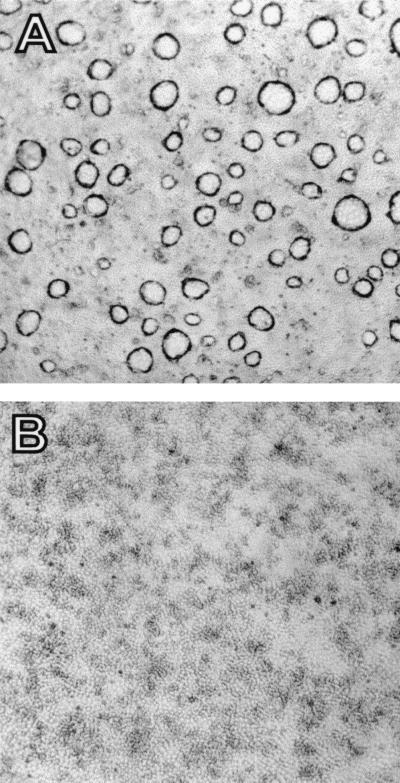
Figure 3.
Antisense anti-Tm-5b oligonucleotides' effect on dome formation in DMSO-induced LA7 cells. (A) Domes develop in LA7 cells induced to differentiate with 1.5% DMSO. (B) Addition of anti-Tm-5b antisense oligonucleotides causes inhibition of dome formation in DMSO-exposed LA7 cells. (Photographs were taken with a ×4 objective.)
Role of Maspin in Dome Formation.
Maspin, a secreted protein acting as a serine protease inhibitor (26, 28), is important for mammary gland alveolar development (29). We investigated whether down-regulation of maspin expression was sufficient for dome formation by exposing the 106A10 cells to antisense oligonucleotides to maspin mRNA. This treatment failed to induce dome formation. We performed the same experiment by using an antibody to maspin, and we observed that the exposure of the 106A10 cells to the antimaspin antibody causes the formation of very few and very small and flat-dome-like structures after a long exposure time of the cells to the antibody; however, a modification of the 106A10 cell morphology was apparent (data not shown). The 106A10 cells, which are usually fusiform (spindle-like), assumed a cuboidal shape, indicating that maspin protein inhibition may alter cell-to-cell contact and adhesion.
We then investigated whether the expression of gene rat8 (which is not present in the 106A10 cells), also was required, because, as we reported previously, the rat8 protein plays a key role in the formation of domes (3). To do so, we repeated the experiment by using 106A10 cells transfected with rat8 cDNA (3), selecting a clone (106/E41–88) highly expressing rat8 mRNA. This clone by itself does not form domes and expresses the maspin protein, as seen by Western blot analysis (data not shown). Maspin protein was blocked by an antimaspin antibody added to the cells in the absence of DMSO. The 106/E41–88 cells did form domes when cultured with the blocking antibody, as shown in Fig. 4A, but did not when the antibody was absent (Fig. 4B) or the antibody was inactivated by exposure to high temperature (water bath at 95°C for 5 min; data not shown). In addition, domes were not formed when the cells were exposed to the antimaspin antibody in the presence of rat8 antisense oligonucleotides (Fig. 4C), confirming that rat8 protein expression is essential for dome formation. The whole experiment was repeated six times by using four different cultures of 106/E41–88 and two different maspin antibody commercial preparations.
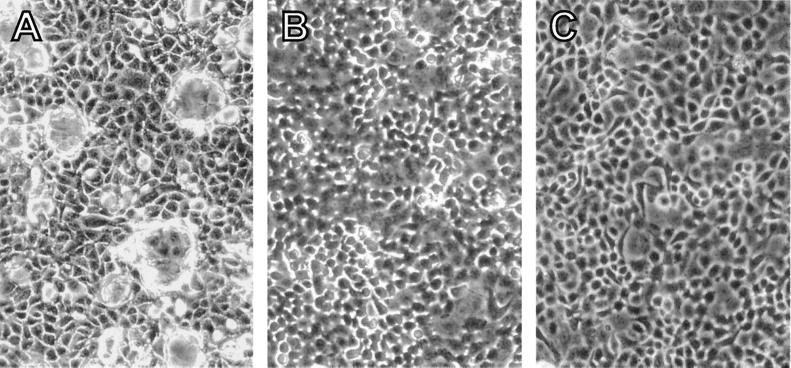
Figure 4.
Antibody antimaspin effect on dome formation of 106E41–88 cells. (A) 106/E41–88 cells were cultured with an antimaspin antibody (1:50 dilution). The same cells were left untreated (B) or exposed to the antimaspin antibody in the presence of rat8 antisense oligonucleotides (C). (Photographs were taken with a ×20 objective.)
Maspin Inhibition Allows ENaC Activation.
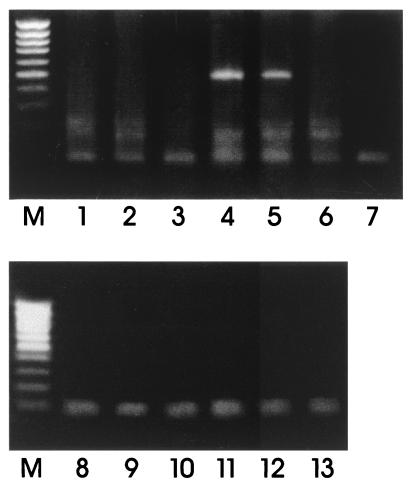
To investigate the mechanism by which maspin inhibits dome formation, we studied its effect on the expression of the ENaC β-subunit, which also has been shown to be essential for the formation of domes (4). Information from the literature suggests that the action of a serine protease is required for the activation of the amiloride-sensitive ENaC in the Xenopus kidney epithelial A6 cells (30, 31) and in a mouse cortical collecting-duct cell line (32). To test whether, in our system, maspin protein may block the expression of the channel and whether the antiserum relieves the block, we performed RT-PCR analysis on 106/E41–88 cDNA to detect the expression of the amiloride-sensitive ENaC β-subunit after exposure of the cells to the antimaspin antibody. RT-PCRs were performed by using primers designed on the coding region of the amiloride-sensitive ENaC β-subunit as described (4). A fragment of correct size (base pair 482) was obtained from DMSO-induced LA7 cDNA, as expected (Fig. 5, lane 4) and from the maspin antibody-treated 106/E41–88 cDNA (Fig. 5, lane 5). The fragment obtained from the 106/E41–88 cDNA was sequenced, showing its identity to the ENaC β-subunit. No amplification was observed when PCR was performed on the same samples after DNase treatment, before retrotranscription, indicating that the samples did not contain DNA (Fig. 5, lanes 8–13).
Figure 5.
Antibody antimaspin effect on ENaC β-subunit expression in 106/E41–88 cells. RT-PCRs were performed with the amiloride-sensitive ENaC β-subunit-specific primers by using cDNA of uninduced and DMSO-induced 106A10 cells (lanes 1 and 2), uninduced and DMSO-induced LA7 cells (lanes 3 and 4), uninduced 106E41–88 rat8-transfected cells exposed to antimaspin antibody (lane 5), and uninduced 106E41–88 not exposed to the antimaspin antibody (lane 6). Lane 7, PCR-negative control. A PCR, using the same primers, was performed on the same samples after DNase treatment and before retrotranscription (lanes 8–13).
Discussion
The formation of domes in the presence of DMSO is dependent on a complex genetic regulation, which involves both functional and structural modifications of the cell surface. The functional modifications are due to the expression of the gene rat8, the gene encoding for the β-subunit of the amiloride-sensitive ENaC, gene 133, and their possible interactions (3, 4). We have demonstrated previously that the expression of rat8 and the β-subunit of ENaC are both required for dome formation, whereas the activity of gene 133 inhibits the expression of the ENaC β-subunit and prevents the formation of domes. We also have demonstrated that when gene 133 is inhibited by DMSO or by the antisense oligonucleotides to its mRNA, dome formation follows (4). We showed by fluorescence microscopy that structural modifications of the cell also are required for dome formation, as indicated by strong expressions of E cadherin and α6β1-integrin at the cell surface and of cytokeratin 8 in DMSO-induced LA7 cells (4).
In this paper we report the participation in this process of two other proteins that influence the cell surface: Tm-5b, a component of the cytoskeleton, and maspin, a serine proteases inhibitor. Considering Tm-5b first, its expression is required because we showed that antisense oligonucleotides against Tm-5b mRNA abolish dome formation in LA7 cells in the presence of DMSO. Tm-5b is a low-molecular-weight isoform of the tropomyosin protein family present in nonmuscle cells. Tm-5b is expressed in specialized epithelial cells and is localized at the adhesion belts that organize cell-to-cell contacts (33). Tropomyosin has an important role in maintaining the differentiated state of many cell types, because its down-regulation, together with that of microfilament-associated proteins, is observed in transformed cells, both in fibroblasts and epithelia (34–36). Tropomyosin down-regulation also has been observed in breast malignant cells and breast carcinoma cell lines (36, 37). Moreover, forced expression of several isoforms of tropomyosin in cancer cells can suppress their growth or induce a more differentiated cellular morphology (38, 39).
It seems likely that the expression of Tm-5b during dome formation is required for the accompanying changes of the cell surface; Tm-5b may act by stabilizing and maintaining the connections mediated by E cadherin, which is involved in establishing the tight junctions between adjacent cells and/or improve their sealing capacity.
With regard to maspin, we show that it is expressed strongly in the non-dome-forming 106 cells and only weakly in LA7 cells induced by DMSO. Given its important role in mammary gland development (29) and knowing its role in reducing the mammary cell ability to form tubular structures (40), we considered maspin overexpression as a possible cause of dome formation inhibition in 106A10 cells. However, exposure of 106A10 cells to maspin antisense oligonucleotides did not cause induction of domes. This result could be explained in two ways: either the antisense oligonucleotides were not effective in abolishing maspin protein synthesis, or dome formation needs the concomitant expression of gene rat8 in 106A10 cells. To distinguish between these two possibilities, knowing the important role of the rat8 gene in dome development and knowing that maspin is a secreted protein acting at the cell surface (28, 41, 42), the effect of maspin was determined by incubating a clone of 106 cells—transfected with rat8 cDNA that strongly expressed rat8 (clone 106E41–88)—with an antimaspin antibody in the absence of DMSO. The results show that the rat8-expressing 106 cells, when treated with an antimaspin antibody, were induced to dome development. Simultaneous exposure of these cells to the antimaspin antibody and rat8 antisense oligonucleotides suppressed the formation of domes, showing that the expression of rat8 and the inhibition of maspin both are required for dome formation. The dome-forming cultures expressed the ENaC β-subunit.
This result can be attributed to the inhibitory action of maspin on a surface serine protease that may regulate the expression of the β-subunit of the ENaC (30), perhaps by acting on proteins of the cell surface. In fact, maspin protein is very important in normal mammary gland development (29), which depends on proper interactions between epithelial cells and the extracellular matrix (43–45). In addition, it has been demonstrated that disruption of cell adhesion disturbs normal mammary gland morphogenesis, development, and differentiation (46). It has been proposed that an important role of maspin in mammary gland development is to regulate cell adhesion and motility, possibly by reducing the amount of β1-integrin at the cell surface (29); β1-integrin perturbation also alters the normal mammary gland development in the mouse (47, 48). These results are in agreement with our observation that β1-integrin plays an important role in dome formation (4). Our results also can explain another observation—the inhibition of alveolar development in the rat mammary gland during pregnancy by maspin overexpression, which results in many precursors of alveoli remaining without a lumen (29). This effect may be explained by maspin repression of ENaC β-subunit expression (which may be needed for lumen formation), as it was found in our cultures. This possibility is supported by the observation that the β-subunit of ENaC is expressed strongly in the normal rat lactating mammary gland, as shown by in situ hybridization (I.Z., unpublished observation).
The role of maspin, which we have observed here, appears to be different from its role as tumor suppressor, which has been widely reported (49, 50). The two roles seem contradictory, because in our experiments maspin acts as an inhibitor of mammary gland differentiation. However, it has been shown that maspin has several different effects, including inhibition of angiogenesis, which, by itself, can explain its antitumoral role. In addition, maspin may inhibit a crucial step in cell-to-cell interaction that is required for the establishment of connections important both in alveolar differentiation of normal mammary cells and in infiltration of cancer cells, because it inhibits tumor cell invasion and cell motility (42).
In conclusion, our results allow us to interpret the mechanism for dome formation as follows. It is caused by two changes: the activation of ENaC through its β-subunit expression, which regulates the passage of ions and water through the cells, and changes of the composition of the cell surface to give rise to tight cell–cell connections. In the absence of the inducer DMSO, the expression of the β-subunit of the ENaC is inhibited in both LA7 and 106A10 cells by the activity of gene 133, as well as in 106A10 cells by maspin. Through still undetermined pathways in both cases domes are not formed. The inducer DMSO acts in LA7 cells by inhibiting the expression of gene 133 so the β-subunit of ENaC becomes expressed. The changes of the cell surface may be initiated by the expression of Tm-5b with that of β1-integrin, followed by E cadherin and cytokeratin 8, and may be inhibited by maspin. All these changes require the expression of rat8 protein, which may interact with the other proteins, forming a complex, as it does on the surface of human B lymphocytes.
Acknowledgments
We especially would like to acknowledge Prof. R. Montesano for his important advice and contributions in this work, Dr. C. Temm-Grove for her help and suggestions, and Prof. P. Neri for his input. This work was funded partially by grants from Associazione Italiana per la Ricerca sul Cancro, P. F. “Biotecnologie” Consiglio Nazionale delle Ricerche (to P.V.), Progetto Strategico Consiglio Nazionale delle Ricerche “Tecnologie di base della postgenomica” (to P.V.), and Ministero dell'Università e della Ricerca Scientifica e Tecnologica (to L.B. and P.V.). D.A. is supported by “Mario e Valeria Rindi” Postdoctoral Fellowship from Fondazione Italiana per la Ricerca sul Cancro. This is manuscript no. 52 of the Genoma 2000/ITBA Project funded by Cariplo.
Abbreviations
- ENaC
epithelial sodium channel
- Tm-5b
tropomyosin-5b
- 2D
two-dimensional
- RT-PCR
reverse transcription–PCR
References
- 1.Dulbecco R, Bologna M, Unger M. Proc Natl Acad Sci USA. 1979;76:1256–1260. doi: 10.1073/pnas.76.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett D C, Peachey L A, Durbin H, Rudland P S. Cell. 1978;15:283–298. doi: 10.1016/0092-8674(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 3.Zucchi I, Montagna C, Susani L, Vezzoni P, Dulbecco R. Proc Natl Acad Sci USA. 1998;95:1079–1084. doi: 10.1073/pnas.95.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zucchi I, Montagna C, Susani L, Montesano R, Affer M, Zanotti S, Redolfi E, Vezzoni P, Dulbecco R. Proc Natl Acad Sci USA. 1999;96:13766–13770. doi: 10.1073/pnas.96.24.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayzer D J, Brinson E, Runge M S. Gene. 1992;117:277–278. doi: 10.1016/0378-1119(92)90739-c. [DOI] [PubMed] [Google Scholar]
- 6.Deblandre G A, Marinx O P, Evans S S, Majjaj S, Leo O, Caput D, Huez G A, Wathelet M G. J Biol Chem. 1995;270:23860–23866. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi S, Doss C, Levy S, Levy R. J Immunol. 1990;145:2207–2213. [PubMed] [Google Scholar]
- 8.Canessa C M, Schild L, Buell G, Thorens B, Gautschi I, Horisberger J D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 9.Taylor V, Suter U. Gene. 1996;175:115–120. doi: 10.1016/0378-1119(96)00134-5. [DOI] [PubMed] [Google Scholar]
- 10.Futcher B, Latter G I, Monardo P, McLaughlin C S, Garrels J I. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gygi S P, Rochon Y, Franza B R, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varshvsky A. Proc Natl Acad Sci USA. 1996;94:13057–13062. [Google Scholar]
- 13.Dulbecco R, Okada S. Proc R Soc London B Biol Sci. 1980;208:399–408. doi: 10.1098/rspb.1980.0058. [DOI] [PubMed] [Google Scholar]
- 14.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Bini L, Magi B, Marzocchi B, Arcuri F, Tripodi S, Cintorino M, Sanchez J C, Frutiger S, Hughes G, Pallini V, et al. Electrophoresis. 1997;18:2832–2841. doi: 10.1002/elps.1150181519. [DOI] [PubMed] [Google Scholar]
- 16.Bjellqvist B, Pasquali C, Ravier F, Sanchez J C, Hochstrasser D. Electrophoresis. 1993;14:1357–1365. doi: 10.1002/elps.11501401209. [DOI] [PubMed] [Google Scholar]
- 17.Görg A, Postel W, Gunther S. Electrophoresis. 1988;9:531–546. doi: 10.1002/elps.1150090913. [DOI] [PubMed] [Google Scholar]
- 18.Hochstrasser D F, Harrington M G, Hochstrasser A C, Miller M J, Merril C R. Anal Biochem. 1988;173:424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 19.Oakley B R, Kirsch D R, Morris N R. Anal Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- 20.Bjellqvist B, Hughes G J, Pasquali C, Paquet N, Ravier F, Sanchez J C, Frutiger S, Hochstrasser D. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 21.Williams K R, Stone K L. In: Techniques in Protein Chemistry VI. Crabb J W, editor. San Diego: Academic; 1995. pp. 143–152. [Google Scholar]
- 22.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Rio M C, Bellocq J P, Gairard B, Rasmussen U B, Krust A, Koehl C, Calderoli H, Schiff V, Renaud R, Chambon P. Proc Natl Acad Sci USA. 1987;84:9243–9247. doi: 10.1073/pnas.84.24.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maggi A, Susanna L, Bettini E, Mantero G, Zucchi I. Mol Endocr. 1989;3:1165–1170. doi: 10.1210/mend-3-7-1165. [DOI] [PubMed] [Google Scholar]
- 26.Zou Z, Anisowicz A, Hendrix M J, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 27.Applel R D, Plagi P M, Walther D, Vargas J R, Sanchez J C, Ravier F, Hochstrasser D. Electrophoresis. 1997;15:2735–2748. doi: 10.1002/elps.1150181507. [DOI] [PubMed] [Google Scholar]
- 28.Pemberton P A, Tipton A R, Pavloff N, Smith J, Erickson J R, Mouchabeck Z M, Kiefer M C. J Histochem Cytochem. 1997;45:1697–1706. doi: 10.1177/002215549704501213. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Magit D, Botteri F, Shi H Y, He K, Li M, Furth P, Sager R. Dev Biol. 1999;215:278–287. doi: 10.1006/dbio.1999.9442. [DOI] [PubMed] [Google Scholar]
- 30.Vallet V, Chraibi A, Gaeggeler H P, Horisberger J D, Rossier B C. Nature (London) 1997;389:607–605. doi: 10.1038/39329. [DOI] [PubMed] [Google Scholar]
- 31.Vuagniaux G, Vallet V, Jaeger N F, Pfister C, Bens M, Farman N, Courtois-Coutry N, Vandewalle A, Rossier B C, Hummler E. J Am Soc Nephrol. 2000;5:828–834. doi: 10.1681/ASN.V115828. [DOI] [PubMed] [Google Scholar]
- 32.Temm-Grove C J, Jockusch B M, Weinburger R P, Schevzov G, Helfman D M. Cell Motil Cytoskeleton. 1998;40:393–407. doi: 10.1002/(SICI)1097-0169(1998)40:4<393::AID-CM7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Hendricks M, Weintraub H. Proc Natl Acad Sci USA. 1981;78:5633–5637. doi: 10.1073/pnas.78.9.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanderkerckhove J, Bauw G K, Vancompernolle B, Honore B, Celis J. J Cell Biol. 1990;111:95–102. doi: 10.1083/jcb.111.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya B, Prasad G L, Valverius E M, Salomon D S, Cooper H L. Cancer Res. 1990;50:2105–2112. [PubMed] [Google Scholar]
- 36.Franzen B, Linder S, Uryu K, Alaiya A A, Hirano T, Kato H, Auer G. Brit J Cancer. 1996;73:903–913. doi: 10.1038/bjc.1996.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad G L, Fuldner R A, Cooper H L. Proc Natl Acad Sci USA. 1993;90:7039–7043. doi: 10.1073/pnas.90.15.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takenega K, Masuda A. Cancer Lett. 1994;87:47–53. doi: 10.1016/0304-3835(94)90408-1. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Volpert O, Shi Y H, Bouck N. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 40.Hendrix M J C. Nat Med. 2000;6:374–376. doi: 10.1038/74624. [DOI] [PubMed] [Google Scholar]
- 41.Sheng S, Carey J, Seftor E A, Dias L, Hendrix M J, Sager R. Proc Natl Acad Sci USA. 1996;93:11669–11674. doi: 10.1073/pnas.93.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talhouk R S, Chin J R, Unemori E N, Werb Z, Bissell M J. Development. 1991;112:439–449. doi: 10.1242/dev.112.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talhouk R S, Bissell M J, Werb Z. J Cell Biol. 1992;118:1271–1282. doi: 10.1083/jcb.118.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sympson C J, Talhouk R S, Alexander C M, Chin J R, Clift S M, Bissell M J, Werb Z. J Cell Biol. 1994;125:681–693. doi: 10.1083/jcb.125.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faraldo M M, Deugnier M A, Thiery J P, Glukhova M A. Adv Exp Med Biol. 2000;480:169–174. doi: 10.1007/0-306-46832-8_21. [DOI] [PubMed] [Google Scholar]
- 47.Faraldo M M, Deugnier M A, Lukashev M, Thiery J P, Glukhova M A. EMBO J. 1998;17:2139–2147. doi: 10.1093/emboj/17.8.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klinowska T C, Soriano J V, Edwards G M, Oliver J M, Valentijn A J, Montesano R, Streuli C H. Dev Biol. 1999;215:13–32. doi: 10.1006/dbio.1999.9435. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, Sheng S, Maass N, Sager R. Mol Med. 1997;1:49–59. [PMC free article] [PubMed] [Google Scholar]
- 50.Seftor R E, Seftor E A, Sheng S, Pemberton P A, Sager R, Hendrix M J. Cancer Res. 1998;58:5681–5685. [PubMed] [Google Scholar]