Abstract
Background
Taenia pisiformis is one of the most common intestinal tapeworms and can cause infections in canines. Adult T. pisiformis (canines as definitive hosts) and Cysticercus pisiformis (rabbits as intermediate hosts) cause significant health problems to the host and considerable socio-economic losses as a consequence. No complete genomic data regarding T. pisiformis are currently available in public databases. RNA-seq provides an effective approach to analyze the eukaryotic transcriptome to generate large functional gene datasets that can be used for further studies.
Methodology/Principal Findings
In this study, 2.67 million sequencing clean reads and 72,957 unigenes were generated using the RNA-seq technique. Based on a sequence similarity search with known proteins, a total of 26,012 unigenes (no redundancy) were identified after quality control procedures via the alignment of four databases. Overall, 15,920 unigenes were mapped to 203 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Through analyzing the glycolysis/gluconeogenesis and axonal guidance pathways, we achieved an in-depth understanding of the biochemistry of T. pisiformis. Here, we selected four unigenes at random and obtained their full-length cDNA clones using RACE PCR. Functional distribution characteristics were gained through comparing four cestode species (72,957 unigenes of T. pisiformis, 30,700 ESTs of T. solium, 1,058 ESTs of Eg+Em [conserved ESTs between Echinococcus granulosus and Echinococcus multilocularis]), with the cluster of orthologous groups (COG) and gene ontology (GO) functional classification systems. Furthermore, the conserved common genes in these four cestode species were obtained and aligned by the KEGG database.
Conclusion
This study provides an extensive transcriptome dataset obtained from the deep sequencing of T. pisiformis in a non-model whole genome. The identification of conserved genes may provide novel approaches for potential drug targets and vaccinations against cestode infections. Research can now accelerate into the functional genomics, immunity and gene expression profiles of cestode species.
Introduction
More than 70 nominal species having been attributed to the ancient genus of Taenia [1], and approximately 42 valid species and three subspecies are currently recognized [2]. Taeniidae have two genera, Echinococcus and Taenia. Echinococcus comprises the tapeworm family, including E. granulosus and E. multilocularis, which causes morbidity in both humans and livestock [3], [4]. The genus Taenia contains six diverse species, including cestodes and metacestodes, which cause significant health problems in humans and socio-economic losses in the livestock industry. Species of Taenia parasitize in different hosts, including fish, reptiles and mammals. The adult stage of T. pisiformis (Cestoidea; Cyclophyllidea; Taeniidae; Taenia) parasitizes and matures in the small intestine of canids and felines [1], [5]–[7]. Lagomorphs are the most common intermediate hosts of Cysticercus pisiformis tapeworms, which usually live within the liver capsule, greater omentum and mesentery [8], [9]. Both T. pisiformis and C. pisiformis are widely distributed worldwide [7], [10]–[13]. Infections may occur when canines ingest the internal organs of rodents infected with C. pisiformis or when lagomorphs consume food polluted by the infected canines with the proglottids of T. pisiformis. T. pisiformis and C. pisiformis can cause significant health problems and even death [14], [15].
To date, the focus of research in Taeniidae has concentrated mostly on the search for functional genes, genetic variation and immune mechanisms between cestode and their hosts that could lead to medications [16]–[19]. The genome project for T. solium was established by a consortium of key laboratories at the National Autonomous University of Mexico in 2006 [20], but the genome-wide data has not yet been presented. Almeida et al. (2009) analyzed the transcriptome of T. solium cysticerci using open reading frames (ORESTES) [21]. A total of 1,520 high quality expressed sequence tags (ESTs) were generated from 20 ORESTES cDNA mini-libraries [21], and a genome-wide sequencing project for E. multilocularis is currently being carried out in a cooperation between the Parasite Sequencing Unit of the Wellcome Trust Sanger Centre and Brehm et al. [22]. The whole sequenced genomes of E. granulosus, E. multilocularis, and Hymenolepis microstoma have been presented on the National Center of Biotechnology Information (NCBI) website. The ESTs of E. granulosus and E. multilocularis are available on the Sanger database (http://www.sanger.ac.uk/), and the transcriptome of E. multilocularis is available through the sequence read archive (SRA) of the NCBI database (http://www.ncbi.nlm.nih.gov/sra/). Genome and transcriptome analyses provide powerful resources for the study of cestodes in terms of biochemistry, neurobiology, pharmacotherapy, vaccine development, and host-parasite interrelations. Thus, it is important to improve the genome-wide transcriptome exploration of T. pisiformis. However, research into T. pisiformis and C. pisiformis has been confined to their etiology, epidemiology and pharmacotherapy alone.
High-throughput sequencing (RNA-seq) provides an approach to analyze the T. pisiformis transcriptome in unparalleled depth and sensitivity. In this study, we performed the first de novo transcriptome analysis of T. pisiformis, which obtained high coverage and depth of gene content. Furthermore, the transcriptome of T. pisiformis was compared with the ESTs of T. solium and Eg+Em (conserved ESTs between E. granulosus and E. multilocularis) with the GO and COG functional classification systems. The intersection of common genes that were conserved between T. pisiformis, T. solium and Eg+Em was analyzed.
To the best of our knowledge, this is the first study of the characteristic transcriptome of T. pisiformis using Solexa/Illumina sequencing technology. The results are expected to determine antigens, give insights into the biochemistry and neurobiology of T. pisiformis, reveal the gene distributions characteristic of four Taeniidae species and accelerate research into functional genomics and host-parasite interrelations in cestodes.
Results
Annotation of adult T. pisiformis transcriptome data
1) Coverage and quality of consensus sequences
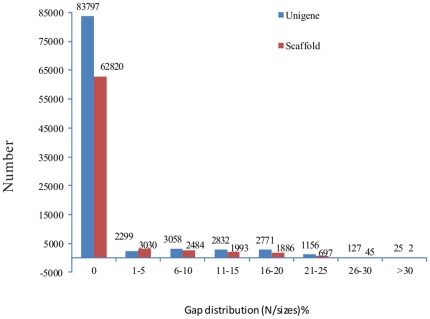
RNA was extracted from the whole organism of an adult T. pisiformis. Approximately 2.67 million clean reads and 240 million total nucleotides were obtained using Solexa/Illumina RNA-seq deep sequencing. All clean reads were submitted to the SRA database at NCBI (accession no.: SRA037310). The SQ20 (sequencing quality 20) refers to a quality score that is greater than the proportion of bases over 20 (an error probability of 0.01). The SQ20 and GC percentage were 89.19% and 50.8%, respectively. The length percentage of contigs was highest within the range of 75–100 bp (66.5%), and scaffolds and unigenes were within 100–500 bp (84.62% and 79.76%, respectively). Overall, 72,957 non-redundant consensus sequences were obtained through the removal of partial overlapping sequences, with an average unigene length of 398 bp and an N50 of 462 bp. All unigenes were provided as Dataset S1. It was determined that 87.23% (83,797/96,065) scaffolds and 86.11% (62,820/72,957) unigene consensus sequences had no gap (Figure 1). Only 25 scaffolds and two unigenes exceeded the 3% gap (Figure 1). In order to evaluate the quality of assembly, unigenes were aligned with the clean reads of T. pisiformis. The results showed that 99.79% (72,802/72,957) unigenes mapped to clean reads. The high mapped scores between unigenes and clean reads, and no gap percent in scaffolds and unigenes, suggested that the SOAPdenovo assembly was reliable.
Figure 1. Gap distribution of assembled scaffolds and unigenes of adult Taenia pisiformis transcriptome.
Gap distribution (N/size) %: gap percentage (N amount/sequence length) distribution. 87.23% of scaffold (83,797/96,065) and 86.11% of unigene (62,820/72,957) consensus sequences had no gap.
RACE PCR was utilized to validate the quality of the adult T. pisiformis transcriptome. Specifically, four unigene consensus sequences were randomly selected as templates. As a result, the four antigens obtained the full-length cDNA clones using RACE PCR, including homologous antigen 18KD (GenBank: JN247398), cC1 (GenBank: JN247399), TP1 (GenBank: JN228964) and TPP2 (GenBank: JN228965).
2) Functional annotation of unigenes in adult T. pisiformis
In order to detect known gene sequences in existing species, the RNA-seq data set of T. pisiformis was successively compared with the Nr, Swiss-prot, COG and KEGG databases successively, using the BLASTx program. A number of unigene consensus sequences unambiguously matched the previously annotated genes or overlapping annotated ORFS when aligned through the four respective databases (Table 1). And the respective annotations were presented in Dataset S2 (Nr), Dataset S3 (Swiss-prot), Dataset S4 (KEGG), and Dataset S5 (COG), respectively, via BLAST (basic local alignment on search tool).
Table 1. Summary of sequence assembly and annotations of the Taenia pisiformis transcriptome.
| Sequences (n) | Mean length (bp) | N50 (bp) | Annotations (n) | Functional classification | |
| Clean reads (paired-end) | 2.67 million | - | - | - | - |
| All assembled contigs | 330, 316 | 154 | 169 | - | - |
| All assembled scaffords | 96,065 | 331 | 414 | - | - |
| All assembled unigenes | 72,957 | 398 | 462 | - | - |
| Gene against Nr | 25,701 | - | - | 201,908 | - |
| GO for Nr protein hits | 7,706 | - | - | 56,217 | 4,580 GO terms |
| Gene against Swiss-prot | 19,564 | - | - | 148,048 | - |
| Gene against COG | 7,760 | - | - | 47,241 | 25 molecular families/98 categories |
| Gene against KEGG | 15,920 | - | - | 134,829 | 203 KEGG pathways |
| CDS against four database | 25,633 | - | - | 25,633 | - |
| All annotated unigenes | 26,012 | - | - | - | - |
N50 = median length of all non-redundant consensus sequences, mean = average length of all consensus sequences.
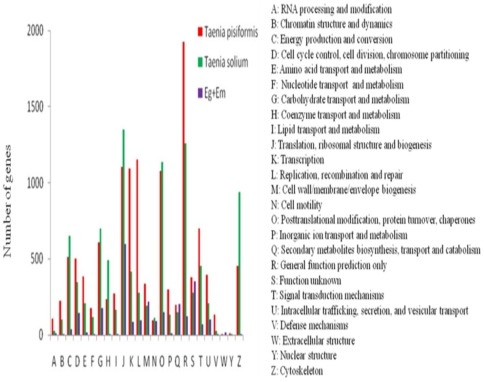
After alignment through the COG database, the functions of 7,760 unigenes were classified into at least 25 molecular families and 98 functional-categories ( Figure 2 ). Among these molecular families, R (general function prediction only) included only 7.04% (1,830/26,012) unigenes. The next commonest, L (replication, recombination and repair), J (translation, ribosomal structure and biogenesis), K (transcription) and O (post-translational modification, protein turnover, chaperones) had 4.30% (1,119), 4.02% (1,046), 4.00% (1,040) and 3.96% (1,029) unigenes, respectively. W (extracellular structures) had two unigenes. Apart from these molecular families, S (function unknown) had 0.47% (322) unigenes. Of the 98 functional-categories, the four largest categories also included ‘general function prediction’, ‘post-translational modification, protein turnover and chaperones’, ‘translation, ribosomal structure and biogenesis’ and ‘replication, recombination and repair’.
Figure 2. COG functional annotations of putative proteins among unigenes of four species in Taeniidae.
Overall, 459 ESTs of Eg+Em (common genes in Echinococcus granulosis and Echinococcus multilocularis), 7,760 unigenes of Taenia pisiformis, and 6,852 ESTs of Taenia solium were classified into at least 25 molecular families.
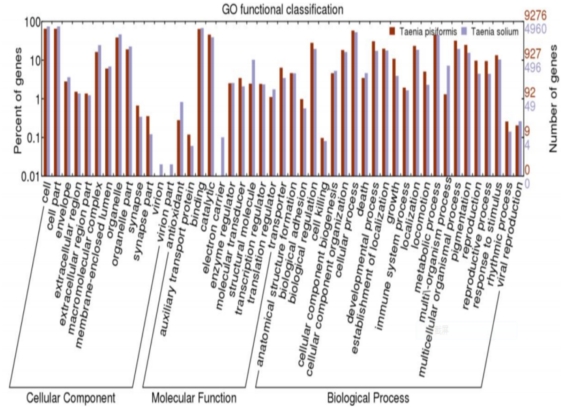
An analysis of GO function provided a GO functional classification annotation for differentially expressed genes, as well as an enrichment analysis for differentially expressed genes. Of the most significant BLASTx hits against the Nr known species dataset, a total of 7,706 unigenes were assigned GO term annotations using BLAST2GO ( Table 1 ). In order to categorize the genome-wide transcriptome of T. pisiformis functionally, these GO terms were summarized into the three main GO categories and 47 sub-categories. GO has three functional categories: molecular function (including 23 sub-categories), cellular components (including 11 sub-categories) and biological process (including 13 sub-categories) ( Figure 3 ). Biological process made up the majority of the GO annotations (29,831, 53.06% of the total), followed by molecular function (16,684, 29.68%) and cellular components (9,702, 17.26%).
Figure 3. GO annotations of unigenes in Taenia pisiformis transcriptome and ESTs in Taenia solium.
In total, 7,706 unigenes of Taenia pisiformis and 4,960 ESTs of Taenia solium obtained 88,471 GO annotations.
Further biological process categories were prominently represented in ‘cellular process’, ‘metabolic process’ and ‘multicellular organismal process’, which indicated that some important cellular activities may occur in T. pisiformis. In addition, ‘immune system process’, ‘metabolic process’, and ‘response to stimulus’ related to the immune responses and immune defenses of T. pisiformis. In the molecular function category, ‘binding’ and ‘catalytic activity’ represented the most abundant classification. Accordingly, ‘cell’, ‘cell part’, and ‘organelle’ were represented in the cellular component category, while 1,615 annotations belonged to the extracellular region.
3) KEGG pathway of unigene consensus sequences
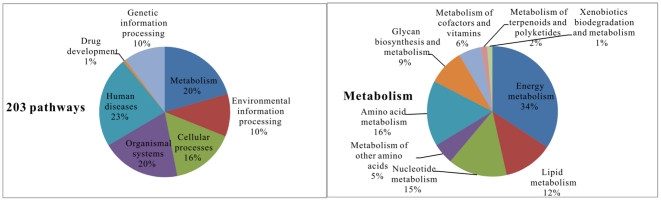
In order to identify the active biological pathways in T. pisiformis, a total of 15,920 unigenes were assigned to 203 KEGG pathways (Dataset S6) from the KEGG database. The KEGG pathways embodied metabolism (9 members), genetic information processing (16 members), environmental information processing (17 members), cellular processes (13 members), organismal systems (34 members), drug development (2 members) and human diseases (37 members) (Figure 4).
Figure 4. A total of 15,920 unigenes of adult Taenia pisiformis transcriptome were assigned to 203 KEGG pathways.
The KEGG pathways embodied in metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, drug development and human diseases.
In order to gain further insights into the biochemistry and physiology of T. pisiformis, we selected the glycolysis/gluconeogenesis pathway of energy metabolism within the metabolism and axon guidance pathway (Dataset S7) of organismal systems for further analysis. The pathway of glycolysis/gluconeogenesis relates to energy generation, and 215 unigenes were related to this pathway that mapped to 32 enzyme genes. Three key enzymes in glycolysis/gluconeogenesis, hexokinase, phosphofructokinase and pyruvate kinase, were mapped by 21, 12, and 21 unigenes, respectively ( Table 2 ). No unigenes mapped to pyruvic decarboxylase, malic dehydrogenase, and ADP-forming acetyl-COA synthetase ( Table 2 ).
Table 2. The key genes of the glycolysis/gluconeogenesis and the axon guidance pathways for Taenia pisiformis.
| Genes name | Ko | Definition | Disease |
| HK | K00844 | hexokinase [EC:2.7.1.1] | Anemia due to disorders of glycolytic enzymes (H00664) |
| PK | K00873 | pyruvate kinase [EC:2.7.1.40] | - |
| ALDO | K01623 | fructose-bisphosphate aldolase, class I [EC:4.1.2.13] | Hereditary fructose intolerance (H00071) |
| E6.2.1.13 | K01905 | acetyl-CoA synthetase (ADP-forming) [EC:6.2.1.13] | - |
| E4.1.1.1, pdc | k01568 | pyruvate decarboxylase [E4.1.1.1] | - |
| NTN1 | K06843 | netrin 1 | - |
| EFNB | K05463 | ephrin-B | Craniosynostosis (H00458) |
| SLIT1 | K06838 | Slit 1 | - |
| SLIT2 | K06839 | Slit 2 | - |
| SLIT3 | K06850 | Slit 3 | - |
| SEMA4 | K06521 | Semaphoring 4 | Cone-rod dystrophy and cone dystrophy ( H00481)Retinitis pigmentosa (RP)( H00527) |
| SEMA5 | K06841 | Semaphoring 5 | Cri du chat syndrome (H00764) |
| SEMA6 | K06842 | Semaphoring 6 | - |
| DCC | K06765 | deleted in colorectal carcinoma | Gastric cancer (H00018), Colorectal cancer (H00020), Cancer of the anal canal (H00044) |
| UNC5 | K07521 | netrin receptor unc-5 | - |
| EPHA1 | K05102 | Eph receptor A1 [EC:2.7.10.1] | - |
| EPHB1 | K05110 | Eph receptor B1 [EC:2.7.10.1] | - |
| ROBO1 | K06753 | roundabout, axon guidance receptor 1 | - |
| ROBO2 | K06754 | roundabout, axon guidance receptor 2 | - |
| ROBO3 | K06755 | roundabout, axon guidance receptor 3 | - |
The other important pathway was axon guidance, which is one of the most important organismal systems for neurotransmission. During the development of the nervous system, axons are guided along specific pathways by different classes of guidance cues within the extracellular environment. Attractive and repulsive guidance molecules play vital roles in axon path-finding through long-range or short-range modes. A total of 262 unigenes were mapped to this pathway. There are four highly conservative axon guidance molecular protein families, netrins (netrin 1), slits (slit1, slit2, and slit3), semaphorins (sema4D, sema5, and sema6), and ephrins (ephrin E) ( Table 2 ). The receptors of netrin 1 contain DCC and UNC-5, which mediate axon repulsion (via Ca2+ concentration) and axon outgrowth, respectively. The receptors for slit1, slit2, and slit3 include Robo1, Robo2, and Robo3, which mediate axon attraction and axon repulsion. The common receptor of sema4D, sema5, and sema6 is plexin B, which mediates axonal repulsion. Eph A and Eph B are receptors for ephrin E, which mediate axonal attraction and axonal repulsion. Through analyzing the glycolysis/gluconeogenesis pathway and the signaling pathway for axonal guidance, we achieved a more in-depth understanding of the biochemistry of adult T. pisiformis.
4) Alignment CDS of unigene consensus sequences
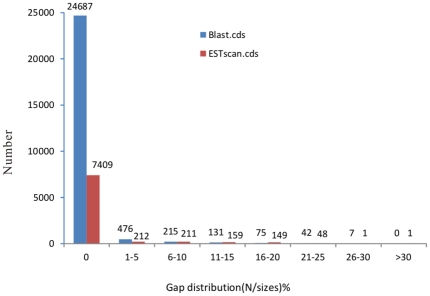
The results showed that among the annotated unigenes, 25,633 coding sequences (CDS) were obtained by the BLASTx algorithm, where one unigene corresponded to one CDS. In particular, 519 CDS perfectly matched the known genes from 10 other tapeworm species. However, the failed-hit unigenes were predicted by the ESTScan software and 8,190 CDS were obtained. The orientation of all CDS was 5′-3′. Furthermore, 97.09% (24,887/25,633) blast CDS and 90.46% (7,409/8,190) ESTScan CDS had no gap ( Figure 5 ), which demonstrated the high quality of the assemblies.
Figure 5. Gap distribution of assembled blast CDS and ESTscan CDS of adult Taenia pisiformis transcriptome.
Gap distribution (N/size) %: gap percentage (N amount/sequence length) distribution. 97.09% (24887/25633) blast CDS and 90.46% (7409/8190) ESTScan CDS had no gap.
Overall, those CDS belonged to different species, including parasites, mammals, aquatic animals and others. Twenty-seven CDS were aligned to the T. pisiformis genome, including mitochondrial genes and antigen genes, such as NADH dehydrogenase subunit 1, subunit 2, subunits 4 to 6, cytochrome b, cathepsin L-like protease, and cysteine protease. The tapeworm species, T. solium, T. saginata, T. taeniaeformis, T. asiatica, T. hydatigena, T. ovis, E. granulosus, E. multilocularis, E. ortleppi, and M. corti, showed a high similarity with T. pisiformis. Among these 10 tapeworm species, apart from normal physiological activities and the structure of their organs, a number of antigens that had been reported were aligned, particularly in T. asiatica, T. solium and E. granulosus. For example, TSES33, Tso31d, GP50, GP50c precursor, Tso22b, cC1, T24, cathepsin L-like cysteine proteinase, heat shock protein (HSP) and paramyosin had been reported in other cestode species, and were present in the CDS of unigene consensus sequences in T. pisiformis. Otherwise, there were some CDS of oncosphere proteins, such as Tso31a, Tso22b, antigen A, and Tm18, which could be used to protect intermediate host (rabbit) as vaccine antigen.
In addition, some CDS of coding antioxidants in eukaryotic cells were identified from the Nr, Swiss-prot, COG and KEGG databases, including thioredoxin peroxidase, glutathione peroxidase, peroxidase, superoxide dismutase, and glutathione reductase. Furthermore, the parasite species not only included cestode species, but also protozoa (Leishmania infantum), nematodes (such as Brugia malayi, Caenorhabditis briggsae, C. elegans, Necator americanus, Ancylostoma caninum, and A. duodenale) and trematodes (such as Schistosoma mansoni, S. haematobium, S. japonicum).
Analysis of comparative transcripts among four Taeniidae species
1) Common genes
In total, 10,983 ESTs of E. granulosis and 12,701 ESTs of E. multilocularis were downloaded from the databases found at ftp://ftp.sanger.ac.uk/pub/pathogens/Echinococcus/granulosis/ESTs/fasta.gz and ftp://ftp.sanger.ac.uk/pub/pathogens/Echinococcus/multilocularis/ESTs/fasta.gz, respectively. As described in the Methods section, 1,058 common ESTs (Eg+Em) were obtained. Then, the 1,058 ESTs of Eg+Em, 72,957 unigenes of T. pisiformis, and 30,700 ESTs (http://www.ncbi.nlm.nih.gov/nucest/?term=Taenia%20solium) were aligned using BLASTx. Six-hundred common genes (M) were found to be conserved between T. pisiformis, T. solium, E. granulosis and E. multilocularis.
2) COG and GO functional classification of T. pisiformis, T. solium and Eg+Em
The 1,058 ESTs of Eg+Em, 72,957 unigenes of T. pisiformis, and 30,700 ESTs of T. solium were aligned with the COG and Nr database and categorized by the COG and GO functional classification systems, respectively. After alignment through the COG database, 459 ESTs of Eg+Em (Dataset S8), 7,760 unigenes of T. pisiformis, and 6,852 ESTs of T. solium unigenes (Dataset S9) were classified into at least 25 molecular families (Figure 2). Functional classifications concentrated on J (translation, ribosomal structure and biogenesis), O (second metabolites biosynthesis, transport and catabolism), R (general function prediction only), and S (function unknown) (Figure 2). Overall, 1,058 ESTs of Eg+Em could not be given a GO functional classification in the Nr database; 7,706 unigenes of T. pisiformis and 4,960 ESTs (Dataset S10) of T. solium obtained the GO annotations according to Nr database (Figure 3). Biological process made up the majority of the GO annotations (47,022, 53.15% of the total) in T. pisiformis and T. solium, followed by cellular component (27,567, 31.16%) and molecular function (13,882, 15.69%). The functions determined by the GO classification mostly focused on cell, cell part, organelle, binding, catalytic activity, cellular process, localization, and metabolic process. Three GO terms only existed in T. solium: virion (4 ESTs), virion part (4 ESTs), and electron carrier (8 ESTs).
3) M genes (T. pisiformis/T. solium/E. granulosis/E. multilocularis)
We obtained 600 common genes (M) that existed in four species of Taeniidae. M was aligned in the KEGG database using BLASTx; 109 genes of M obtained 118 annotations (Dataset S11) and 21 pathways (Dataset S12). A series of key genes were identified that were closely related to the biochemistry of cestode, such as the G protein-coupled receptor 128 (ko08464), formin 2 (ko12821), survival motor neuron protein (ko13129), MKP (ko04459) of the MAPK signaling pathway, WASP (ko05747) in the adherens junction and chemokine signaling pathway, SF3a (ko12826) and FBP11 (ko12821) in splicesomes. In 21 pathways, approximately 36.7% genes were conserved in amoebas and Vibrio cholerae. The intestinal mucin was found in these two pathways. Furthermore, the adenomatosis polyposis coli (APC) protein was present in the ‘Regulation of actin cytoskeleton’, ‘Wnt signaling pathway’, ‘Colorectal cancer’, ‘Pathways in cancer’, and ‘Endometrial cancer’ ( Table 3 ).
Table 3. M (common genes in T. pisiformis, T. solium, E. granulosis, and E. multilocularis) was aligned KEGG database using BLASTx, and 109 genes obtained 21 pathways.
| Pathway | Count (109 genes) | Pathway ID |
| Amoebiasis | 20 | ko05146 |
| Vibrio cholerae infection | 20 | ko05110 |
| Dorso-ventral axis formation | 18 | ko04320 |
| Salivary secretion | 14 | ko04970 |
| Regulation of actin cytoskeleton | 9 | ko04810 |
| Pathogenic Escherichia coli infection | 4 | ko05130 |
| Spliceosome | 4 | ko03040 |
| Shigellosis | 4 | ko05131 |
| Fc gamma R-mediated phagocytosis | 3 | ko04666 |
| Chemokine signaling pathway | 3 | ko04062 |
| Bacterial invasion of epithelial cells | 3 | ko05100 |
| Adherens junction | 3 | ko04520 |
| Basal cell carcinoma | 2 | ko05217 |
| Wnt signaling pathway | 2 | ko04310 |
| Systemic lupus erythematosus | 2 | ko05322 |
| Endometrial cancer | 2 | ko05213 |
| Pathways in cancer | 2 | ko05200 |
| Colorectal cancer | 2 | ko05210 |
| Focal adhesion | 1 | ko04510 |
| MAPK signaling pathway | 1 | ko04010 |
| RNA degradation | 1 | ko03018 |
Discussion
The characterization of the transcriptome is essential for deciphering the functional complexity of the genome and to obtain a better understanding of cellular activities in organisms, including growth, development, disease, and immune defense [23]. The RNA-seq approach has not only proven to be an efficient method for transcriptome profiling analyzes [24], but is also effective in clarifying transcriptome complexity. In this study, the transcripts of T. pisiformis indicated that de novo short-read assembly and the Solexa/Illumina platform offered a larger number of distinct reads and increased physical coverage as a result of long fragment lengths. Those long fragments could be used to characterize gene expression, discover and identify new genes, and research metabolic pathways in non-reference whole genomes.
The great numbers of clean reads and unigenes resulted in a relatively deep coverage. Assembly results showed that the mean length of unigenes was much longer than that of contigs. In addition, the mean length of T. pisiformis unigenes was longer than those assembled in previous studies, such as Eucalyptus grandis (247 bp) [25]. The percentages of no gaps and high matching scores between unigenes and clean reads indicated high quality and validity level of the SOAPdenovo assembly. Annotated unigenes were assigned with not only gene or protein name descriptions, but also putative conserved domains, GO terms, and metabolic or signaling pathways (only in the KEGG database). The annotations of unigenes provided the biological functions, metabolic and signaling pathways of candidate genes in a given time. We therefore gained a better understanding of the gene expression of T. pisiformis in the terminal host, and searched for antigens of C. pisiformis in intermediate hosts. Functional annotations in the Nr database laid a foundation for the analysis of gene ontology. All annotated unigenes in the Swiss-prot database played an important role in functional annotations.
A large number of unigenes were assigned to a wide range of COG classifications, which indicated that our RNA-seq data represented a wide diversity of transcripts. COGs consist of protein sequences encoded in 21 complete genomes, including bacteria, algae and eukaryotes, and were built on classifications according to phylogenetic relationships [26]. Each COG consists of individual proteins or groups of analogs from at least three lineages and thus corresponds to an ancient conserved domain [26]. In 25 molecular families, H (coenzyme transport and metabolism), L (replication, recombination and repair), U (intracellular trafficking secretion, and vesicular transport) and V (defense mechanisms) played a significant role, and were closely associated with the immunology and physiology of T. pisiformis. The analysis of gene expression levels, a GO classification of genetic functions and prediction of protein metabolic pathways, predicted the screening of differentially expressed genes, GO functional classification of different genes and positioning of metabolic pathways. These sequence data and statistical analysis have provided abundant information on T. pisiformis infections, enabling a better understanding of antigens and the basic functional distribution of the gene.
At present, there are just 26 ESTs in NCBI, and only three antigens of T. pisiformis have been registered on NCBI: T24 (GenBank: GU321333.1), cathepsin L-like cysteine protease (GenBank: JF798507.1), and cysteine protease (GenBank: JF718743.1). The substantial amounts of antigens were aligned and had clusters of orthologous group with other tapeworm species. For example, Tso31d [27], Tm18 [27], cC1 [28], and paramyosin [29] could be used as vaccine antigens; GP50 [30], cathepsin L-like cysteine proteinase [31], and heat shock protein [32] could be used as effective diagnostic antigens. In addition, vast antigens of oncosphere can be found in annotated and no hit unigenes for protecting intermediate host. T. pisiformis also has a homologous CDS with Caenorhabditis elegans, Ancylostoma caninum, Schistosoma mansoni and Leishmania infantum. This indicates that RNA-seq data indeed had the necessary depth, coverage and diversity. Therefore, we could screen mass new genes for T. pisiformis from those annotated CDS. Paramyosin is not only utilized as an antigen, but may also have other biological functions in T. pisiformis. For example, the metacestodes have elaborate means of evading complement-mediated destruction, including paramyosin (restraint C1q) [33]. Antioxidant like the CDS of thioredoxin peroxidase (TPX) plays a role in protecting against oxidative damage. Additionally, recombinant EgTPx may be useful for the screening of specific inhibitors that could serve as new drugs for the treatment of hydatid disease [34]. Thus, we predict that those key genes may play an important role in life activites of T. pisiformis in the small intestine of dogs, which need be analyzed in further study.
Functional unigenes closely relate to the metabolic or signaling pathways, which play an important role in life history of T. pisiformis. In this study, we analyzed the glycolysis/gluconeogenesis and axon guidance pathways. Phosphofructokinase is not only a key enzyme in the pathway of glycolysis/gluconeogenesis, but also the biochemical basis of the long-term utilization of antimony for the treatment of schistosomiasis [35]. Therefore, further research into analogous enzymes could provide new potential pharmacotherapeutic targets for the treatment of cestode infections. Meanwhile, we suppose that anerobic glycolysis is the principal way that cestodes obtain energy in the intestinal tract of a host. Cestodes have a developed nervous system that plays an important role in multiple aspects of life activity, such as growth and development, muscle movement, metabolism of salt and water, and reproduction. The secretory vesicles of nerve cells play a neural regulatory role via axonal guidance [36]. Therefore, axon guidance represents a key stage in the formation of neuronal networks. Netrins, slits, semaphorins, and ephrins are the highly conservative protein families that are involved in axonal guidance, which afford a research basis for the neurobiology of T. pisiformis. These guidance cues are read by growth cone receptors, and signal transduction pathways downstream of these receptors converge onto the Rho GTPases to elicit changes in cytoskeletal organization that determine the direction that the growth cone will turn [37]. As cestodes live in the small intestine of dogs, two aspects of the firm attachment to the host small intestine and high fertility rates are very important to T. pisiformis. Sensory organs in tapeworm have gradually diminished due to life in the host small intestine. Thus, the developed nervous system plays an important role in the life activities of T. pisiformis.
T. pisiformis, T. solium, E. granulosis, and E. multilocularis are important species of tapeworm that damage public health and bring huge economic losses in developing countries [38]–[40]. In particular, T. solium, E. granulosis, and E. multilocularis can severely damage human health. Thus, prophylaxis and treatment against these tapeworms are particularly important. A large number of COG and GO functional annotations were related to the basic life activities of the cestode. Functional distribution characteristics were consequently obtained from these data. Some of common genes (M) from four species of Taeniidae in the KEGG pathway were identified to be related to cestode biochemistry. The G protein-coupled receptor [41] and survival motor neuron protein [42] mediate signal transduction. MKP is a dual-specificity phosphatase family that is involved in the MAPK signaling pathway, and includes MKP-2 that dephosphorylates and inactivates mitogen-activated protein kinases (MAPKs) [43].
Interestingly, we found two common genes in four cestode species through the analysis of comparative transcripts for intestinal mucin and APC genes. Amoebiasis and cholera are serious human gastrointestinal diseases caused by Entamoebahistolytica and Vibrio cholerae, respectively [44], [45]. Lee et al. (2010) indicated that Gymnophalloides seoi antigen upregulates the expression of Toll-like receptor 2 and mucin-related 2 by human intestinal epithelial cells, which reflects a helminth-induced, IFN-c–dependent, and innate mucosal immune mechanism in this human intestinal cell line [46]. In addition, APC participates in the Wnt signaling pathway. When spontaneous mutations occur that means APC cannot play a normal physiological function, the individual is at a higher risk of cancers, including colorectal cancer [47]. The APC gene has been reported in canines in the NCBI database, whereas intestinal mucin is not conserved in canines. Scholl (2003) argued that horizontal gene transfer between parasites and hosts, sometimes involving viral-bacterial-parasite-host chains, might be of great consequence for the evolution of hosts and parasites [48]. We cannot as yet whether the APC gene of T. pisiformis has been transferred from dogs, but this will undergo further analysis. Thus, there is major merit in undertaking comparative functional genomics, transcriptomic and proteomic investigations to establish whether intestinal mucin 2 and APC play a similar role in the development of these four cestode species and their transition to parasitism in canine intestines. These genes may be putative parasitism-related genes in cestodes that are adapted to avoid host immunity. Further gene expression profiling and experimental validation will be needed to test the functional role of these new T. pisiformis genes.
To the best of our knowledge, this is the first study that uses a de novo short-read assembly and the Solexa/Illumina platform to generate transcriptome (RNA-seq) data for T. pisiformis. In this study, we obtained 72,957 assembled unigenes, and 26,012 unigenes of them acquired the annotations. The identification of metabolic and signaling pathways in the present study will accelerate the understanding of the pathways of energy, anti-oxidation, immune system formation and development. This study demonstrates that this form of sequencing platform can be used as a rapid and cost-effective method for the analysis of non-model whole genomes. We utilized the unigenes of T. pisiformis to align against ESTs of T. solium, E. granulosis, and E. multilocularis. Functional distribution characteristics and common genes sets were obtained. We believe that this transcriptome dataset will serve as an important public information platform for T. pisiformis, which will accelerate research into gene expression, genomics, and functional genomics of cestodes. The identification of these common genes will provide novel platform for the development of vaccine candidates and drug targets.
Materials and Methods
Parasite material
Under supervision of a licensed veterinarian, the larvae (C. pisiformis) were collected (during a routine autopsy) from the gastric omentum majus of two dead New Zealand white rabbit naturally infected by this tapeworm in a farm, Sichuan, China. After morphological identification, 20 C. pisiformis from the rabbits were used for the dog infection. Adult worms (T. pisiformis) were taken out of small intestine, and then washed in warm physiological saline for three times to avoid contamination before they were frozen immediately and stored in liquid nitrogen. All animals were handled in strict accordance with animal protection law of the People's Republic of China (a draft of an animal protection law in China released on September 18, 2009). All study protocols (including the collection of the larvae from the rabbits, and the infection and sacrifice of the dogs) were reviewed and approved by the National Institute of Animal Health Animal Care and Use Committee at Sichuan Agricultural University, China (approval ID number 2009-013, approved for three years beginning 06/2/2009).
RNA isolation and Illumina sequencing
Total RNA was isolated from single adult T. pisiformis (2.5 g, including scoles, neck, and strobila) using Trizol reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol. Total RNA of independent adult T. pisiformis were stored at −80°C until their use. The RNA quality was verified by an Agilent 2100 RNA Nanochip (Agilent, Santa Clara, CA, USA) in terms of concentration (2.57 ng/µl), RNA integrity number (RIN: 5.3) and the 28S∶18S ratio (1.0). A total of 244.15 µg of RNA was pooled from adult T. pisiformis for the preparation of the cDNA library.
The OligoTex mRNA mini kit (Qiagen) was used to isolate poly (A) mRNA after total RNA was collected from adult T. pisiformis according to the manufacturer's protocol. Fragmentation buffer was added to interrupt mRNA to short fragments (100–400 bp). Following the agarose gel electrophoresis (2% TAE-agarose gel), a range of cDNA fragments (200±25 bp) were excised from the gel, and selected for the PCR amplification as templates using the SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen, Camarillo, CA) following the manufacturer's protocol, and PCR Primer PE 2.0 (Illumina, San Diego, CA). Illumina HiSeq™ 2000 was applied to sequencing at the Beijing Genomics Institute (BGI)-Shenzhen, Shenzhen, China (http://www.genomics.cn/index.php) according to the manufacturer's instructions (Illumina).
De novo assembly
Prior to assembly, the high-quality clean reads were obtained from raw reads by removing adaptor sequences, duplication sequences, reads that contained more than 10% “N” rates (the “N” character representing ambiguous bases in reads), and low-quality reads containing more than 10% bases with Q-value≤20. Then, 21-bp K-mers was utilized in the assembly of the short reads when using the SOAPdenovo program (http://soap.genomics.org.cn/soapdenovo.html) [49]. The detailed process of the assembly is described as supplemental Dataset S13. Finally, non-redundant unigenes were obtained with as long as length as possible.
Pipeline of bioinformatics analysis
BLASTx alignment (e-value<10−5) between unigenes and protein databases was performed, such as NCBI non-redundant protein (Nr) database (http://www.ncbi.nlm.nih.gov), Swiss-prot protein database (http://www.expasy.ch/sprot), the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg), and the Cluster of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG). The sequence direction of unigenes, and expression and functional annotation of unigenes were then decided by the best alignment results. BLASTx first aligned unigene sequences in four protein databases (e-value<10−5), which retrieved proteins with the highest sequence similarity with the given unigenes along with their protein functional annotations. Annotated unigenes were classified by Gene Ontology (GO) that was aligned by Nr database. With Nr annotations, Blast2GO program [50] was used to obtain GO annotations of unigenes. After receiving GO annotations for all unigenes, WEGO software [51] classified them to understand the distribution of gene functions of the species macroscopically. In addition, there had a high density of coding sequences (CDS) in comparison to most eukaryotes. Unigenes aligned to databases with higher priority did not enter the next circle. The alignments ended when all circles were finished. Both the nucleotide sequences (5′-3′) and amino sequences of the unigene coding regions were obtained. Unigenes that could not be aligned to any database were scanned by ESTScan [52] to obtain the nucleotide (5′-3′) and amino sequences of the coding regions.
Assessment to quality and reliability of transcriptome of adult T. pisiformis
Unigenes were aligned with clean reads of the T. pisiformis transcriptome. The map percentages were obtained between them, which assisted the analysis of the quality of the assembly. Furthermore, the CDS of four antigens were amplified by RACE PCR using cDNA of adult T. pisiformis (primers and annealing temperatures are shown in Table S1). Oligonucleotide primers of four antigens were designed using Primer software version 5.0.
Comparative transcripts analysis
Firstly, the ESTs of E. granulosus (ftp://ftp.sanger.ac.uk/pub/pathogens/Echinococcus/granulosis/ESTs/fasta.gz) and E. multilocularis (ftp://ftp.sanger.ac.uk/pub/pathogens/Echinococcus/multilocularis/ESTs/fasta.gz) were translated into amino acid sequences respectively, before the two tapeworm species were aligned with the BLASTx algorithm (e-value<10−5). Consequently, we obtained the common ESTs in E. granulosus and E. multilocularis that were termed Eg+Em. Secondly, the Eg+Em, unigenes of T. pisiformis, and ESTs of T. solium (http://www.ncbi.nlm.nih.gov/nucest?term=Taenia%20solium) were classified in COG and GO functional terms. The mutual-best matches among Eg+Em, T. pisiformis, and T. solium were carried out using the BLASTx algorithm. Finally, the common genes (termed M) that were conserved between T. pisiformis, T. solium, E. granulosus, and E. multilocularis were aligned to the KEGG database using the BLASTx algorithm.
Supporting Information
Oligonucleotide primers for RACE-PCR of four antigens. The 3′- and 5′-end of the gene was amplified by RACE PCR using the five oligonucleotide primers (reverse transcription, first-round nested PCR, and second-round nested-PCR) and TIANscript RT Kit (TianGenBioteh CO., LTD, Beijing), according to the manufacturer's manual.
(DOC)
A total of 72,957 unigenes were generated using the RNA-seq technique or Taenia pisiformis . The serial numbers of unigenes were used in keeping with the Dataset S2 to S7, S10 to S
(RAR)
The transcriptome annotations of Taenia pisiformis in non-redundant protein (Nr) database. Overall, 25,701 unigenes and 201,908 subject functional annotations were obtained from the Nr database.
(XLS)
The transcriptome annotations of Taenia pisiformis in Swiss-prot database. A total of 19,564 unigenes and 148,048 subject functional annotations were obtained from the Swiss-prot database.
(XLS)
The transcriptome annotations of Taenia pisiformis in KEGG database. Overall, 15,920 unigenes and 134,829 subject functional annotations were obtained from the KEGG database.
(XLSX)
The transcriptome annotations of Taenia pisiformis in COG database. A total of 7,760 unigenes and 47,241 subject functional annotations were obtained from the COG database.
(XLS)
The 203 KEGG pathways of Taenia pisiformis . In order to identify the active biological pathways in T. pisiformis, a total of 15,920 unigenes were assigned to 203 KEGG pathways.
(HTM)
The glycolysis/gluconeogenesis and axon guidance pathway of Taenia pisiformis . There were three key enzymes in glycolysis/gluconeogenesis: hexokinase (EC 2.7.1.1), phosphofructokinase (EC 4.1.2.13), and pyruvate kinase (EC 2.7.1.40). Additionally, there were four highly conservative axon guidance molecular families, netrins (netrin 1), slits (slits 1, slits 2, and slits 3), semaphorins (sema4D, sema5, and sema6), and ephrins (ephrin E).
(DOCX)
The ESTs of Eg+Em were classified into at least 25 molecular families against COG database.
(RAR)
The ESTs of Taenia solium were classified into at least 25 molecular families against COG database.
(RAR)
The ESTs of Taenia solium obtained the GO annotations according to Nr database.
(RAR)
Annotation of M (common genes of Taenia pisiformis , Taenia solium , Echinococcus granulosis and Echinococcus multilocularis ). The genes annotations of M (T. pisiformis, T. solium, E. granulosis, and E. multilocularis) against the KEGG database.
(RAR)
21 KEGG pathways of Taenia pisiformis , Taenia solium , Echinococcus granulosis and Echinococcus multilocularis . M was aligned in the KEGG database using BLASTx, and 109 genes obtained 21 pathways.
(HTM)
The process of the SOAPdenovo assembly.
(DOCX)
Acknowledgments
We thank Shenzhen Welltec Gene Tecnology Co., Ltd., for assistance in raw data processing, and Ning Yan, Runhui Zhang, and Wanpeng Zheng for related bioinformatics analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grant from the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) (grant no. IRT0848). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jia WZ, Yan HB, Guo AJ, Zhu XQ, Wang YC, et al. Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics. 2010;11:447. doi: 10.1186/1471-2164-11-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoberg EP. Phylogeny of Taenia: Species definitions and origins of human parasites. Parasitology International. 2006;55:23–30. doi: 10.1016/j.parint.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 3.McManus DP, Zhang WB, Li J, Bartley PB. Echinococcosis. Lancet. 2003;362:1295–1304. doi: 10.1016/S0140-6736(03)14573-4. [DOI] [PubMed] [Google Scholar]
- 4.Thompson RCA. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. 2008;119:439–446. doi: 10.1016/j.exppara.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 5.Bagrade G, Kirjusina M, Vismanis K, Ozoliņs J. Helminth parasites of the wolf Canis lupus from Latvia. J Helminthol. 2009;83:63–68. doi: 10.1017/S0022149X08123860. [DOI] [PubMed] [Google Scholar]
- 6.Lahmar S, Sarciron ME, Rouiss M, Mensi M. Echinococcus granulosus and other intestinal helminths in semi-stray dogs in Tunisia: infection and re-infection rates. Tunis Med. 2008;86:657–664. [PubMed] [Google Scholar]
- 7.Saeed I, Maddox-Hyttel C, Monrad J, Kapel CM. Helminths of red foxes (Vulpes vulpes) in Denmark. Vet Parasitol. 2006;139:1–3. doi: 10.1016/j.vetpar.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Owiny JR. Cysticercosis in laboratory rabbits. Contemp Top Lab Anim Sci. 2001;40:45–48. [PubMed] [Google Scholar]
- 9.Wang M. veterinary parasitology. Beijing: China Ariculture Press; 2004. [Google Scholar]
- 10.Martínez-Moreno FJ, Hernández S, López-Cobos E, Becerra C, Acosta I, et al. Estimation of canine intestinal parasites in Córdoba (Spain) and their risk to public health. Vet Parasitol. 2007;143:7–13. doi: 10.1016/j.vetpar.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Foronda P, Valladares B, Lorenzo-Morales J, Ribas A, Feliu C, et al. Helminths of the wild rabbit (Oryctolagus cuniculus) in Macaronesia. J Parasitol. 2003;89:952–957. doi: 10.1645/GE-3048. [DOI] [PubMed] [Google Scholar]
- 12.Allan JC, Craig PS, Sherington J, Rogan MT, Storey DM, et al. Helminth parasites of the wild rabbit Oryctolagus cuniculus near Malham Tarn, Yorkshire, UK. J Helminthol. 1999;73:289–294. doi: 10.1017/s0022149x99000487. [DOI] [PubMed] [Google Scholar]
- 13.Zhou YX, Du AF, Zhang XJ, Wu YM, Tong FY, et al. Research of harmfulness of Cysticercus pisiformis in rabbit. Journal of Zhejiang agricultural science. 2008;3:372–373. [Google Scholar]
- 14.Rajasekariah GR, Rickard MD, O'Donnell IJ. Taenia pisiformis: protective immunization of rabbits with solubilized oncospheral antigens. Experimental Parasitology. 1985;59:321–327. doi: 10.1016/0014-4894(85)90087-6. [DOI] [PubMed] [Google Scholar]
- 15.Sun XL, Chen HT, Cai XP. A histopathologic study on cysticercus pisiformis infected rabbits. Acta Veterinaria Et Zootechnica Sinica. 2008;39:1100–1106. [Google Scholar]
- 16.Amin Pour A, Hosseini SH, Shayan P. Comparative genotyping of Echinococcus granulosus infecting buffalo in Iran using cox1 gene. Parasitol Res. 2011;108:1229–1234. doi: 10.1007/s00436-010-2170-x. [DOI] [PubMed] [Google Scholar]
- 17.Ragunathan L, Kalivaradhan SK, Ramadass S, Nagaraj M, Ramesh K. Helminthic infections in school children in puducherry, South India. J Microbiol Immunol Infect. 2010;43:228–232. doi: 10.1016/S1684-1182(10)60036-9. [DOI] [PubMed] [Google Scholar]
- 18.Atluri VS, Singhi PD, Khandelwal N, Malla N. 2D-PAGE analysis of Taenia solium metacestode 10–30 kDa antigens for the serodiagnosis of neurocysticercosis in children. Acta tropica. 2011;18:165–169. doi: 10.1016/j.actatropica.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Du W, Hu F, Yang Y, Hu D, Hu X, et al. Molecular cloning, characterization, and immunolocalization of two lactate dehydrogenase homologous genes from Taenia solium. Parasitol Res. 2011;109:567–574. doi: 10.1007/s00436-011-2285-8. [DOI] [PubMed] [Google Scholar]
- 20.Aguilar-Diaz H, Bobes RJ, Carrero JC, Camacho-Carranza R, Cervantes C, et al. The genome project of Taenia solium. Parasitology International. 2006;55:S127–S130. doi: 10.1016/j.parint.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Almeida CR, Stoco PH, Wagner G, Sincero TC, Rotava G, et al. Transcriptome analysis of Taenia solium cysticerci using Open Reading Frame ESTs (ORESTES). Parasit Vectors. 2009;2:35. doi: 10.1186/1756-3305-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brehm K. Echinococcus multilocularis as an experimental model in stem cell research and molecular host-parasite interaction. Parasitology. 2010;137:537–555. doi: 10.1017/S0031182009991727. [DOI] [PubMed] [Google Scholar]
- 23.Xiang LX, He D, Dong WR, Zhang YW, Shao JZ. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics. 2010;11:472. doi: 10.1186/1471-2164-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu T, Lu G, Fan D, Zhu C, Li W, et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–1249. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novaes E, Drost DR, Farmerie WG, Pappas GJJ, Grattapaglia D, et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi C, Yang H, Wei C, Yu O, Zhang Z, et al. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12:131. doi: 10.1186/1471-2164-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundström J, Salazar-Anton F, Sherwood E, Andersson B, Lindh J. Analyses of an expressed sequence tag library from Taenia solium, Cysticerca. Plos Negl Trop Dis. 2010;4:e919. doi: 10.1371/journal.pntd.0000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo YJ, Sun SH, Zhang Y, Chen ZH, Wang KY, et al. Protection of pigs against Taenia solium cysticercosis using recombinant antigen or in combination with DNA vaccine. Vaccine. 2004;22:3841–3847. doi: 10.1016/j.vaccine.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Solís CF, Ostoa-Saloma P, Lugo-Martínez VH, Johnston SA, Laclette JP. Genetic vaccination against murine cysticercosis by using a plasmid vector carrying Taenia solium paramyosin. Infect Immun. 2005;73:1895–1897. doi: 10.1128/IAI.73.3.1895-1897.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bueno EC, Scheel CM, Vaz AJ, Machado LR, Livramento JA, et al. Application of synthetic 8-KD and recombinant GP50 antigens in the diagnosis of neurocysticercosis by enzyme-linked immuosorbent assay. Am J Trop Med Hyg. 2005;72:278–283. [PubMed] [Google Scholar]
- 31.Sako Y, Yamasaki H, Nakaya K, Nakao M, Ito A. Cloning and characterization of cathepsin L-like peptidases of Echinococcus multilocularis metacestodes. Mol Biochem Patasitol. 2007;154:181–189. doi: 10.1016/j.molbiopara.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Ferrer E, González LM, Foster-Cuevas M, Cortéz MM, Dávila I, et al. Taenia solium: characterization of a small heat shock protein (Tsol-sHSP35.6) and its possible relevance to the diagnosis and pathogenesis of neurocysticercosis. Exp Parasitol. 2005;110:1–11. doi: 10.1016/j.exppara.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 33.White ACJ, Robinson P, Kuhn R. Taenia solium cysticercosis: host-parasite interactions and the immune response. Chen Immunol. 1997;66:209–230. [PubMed] [Google Scholar]
- 34.Li J, Zhang WB, Loukas A, Lin RY, Ito A, et al. Functional expression and characterization of Echinococcus granulosus thioredoxin peroxidase suggests a role in protection against xidative damage. Gene. 2004;326:157–165. doi: 10.1016/j.gene.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 35.Li GQ, Xie MQ. Senior parasitology. Beijing: Higher Education Press; 2007. 92 [Google Scholar]
- 36.Liu B. Studies on the pePtidergie nervous systemin cestodes: PHD thesis. 2001. Jilin University, Agriculture department.
- 37.Guan KL, Rao Y. Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci. 2003;4:941–956. doi: 10.1038/nrn1254. [DOI] [PubMed] [Google Scholar]
- 38.Saarma U, Jogisalu I, Moks E, Varcasia A, Lavikanen A, et al. A novel phylogeny for the genus Echinococcus, based on nuclear data, challenges relationships based on mitochondrial evidence. Parasitology. 2009;136:317–328. doi: 10.1017/S0031182008005453. [DOI] [PubMed] [Google Scholar]
- 39.Sáenz B, Fleury A, Chavarría A, Hernández M, Crispin JC, et al. Neurocysticercosis: local and systemic immune-inflammatory features related to severity. Med Microbiol Immunol. 2011 doi: 10.1007/s00430-011-0207-0. in press. [DOI] [PubMed] [Google Scholar]
- 40.Torgerson PR, Deplazes P. Echinococcosis: diagnosis and diagnostic interpretation in population studies. Trends Parasitol. 2009;25:164–170. doi: 10.1016/j.pt.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Borroto-Escuela DO, Tarakanov AO, Guidolin D, Ciruela F, Agnati LF, et al. Moonlighting characteristics of G protein-coupled receptors: Focus on receptor heteromers and relevance for neurodegeneration. IUBMB Life. 2011;63:463–472. doi: 10.1002/iub.473. [DOI] [PubMed] [Google Scholar]
- 42.Ymlahi-Ouazzani Q, J Bronchain O, Paillard E, Ballagny C, Chesneau A, et al. Reduced levels of survival motor neuron protein leads to aberrant motoneuron growth in a Xenopus model of muscular atrophy. Neuroqenetics. 2010;11:27–40. doi: 10.1007/s10048-009-0200-6. [DOI] [PubMed] [Google Scholar]
- 43.Peng DJ, Zhou JY, Wu GS. Post-translational regulation of mitogen-activated protein kinase phosphatase-2 (MKP-2) by ERK. Cell Cycle. 2010;9:4650–4655. doi: 10.4161/cc.9.23.13957. [DOI] [PubMed] [Google Scholar]
- 44.Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci U S A. 2006;103:9298–9303. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin NA, Mount Patrick SK, Estrada TE, Frisk HA, Rogan DT, et al. Active transport of bile acids decreases mucin 2 in neonatal ileum: implications for development of necrotizing enterocolitis. PLOS One. 2011;6:e27191. doi: 10.1371/journal.pone.0027191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee KD, Guk SM, Chai JY. Toll-like receptor 2 and Muc2 expression on human intestinal epithelial cells by Gymnophalloides seoi adult antigen. J Parasitol. 2010;96:58–66. doi: 10.1645/GE-2195.1. [DOI] [PubMed] [Google Scholar]
- 47.Cheadle JP, Krawczak M, Thomas MW, Hodges AK, Al-Tassan N, et al. Different combinations of biallelic APC mutation confer different growth advantages in colorectal tumours. Cancer Res. 2002;62:363–366. [PubMed] [Google Scholar]
- 48.Scholl EH, Thorne JL, McCarter JP, Bird DM. Horizontally transferred genes in plant-parasitic nematodes: a high-throughput genomic approach. Genome Biology. 2003;4:R39. doi: 10.1186/gb-2003-4-6-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Zhu H, Ruan J, Qian W, Fang X, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talón M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 51.Ye J, Fang L, Zheng H, Zhang Y, Chen J, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol. 1999:138–148. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers for RACE-PCR of four antigens. The 3′- and 5′-end of the gene was amplified by RACE PCR using the five oligonucleotide primers (reverse transcription, first-round nested PCR, and second-round nested-PCR) and TIANscript RT Kit (TianGenBioteh CO., LTD, Beijing), according to the manufacturer's manual.
(DOC)
A total of 72,957 unigenes were generated using the RNA-seq technique or Taenia pisiformis . The serial numbers of unigenes were used in keeping with the Dataset S2 to S7, S10 to S
(RAR)
The transcriptome annotations of Taenia pisiformis in non-redundant protein (Nr) database. Overall, 25,701 unigenes and 201,908 subject functional annotations were obtained from the Nr database.
(XLS)
The transcriptome annotations of Taenia pisiformis in Swiss-prot database. A total of 19,564 unigenes and 148,048 subject functional annotations were obtained from the Swiss-prot database.
(XLS)
The transcriptome annotations of Taenia pisiformis in KEGG database. Overall, 15,920 unigenes and 134,829 subject functional annotations were obtained from the KEGG database.
(XLSX)
The transcriptome annotations of Taenia pisiformis in COG database. A total of 7,760 unigenes and 47,241 subject functional annotations were obtained from the COG database.
(XLS)
The 203 KEGG pathways of Taenia pisiformis . In order to identify the active biological pathways in T. pisiformis, a total of 15,920 unigenes were assigned to 203 KEGG pathways.
(HTM)
The glycolysis/gluconeogenesis and axon guidance pathway of Taenia pisiformis . There were three key enzymes in glycolysis/gluconeogenesis: hexokinase (EC 2.7.1.1), phosphofructokinase (EC 4.1.2.13), and pyruvate kinase (EC 2.7.1.40). Additionally, there were four highly conservative axon guidance molecular families, netrins (netrin 1), slits (slits 1, slits 2, and slits 3), semaphorins (sema4D, sema5, and sema6), and ephrins (ephrin E).
(DOCX)
The ESTs of Eg+Em were classified into at least 25 molecular families against COG database.
(RAR)
The ESTs of Taenia solium were classified into at least 25 molecular families against COG database.
(RAR)
The ESTs of Taenia solium obtained the GO annotations according to Nr database.
(RAR)
Annotation of M (common genes of Taenia pisiformis , Taenia solium , Echinococcus granulosis and Echinococcus multilocularis ). The genes annotations of M (T. pisiformis, T. solium, E. granulosis, and E. multilocularis) against the KEGG database.
(RAR)
21 KEGG pathways of Taenia pisiformis , Taenia solium , Echinococcus granulosis and Echinococcus multilocularis . M was aligned in the KEGG database using BLASTx, and 109 genes obtained 21 pathways.
(HTM)
The process of the SOAPdenovo assembly.
(DOCX)