Abstract
Membrane transporters can be major determinants of the pharmacokinetic, safety and efficacy profiles of drugs. This presents several key questions for drug development, including which transporters are clinically important in drug absorption and disposition, and which in vitro methods are suitable for studying drug interactions with these transporters. In addition, what criteria should trigger follow-up clinical studies, and which clinical studies should be conducted if needed. In this article, we provide the recommendations of the International Transporter Consortium on these issues, and present decision trees that are intended to help guide clinical studies on the currently recognized most important drug transporter interactions. The recommendations are generally intended to support clinical development and filing of a new drug application. Overall, it is advised that the timing of transporter investigations should be driven by efficacy, safety and clinical trial enrolment questions (for example, exclusion and inclusion criteria), as well as a need for further understanding of the absorption, distribution, metabolism and excretion properties of the drug molecule, and information required for drug labeling.
Recent progress has been made in understanding the role of membrane transporters in drug safety and efficacy1. In particular, more than 400 membrane transporters in two major superfamilies — ATP-binding cassette (ABC; for review see refs 1–5) and solute carrier (SLC; for review see refs 1,3,6,7) — have been annotated in the human genome. Many of these transporters have been cloned, characterized and localized to tissues and cellular membrane domains in the human body. In drug development, particular attention has been paid to transporters expressed in epithelia of the intestine, liver and kidney, and in the endothelium of the blood– brain barrier. As a result there is now an enormous body of literature that focuses on the interaction of drugs and their metabolites with mammalian transporters present in epithelial and endothelial barriers.
Numerous studies have suggested that transporters play a part in vivo in drug disposition, therapeutic efficacy and adverse drug reactions. The in vivo role of transporters is demonstrated in several animal species, including knockout mice8,9, and by loss-of-function genetic variants in humans4,10,11. These studies have provided considerable information on the in vivo role of many ABC and SLC transporters. Clinical pharmacokinetic drug–drug interaction (DDI) studies have suggested that transporters often work together with drug-metabolizing enzymes (DMEs) in drug absorption and elimination.
A major goal of preclinical drug evaluation is to propose clinical studies that are needed to appropriately label a drug for safe and effective use. For example, in vitro studies of drug interactions with metabolizing enzymes may lead to the design and conduct of DDI studies, or investigations in individuals with genetic polymorphisms of DMEs12,13. In fact, for drug interactions with metabolizing enzymes, the US Food and Drug Administration has developed guidances to assist drug development scientists in conducting informative in vitro and follow-up clinical studies14,15. By contrast, for drug interactions with transporters, recommendations about the appropriate conduct of in vitro and in vivo studies are not generally available, with the exception of drug interactions with multidrug resistance P-glycoprotein (P-gp; also known as MDR1, ABCB1)16. Many questions from pharmaceutical scientists involved in drug development are now being raised. In particular, which transporters are clinically important in drug absorption and disposition (distribution and elimination), and therefore could mediate DDIs? Which methods are suitable for studying in vitro drug interactions with important transporters? What criteria should be used to trigger follow-up clinical studies? What follow-up clinical studies should be conducted?
Against this backdrop, we formed the International Transporter Consortium (ITC) (BOX 1) comprising industrial, regulatory and academic scientists with expertise in drug metabolism, transport and pharmacokinetics. The ITC met by conference calls between the spring of 2007 and the summer of 2009, and held a workshop in Bethesda, Maryland, USA, in october 2008, which was co-sponsored by the Critical Path Initiative of the US Food and Drug Administration and by the Drug Information Association. The focus of the workshop was to identify which transporters, based on current knowledge, are well-established determinants of pharmacokinetics; discuss methodologies to characterize drug–transporter interactions using in vitro and in vivo studies; and propose recommendations that are important for drug development scientists in guiding preclinical and clinical studies of transporter-mediated drug interactions. For this latter point, the key consideration was that in vitro studies of drug–transporter interactions, if positive, would lead to, or inform, clinical studies that are relevant to drug safety or efficacy.
Box 1 | The International Transporter Consortium.
Kathleen M. Giacomini*: Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, 513 Parnassus Avenue, California 94143-0912, USA
Shiew-Mei Huang*: Office of Clinical Pharmacology, Office of Translational Sciences, Center for Drug Evaluation and Research, Food and Drug Administration, 10903 New Hampshire Avenue, Silver Spring, Maryland 20993-0002, USA
Donald J. Tweedie*: Boehringer Ingelheim Pharmaceuticals, 900 Ridgebury Road, PO Box 368, Ridgefield, Connecticut 06877, USA
Leslie Z. Benet: Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, 513 Parnassus Avenue, California 94143-0912, USA
Kim L. R. Brouwer: Division of Pharmacotherapy and Experimental Therapeutics, UNC Eshelman School of Pharmacy, The University of North Carolina at Chapel Hill, CB #7355, Chapel Hill, North Carolina 27599-7355, USA
Xiaoyan Chu: Drug Metabolism and Pharmacokinetics, Merck & Co., 126 East Lincoln Avenue, Rahway, New Jersey 07065-464, USA
Amber Dahlin: Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, 513 Parnassus Avenue, California 94143-0912, USA
Raymond Evers: Drug Metabolism and Pharmacokinetics, Merck & Co., 126 East Lincoln Avenue, Rahway, New Jersey 07065-464, USA
Volker Fischer: Drug Metabolism, Pharmacokinetics and Bioanalysis, Abbott, 100 Abbott Park, Illinois 60068, USA
Kathleen M. Hillgren: Drug Disposition, Lilly Research Laboratories, Lilly Corporate Center, Indianapolis, Illinois 46285, USA
Keith A. Hoffmaster: Novartis Institutes for BioMedical Research, Metabolism and Pharmacokinetics, 250 Massachusetts Avenue, Cambridge, Massachusetts, USA
Toshihisa Ishikawa: Omics Science Center, RIKEN Yokohama Institute, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan
Dietrich Keppler: German Cancer Research Center, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany
Richard B. Kim: Department of Medicine, University of Western Ontario, London, Ontario N6A 5C1, Canada
Caroline A. Lee: Pharmacokinetics, Metabolism and Dynamics, Pfizer Global Research and Development, La Jolla Laboratories, 10724 Science Center Drive, San Diego, California 92121, USA
Mikko Niemi: Department of Clinical Pharmacology, University of Helsinki and Helsinki University Central Hospital, PO Box 340, FIN-00029 HUS, Helsinki, Finland
Joseph W. Polli: Drug Metabolism and Pharmacokinetics, PO Box 13398, GlaxoSmithKline, Research Triangle Park, North Carolina 27709, USA
Yuichi Sugiyama: Department of Molecular Pharmacokinetics, Graduate School of Pharmaceutical Sciences, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan
Peter W. Swaan: Department of Pharmaceutical Science, University of Maryland, 20 Penn Street, Rm 621, Baltimore, Maryland 21201-1075, USA
Joseph A. Ware: Clinical Pharmacology, Genentech, 460 Point San Bruno Boulevard, South San Francisco, California 94080, USA
Stephen H. Wright: Department of Physiology, College of Medicine, University of Arizona, Tucson, Arizona 85724, USA
Sook Wah Yee: Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, 513 Parnassus Avenue, California 94143-0912, USA
Maciej J. Zamek-Gliszczynski: Drug Disposition, Lilly Research Laboratories, Lilly Corporate Center, Indianapolis, Illinois 46285, USA
Lei Zhang: Office of Clinical Pharmacology, Office of Translational Sciences, Center for Drug Evaluation and Research, Food and Drug Administration, 10903 New Hampshire Avenue, Silver Spring, Maryland 20993-0002, USA
The International Transporter Consortium considers this report as a work in progress, and is highly interested in obtaining feedback. Please send any comments, including areas that have not been included in this report but should be considered in the next version as well as controversial concepts, to the corresponding authors (highlighted by asterisk).
Here, we present the recommendations of the ITC regarding the conduct of transporter assays and data interpretation. This manuscript is divided into three major sections. In Section 1, the key transporters that have a role in drug disposition and response are described. We focused on transporters that have a compelling body of evidence from published studies demonstrating a role of the transporter in pharmacokinetics and DDIs. Many transporters could have been considered in this group; however, after ample discussions, and realizing that the field is dynamic and will change in the future, the ITC decided to focus on seven selected transporters for which all members agreed there is compelling evidence that they are involved in drug absorption, disposition and/or DDIs. In Section 2, current methods for studying drug–transporter interactions are presented together with comments about the limitations of each approach. In Section 3, recommendations and examples of guidelines that should be considered in drug development are presented. The ITC did not reach consensus on every issue, and in this article we have highlighted areas of disagreement or in need of further study. Transporter pharmacology is a rapidly emerging field in drug discovery and development with challenges of overlapping substrate or inhibitor specificities across the transporters, and recommendations based on strong scientific evidence will evolve as new information becomes available.
Section 1: Overview of transporters
A limited number of transporters, including several uptake transporters from the SLC superfamily6 and some ATP-dependent efflux pumps from the ABC superfamily2, have been given priority in the context of this article. Their selection was based on practical considerations and on clinical evidence that these transport proteins influence, to different degrees, drug disposition and/or side effects. FIGURE 1 and TABLES 1,2 describe the selected transporters as well as other can be explained, in part or in full, by modulation of important transporters. Note that clinical data documenting the importance of these transport proteins with respect to drug disposition and/or toxicity continues to emerge. TABLES 1,2 include clinically relevant information with respect to DDIs and genetic polymorphisms. Examples of clinically relevant DDIs that can be explained, in part or in full, by modulation of transporter activity are compiled in TABLE 3. Below, we present an overview of P-gp, breast cancer resistance protein (BCRP; also known as ABCG2); organic cation transporters (OCTs) and organic anion transporters (OATs); and organic anion transporting polypeptides (OATPs). For ease of reading we refer to MDR1/P-gp as P-gp throughout this report. Although evidence is available demonstrating that various members of the multidrug resistance protein (MRP) and the multidrug and toxin extrusion transporter (MATE) families are involved in drug disposition, as mentioned in various sections, because of the need to limit the scope of this manuscript, they are not discussed in detail here. Because of the extensive body of literature obtained over many years on the interaction of drugs with P-gp, as well as the vast experience in studying this transporter in the industry, P-gp will be described in more detail than other transporters. Many issues discussed for P-gp are relevant for other transporters. In this article capitalized letters are used for human genes and proteins (for example, SLCO and OATP), whereas only the initial letter is capitalized for rodent genes and proteins (for example, Abcg2 and Bcrp). The standard human gene nomenclature is listed in TABLES 1,2, but, in general, the transport proteins or gene products responsible for transport function are referred to by the names commonly used in the field.
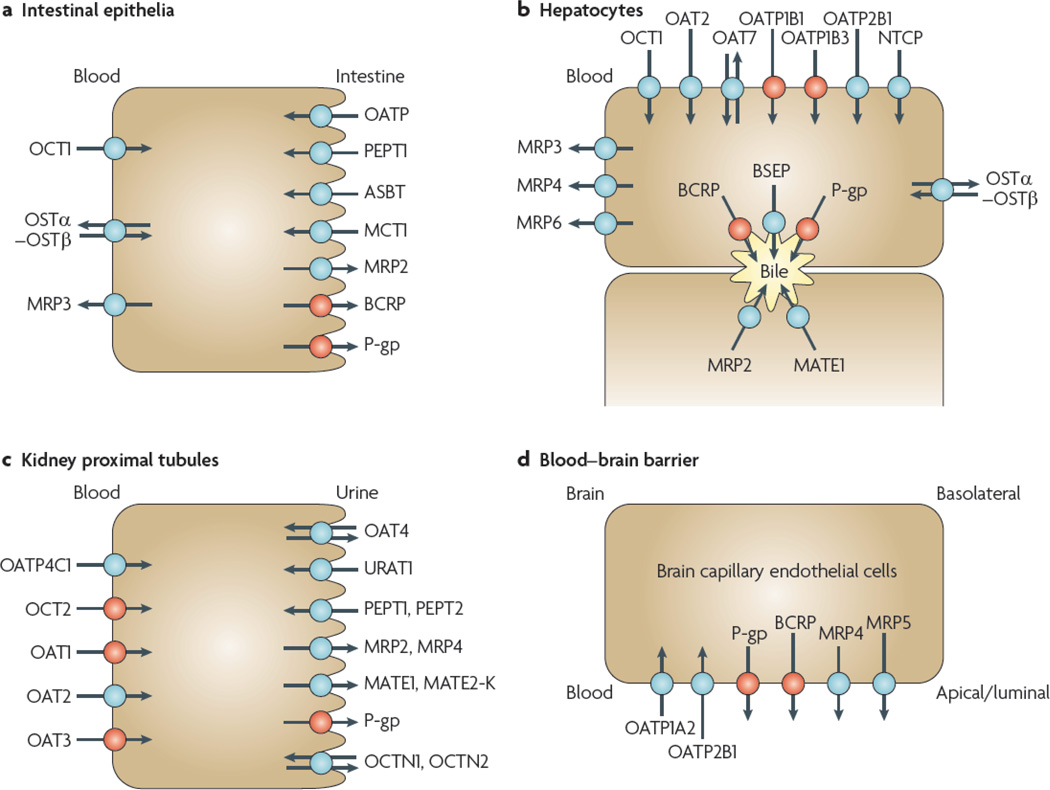
Figure 1. selected human transport proteins for drugs and endogenous substances.
Transporters in plasma membrane domains of intestinal epithelia, hepatocytes, kidney proximal tubules and brain capillary endothelial cells are presented. Those coloured in red indicate that the selected transporters are described in detail in this manuscript. Those coloured in blue indicate that the transport proteins are of importance but are not described in this manuscript. a | Intestinal epithelia contain in their apical (luminal) membrane several uptake transporters including one or more members of the organic anion transporting polypeptide (OATP) family; peptide transporter 1 (PEPT1; SLC15A1); ileal apical sodium/bile acid co-transporter (ASBT; SLC10A2); and monocarboxylic acid transporter 1 (MCT1; SLC16A1). The apical ATP-dependent efflux pumps include multidrug resistance protein 2 (MRP2; ABCC2); breast cancer resistance protein (BCRP; ABCG2); and P-glycoprotein (P-gp; MDR1, ABCB1). The basolateral membrane of intestinal epithelia contains organic cation transporter 1 (OCT1; SLC22A1); heteromeric organic solute transporter (OSTα–OSTβ); and MRP3 (ABCC3). b | Human hepatocyte uptake transporters in the basolateral (sinusoidal) membrane include the sodium/taurocholate co-transporting peptide (NTCP; SLC10A1); three members of the OATP family (OATP1B1 (SLCO1B1), OATP1B3 (SLCO1B3) and OATP2B1 (SLCO2B1)); organic anion transporter 2 (OAT2; SLC22A7) and OAT7 (SLC22A9); and OCT1. Efflux pumps in the hepatocyte basolateral membrane include MRP3, MRP4 (ABCC4) and MRP6 (ABCC6). Apical (canalicular) efflux pumps of the hepatocyte comprise P-gp; bile-salt export pump (BSEP or SPGP; ABCB11); BCRP (ABCG2); and MRP2. In addition, multidrug and toxin extrusion protein 1 (MATE1; SLC47A1) is located in the apical hepatocyte membrane. c | Kidney proximal tubules contain in the apical (luminal) membrane OAT4 (SLC22A11); urate transporter 1 (URAT1; SCL22A12); PEPT1 and PEPT2 (SLC15A2); MRP2 and MRP4; MATE1 and MATE2-K (SLC47A2); P-gp; organic cation/ergothioneine transporter (OCTN1; SLC22A4); and organic cation/carnitine transporter (OCTN2; SLC22A5). Basolateral uptake transporters in proximal tubule epithelia include OATP4C1 (SLCO4C1); OCT2; and OAT1, OAT2 and OAT3 (SLC22A8). d | Apical (luminal) transport proteins of brain capillary endothelial cells contributing to the function of the blood–brain barrier include the uptake transporters OATP1A2 and OATP2B1; and the efflux pumps P-gp, BCRP, MRP4 and MRP5 (ABCC5). Note that localization of transporters to particular membranes and tissues is sometimes controversial; therefore, the International Transporter Consortium erred on the conservative side in only showing the localization of transporters for which good evidence exists.
Table 1.
SLC transporters of emerging clinical importance in the absorption and disposition of drugs
| Transporter/ alias (Gene) |
Selected substrates | Selected inhibitors |
Organs/cells | Comments |
|---|---|---|---|---|
| OATP1B1/O ATP-C, OATP2, LST- 1 (SLCO1B1) |
Bromosulphophthalein, oestrone-3-sulphate, oestradiol-17β-glucuronide, statins*, repaglinide*, valsartan, olmesartan*, bilirubin glucuronide, bilirubin, bile acids |
Saquinavir, ritonavir*, lopinavir*, rifampicin*, cyclosporine* |
Hepatocytes (sinusoidal) |
• Has a role in disposition and excretion • Has clinically relevant polymorphisms • Has a role in clinical drug–drug interactions |
| OATP1B3/O ATP-8 (SLCO1B3) |
Bromosulphophthalein, cholecysto kinin 8, statins*, digoxin, fexofenadine, telmisartan glucuronide, telmisartan*, valsartan, olmesartan, oestradiol-17- β-glucuronide, bile acids |
Rifampicin*, cyclosporine*, ritonavir, lopinavir* |
Hepatocytes (sinusoidal) |
• Has a role in disposition and excretion |
| OAT1 (SLC22A6) |
Para-aminohippurate, adefovir, cidofovir, zidovudine*, lamivudine*, zalcitabine*, acyclovir*, tenofovir*, ciprofloxacin*, methotrexate* |
Probenecid*, novobiocin |
Kidney proximal tubule, placenta |
• Has a role in disposition and excretion Has a role in clinical drug–drug interactions |
| OAT3 (SLC22A8) |
Oestrone-3-sulphate, non- steroidal anti-inflammatory drugs, cefaclor, ceftizoxime, furosemide*, bumetanide* |
Probenecid*, novobiocin |
Kidney proximal tubule, choroid plexus, blood–brain barrier |
• Has a role in disposition and excretion • Has a role in clinical drug–drug interactions |
| OCT2 (SLC22A2) |
N-Methylpyridinium, tetraethylammonium, metformin*, pindolol, procainamide, ranitidine amantadine, amiloride, oxaliplatin, varenicline* |
Cimetidine*, pilsicainide, cetirizine*, testosterone, quinidine |
Kidney proximal tubule, neurons |
• Has a role in disposition and excretion • Has clinically relevant genetic polymorphisms • Has a role in clinical drug–drug interactions |
| OATP1A2/O ATP-A (SLCO1A2) |
Oestrone-3-sulphate, dehydroepiandrosterone sulphate, fexofenadine*, bile salts, methotrexate, bromosulphophthalein, ouabain, digoxin, levofloxacin, statins* |
Naringin, ritonavir, lopinavir, saquinavir, rifampicin* |
Brain capillaries endothelia, cholangiocyte s, distal nephron |
• Has role in disposition and excretion |
| OATP2B1/O ATP-B (SLCO2B1) |
Oestrone-3-sulphate, bromosulphophthalein, taurocholate, *statins, fexofenadine, glyburide, taurocholate |
Rifampicin, cyclosporine* |
Hepatocytes (sinusoidal), endothelia |
• Has a role in disposition and excretion • Has a role in clinical drug–drug interactions |
| OCT1 (SLC22A1) |
Tetraethylammonium, N- methylpyridinium, metformin*, oxaliplatin |
Quinine, quinidine, disopyramide |
Hepatocytes (sinusoidal), intestinal enterocytes |
• Has a role in disposition and excretion • Has clinically relevant genetic polymorphisms • Has a role in clinical drug–drug interactions |
| PEPT1 (SLC15A1) |
Glycylsarcosine, cephalexin, cefadroxil, bestatin, valacyclovir, enalapril, aminolevulinic acid, captopril, dipeptides, tripeptides |
Glycyl-proline | Intestinal enterocytes, kidney proximal tubule |
• Has a role in absorption, disposition and excretion • Has a role in clinical drug–drug interactions |
| PEPT2 (SLC15A2) |
Glycylsarcosine, cephalexin, cefadroxil, bestatin, valacyclovir, enalapril, aminolevulinic acid, captopril, dipeptides, tripeptides |
Zofenopril, fosinopril |
Kidney proximal tubule, choroid plexus, lung |
• Has a role in excretion MATE1 |
| (SLC47A1) | Metformin, N- methylpyridinium, tetraethylammonium |
Quinidine, cimetidine, procainamide |
Kidney proximal tubule, liver (canalicular membrane), skeletal muscle |
• Has a role in disposition and excretion • Has a role in clinical drug–drug interactions |
| MATE2-K (SLC47A2) |
Metformin, N- methylpyridinium, tetraethylammonium |
Cimetidine, quinidine, pramipexole |
Kidney proximal tubule |
• Has a role in disposition and excretion |
Can potentially be used for in vivo (clinical) studies.
Table 2.
ABC transporters of emerging clinical importance in the absorption and disposition of drugs
| Transporter/alias (Gene) |
Selected substrates | Selected inhibitors |
Organs/cells | Comments |
|---|---|---|---|---|
| MDR1/P-gp, ABCB1 (ABCB1) | Digoxin*, loperamide*, berberine, irinotecan, doxorubicin, vinblastine, paclitaxel, fexofenadine | Cyclosporine*, quinidine*, tariquidar, verapamil | Intestinal enterocytes, kidney proximal tubule, hepatocytes (canalicular), brain endothelia |
|
| BCRP/MXR (ABCG2) | Mitoxantrone, methotrexate, topotecan, imatinib, irinotecan, statins*, sulphate conjugates, porphyrins | Oestrone-17β-oestradiol, fumitre - morgin C | Intestinal enterocytes, hepatocytes (canalicular), kidney proximal tubule, brain endothelia, placenta, stem cells, mammary glands (lactating) |
|
| BSEP/SPGP, cBAT, ABCB11 (ABCB11) | Taurocholic acid, pravastatin, bile acids | Cyclosporin A, rifampicin, glibenclamide | Hepatocytes (canalicular) |
|
| MRP2/ABCC2, cMOAT (ABCC2) | Glutathione and glucuronide conjugates, methotrexate, etoposide, mitoxantrone, valsartan, olmesartan, glucuronidated SN-38 | Cyclosporine, delaviridine, efavirenz, emtricitabine | Hepatocytes (canalicular), kidney (proximal tubule, luminal), enterocytes (luminal) |
|
| MRP3/ABCC3 (ABCC3) | Oestradiol-17β-glucuronide, methotrexate, fexofenadine, glucuronate conjugates | Delaviridine, efavirenz, emtricitabine | Hepatocytes (sinusoidal), intestinal enterocytes (basolateral) |
|
| MRP4/ABCC4 (ABCC4) | Adefovir, tenofovir, cyclic AMP, dehydroepiandrostero ne sulphate, methotrexate, topotecan, furosemide, cyclic GMP, bile acids plus glutathione | Celecoxib, diclofenac | Kidney proximal tubule (luminal), choroid plexus, hepatocytes (sinusoidal), platelets |
|
| MDR3/ABCB4 (ABCB4) | Phosphatidylcholine, paclitaxel, digoxin, vinblastine | Verapamil, cyclosporine | Hepatocytes (canalicular) |
|
ABC, ATP-binding cassette.
Can potentially be used for in vivo (clinical) studies.
Table 3.
Selected transporter-mediated clinical drug–drug interactions
| Implicated transporter* | Interacting drug | Affected drug | Clinical pharmacokinetic impact on affected drug‡ |
|---|---|---|---|
| Organic anion transporting polypeptides | Cyclosporine | Pravastatin | AUC ↑890% and Cmax ↑678%102,204 |
| Cyclosporine | Rosuvastatin | AUC ↑610%205 | |
| Cyclosporine | Pitavastatin | AUC ↑ 360% and Cmax ↑560%206 | |
| Rifampicin (single dose) | Glyburide | AUC ↑125%207 | |
| Rifampicin (single dose) | Bosentan | Ctrough ↑ 500%208 | |
| Lopinavir/ritonavir | Bosentan | Day 4: Ctrough ↑ 4,700%208; day 10: Ctrough ↑ 400%208 | |
| Lopinavir/ritonavir | Rosuvastatin | AUC ↑107% and Cmax ↑365%209 | |
| Organic anion transporters | Probenecid | Cidofovir | CLr ↓32%210,211 |
| Probenecid | Furosemide | CLr ↓66%210 | |
| Probenecid | Acyclovir | CLr ↓32% and AUC ↑40%210,212 | |
| Organic cation transporters | Cimetidine | Metformin | AUC ↑50% and CLr ↓ 27%213,214 |
| Cimetidine | Pindolol | CLr ↓~34%215 | |
| Cimetidine | Varenicline | AUC ↑29%216 | |
| Cimetidine | Pilsicainide | AUC ↑33%, CLr ↓28%217 | |
| Cimetidine | Pilsicainide | CLr ↓41%218 | |
| Cimetidine | Dofetilide | CLr ↓33%219 | |
| P-glycoprotein | Quinidine | Digoxin | CLr ↓34–48%220,221 |
| Ritonavir | Digoxin | AUC ↑86%222 | |
| Dronedarone | Digoxin | AUC ↑157% and Cmax ↑75%223 | |
| Ranolazine | Digoxin | AUC ↑60% and Cmax ↑46%224 | |
| Breast cancer resistance protein | GF120918 | Topotecan | AUC ↑143%225 |
Implicated transporter refers to the likely transporter; however, because the studies are carried out in vivo it is not possible to assign specific transporters to the drug–drug interaction.
Percent change refers to the difference between the area under the curve (AUC), or Cmax, in the presence and the absence of the inhibitor (interacting drug) normalized to the AUC in the absence of the inhibitor. For clearance values (CLr), the values are normalized for the absence of the inhibitor. Ctrough is the minimum drug concentration observed after administration of a dose of the drug and the concentration prior to the administration of a subsequent dose.
P-glycoprotein
General description
P-gp mediates the ATP-dependent export of drugs from cells. It is expressed in the luminal membrane of the small intestine and blood–brain barrier, and in the apical membranes of excretory cells such as hepatocytes and kidney proximal tubule epithelia. P-gp has an important role in limiting entry of various drugs into the central nervous system. In addition, it also plays a part in the intestinal absorption and in the biliary and urinary excretion of drugs. The level of expression and functionality of P-gp can be modulated by inhibition and induction, which can affect the pharmacokinetics, efficacy, safety or tissue levels of P-gp substrates5,17–21.
Substrate and inhibitor selectivity
Initially discovered as a result of its interaction with multiple anticancer drugs, P-gp is responsible for the efflux across biological membranes of a broad range of therapeutic drugs. Recently, a high-resolution structure of the mouse P-gp has been described, which revealed distinct binding sites for drugs22. A select number of substrates and inhibitors of human P-gp are shown in TABLE 2. P-gp substrates are generally hydrophobic molecules, of which many are cationic. Multiple binding sites for substrates and inhibitors on P-gp have been identified using site-directed mutagenesis23–25.
Methodology for evaluating function
Cell lines that express P-gp and inside-out membrane vesicles prepared from these cell lines can be used to determine whether a drug is a P-gp substrate or inhibitor. As P-gp is localized in the apical plasma membrane in polarized cell monolayers, a high efflux ratio of basal-to-apical to apical-to-basal (the so called B-A/A-B ratio) indicates a potentially significant role for P-gp in transporting drugs across cell monolayers. A high efflux ratio in vitro has been shown to correlate well with studies that have demonstrated a role for P-gp in drug penetration to the central nervous system in mice26,27. By contrast, a high efflux ratio does not always translate into poor oral absorption. The involvement of P-gp in absorption of a drug is more pronounced if the drug has a poor apparent permeability coefficient (Papp), or in cases in which there is interplay between metabolism and efflux.
Mice deficient in Mdr1a or Mdr1a/b are widely used as powerful tools for assessing the role of P-gp in vivo9. Knockout mice can be used as a reference for complete inhibition of P-gp, and represent the ‘worst case’ scenario. Sasongko et al.28 demonstrated that the AUCbrain/ AUCblood ratio of 11C-verapamil in humans increased by less than twofold (88 ± 20%) in the presence of a high intravenous dose of the P-gp inhibitor cyclosporine. By contrast, an almost eightfold (770%) increase was observed in the Mdr1a/b knockout mice compared with control mice. These data can be interpreted in multiple ways, but illustrate the challenge in predicting human transporter-mediated DDIs based on preclinical animal data.
Clinical significance
The clinical significance of a P-gp inhibitor can be investigated in humans by assessing the P-gp-mediated clearance or exposure of a probe substrate in the presence of the inhibitor. Digoxin is transported by P-gp in vitro and should be considered in DDI studies with new molecular entities (NMEs) that are inhibitors of P-gp (TABLE 3, BOX 2). Current clinical data indicate that there are no consistent examples in which inhibition of P-gp in the blood–brain barrier resulted in adverse effects29–31. It is therefore difficult to extrapolate the data obtained for P-gp inhibitors in knockout mice to humans to indicate the potential for a clinically significant DDI at the human blood–brain barrier31.
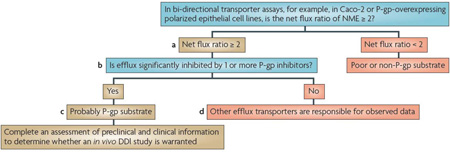
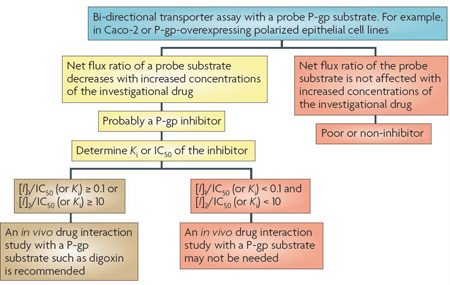
Box 2 | Decision trees for P-lycoprotein or BCRP substrate interactions.
Although the current US Food and Drug Administration draft Drug Interaction Guidance provides a decision tree specifically for P-glycoprotein (P-gp; also known as MDR1, ABCB1)14,15,183, we broadened the decision tree here and in BOX 3 to include both P-gp and breast cancer resistance protein (BCRP; also known as ABCG2). This is because similar in vitro methodologies and criteria are used to suggest clinical drug–drug interaction (DDI) studies for these two transporters. Generally, most P-gp substrates are organic cations or neutral molecules, relatively hydrophobic, and have a range of molecular masses (200 daltons to greater than 1,000 daltons). BCRP substrates tend to overlap with P-gp but also include acids or drug conjugates that are not good substrates for P-gp. A new molecular entity (NME) is considered to be a potential P-gp or BCRP substrate if the efflux ratio — basal to apical (B-A) to apical to basal (A–B) — is ≥ 2 in an epithelial cell system that expresses one or both transporters (see footnote and (a) in the figure). A net flux ratio cut-off higher than 2 or a relative ratio to positive controls may be used to avoid false positives if a ratio of 2 is deemed non-discriminative as supported by prior experience with the cell system used. Additional corroboration that an NME may be a P-gp or BCRP substrate can be achieved with the use of inhibitors; information especially valuable if non-transfected cells are not included as controls for endogenous transport activity. Reduction of the flux ratio by the P-gp (or BCRP) inhibitors should be greater than 50% (see (b) in the figure). If the flux ratio is not reduced by P-gp (or BCRP) inhibitors, then other efflux transporters may be responsible for the observed net flux (see (d) in the figure).
The conclusions from in vitro efflux studies are dependent on the cell line used. For example, for cells heterologously expressing P-gp or BCRP (for example, MDCK or LLC-PK1), in which expression of the transporters can be high, the efflux ratio would be expected to be much higher than 2 for strong substrates. A classification of relevant substrates versus non-substrates will depend on the assay system. Validation studies of in vitro assay systems using known weak and strong substrates and, where possible, correlations with in vivo model systems, should be established. Meaningful efflux ratios are dependent on the transporter expression; thus, results should be related to a reference compound. In cell lines such as Caco-2, which express multiple uptake and efflux transporters, reduction of efflux in the presence of an inhibitor may support efflux by P-gp and/or BCRP, depending on the selectivity of the inhibitor used (for example, cyclosporine inhibits multiple transporters).
If in vitro experiments suggest that the NME is a P-gp and/or BCRP substrate (see (c) in the figure), preclinical and clinical information should be assessed to determine whether a clinical in vivo DDI study is warranted. In particular, the relative contribution of the transporter-mediated pathway to the overall clearance of the drug is the primary determinant of whether an inhibitor will have a major effect on the disposition of the NME. For example, the pharmacokinetics of an NME that has high solubility, high permeability and/or is highly metabolized are less likely to be affected by a co-administered drug that is a P-gp inhibitor15,173,174. Therefore, an in vivo interaction study may not be needed. By contrast, an NME that has poor solubility, limited permeability and is metabolically stable (eliminated primarily as the parent compound) is more likely to demonstrate a pharmacokinetic change in the presence of an inhibitor. The possible clinical consequence of such an interaction needs to be assessed based on careful evaluation of available exposure–response data186,187. The recommended clinical study with a known P-gp inhibitor should consider the therapeutic use of the NME; however, cyclosporine is a reasonable choice, as it is known to inhibit P-gp in vivo. As noted previously, because cyclosporine inhibits multiple transporters, care must be taken in the design and interpretation of DDI studies in which it is used as an inhibitor.
This figure shows a decision tree for P-gp and a similar tree could be used for BCRP. Although flux systems have traditionally been used to determine whether an NME is a substrate of P-gp or BCRP, inside-out vesicles expressing P-gp or BCRP, or transfected cell monolayers grown on solid support with appropriate controls (for example, inhibitors and positive controls) as described in Section 2, also can be used.
Findings from many studies on the effect of ABCB1 polymorphisms20 on P-gp substrates have not been consistently reproduced; therefore, routine application of ABCB1 polymorphism analysis to clinical studies is not warranted at this time. Studies with larger numbers of samples may be needed to clarify the role of ABCB1 polymorphisms in pharmacokinetics and pharmacodynamics32.
BCRP
General description
BCRP is a ‘half ABC transporter’ consisting of 655 amino acids and six transmembrane domains33. BCRP was identified originally as a determinant of multidrug resistance in cancer cell lines in vitro34,35. BCRP is expressed in the gastrointestinal tract, liver, kidney, brain endothelium, mammary tissue, testis and placenta. It has a role in limiting oral bioavailability and transport across the blood–brain barrier, blood– testis barrier and the maternal–fetal barrier of some selected substrates36,37. The physiological functions of BCRP include the extrusion of porphyrins from haematopoietic cells and hepatocytes, as well as the secretion of vitamin B2 (riboflavin) and possibly other vitamins (such as biotin and vitamin K) into breast milk36.
Substrate and inhibitor selectivity
BCRP actively extrudes a broad range of endogenous and exogenous substrates across biological membranes37. TABLE 2 lists selected substrates and inhibitors of human BCRP. High-speed screening and quantitative structure–activity relationship (QSAR) analysis methods suggest that one amine bonded to one carbon of a heterocyclic ring is an important component for drug interactions with BCRP. In addition, fused heterocyclic ring(s) and two substituents on a carbocyclic ring of the fused heterocyclic ring(s) are also important chemical moieties for the interaction with BCRP38–40. Many protein kinase inhibitors such as imatinib (Gleevec; Novartis) carry such structural components. Substrates of BCRP include pitavastatin (Livalo; Kowa Pharmaceuticals America)41 and phytoestrogens, such as genistein, daidzein and coumestrol42.
Methodology for evaluating function
Polarized and non-polarized cell lines expressing BCRP are used to assay BCRP-mediated transport and inhibition43. In addition, membrane vesicles containing BCRP that constitutively express the transporter can also be used43. Prazosin and cimetidine can be used as a positive control in cell lines, whereas several relatively polar, hydrophilic substrates (for example, methotrexate, oestrone 3-sulphate and sulphasalazine) can be used as controls in vesicular transport assays. Since the initial publications of the Bcrp−/−knockout mouse36,37, drug disposition studies in Abcg2−/− knockout mice have delineated an important role for intestinal Bcrp as a rate-determining barrier for the oral bioavailability of several drugs. These include topotecan (hycamtin; GlaxoSmithKline)44 and sulphasalazine (Salazopyrin; Pharmacia), which had 10-fold to 110-fold increase in relative AUC45. BCRP is also a moderate determinant of the bioavailability of nitrofurantoin46, some fluoroquinolones47 and imatinib48.
Clinical significance
Recent clinical studies have demonstrated that subjects with reduced BCRP expression levels, correlating with the Q141K variant, are at increased risk for gefitinib (Iressa; AstraZeneca)-induced diarrhoea and altered pharmacokinetics of 9-aminocamptothecin, diflomotecan, irinotecan (Camptosar; Pfizer), rosuvastatin (Crestor; AstraZeneca), sulphasalazine and topotecan49–55. Inter-individual differences in BCRP function probably contribute to variable bioavailability, exposure (AUC and Cmax), and pharmacological response of drugs that are BCRP substrates. The most significant clinical effects are likely to be for drugs that have a low bioavailability and have a narrow therapeutic index.
OCTs and OATs
General description
A distinct family of proteins within the SLC superfamily is encoded by 22 genes of the human SLC22A family, and includes the electrogenic OCTs (isoforms 1–3) and the oATs (significant isoforms in humans include oAT1–4 and 7, and URAT1)6,7,56,57. The genes encoding OCT1–3 and OATs encode proteins that are 542–556 amino acids long with 12 predicted transmembrane-spanning domains57. The tissue distribution and localization of OCTs and OATs are summarized in TABLE 1 and FIG. 1. There are several published reviews of the molecular characteristics, expression and function of OCTs7,57–60 and OATs57,61–63.
Substrate and inhibitor selectivity
TABLe 1 summarizes various compounds that interact with human OCTs and OATs. OCTs transport relatively hydrophilic, low molecular mass organic cations. Properties of inhibitors of OCT1 and OCT2 have been identified and include a net positive charge and high lipophilicity59,64. OAT1, OAT3 and OAT4 support exchange of intracellular 2-oxoglutarate for extracellular substrate62. OAT1 and OAT3 mediate the basolateral entry step in renal secretion of many organic anions, and have distinct selectivities for different structural classes of type I organic anions. That is, monovalent (or selected divalent) anions that are less than 500 daltons58; although OAT3 can also transport some positively charged drugs such as cimetidine.
Methodology for evaluating function
In vitro assays that are commonly used to characterize OCT function include Xenopus laevis oocytes, membrane vesicles and cell lines derived from various tissues including proximal tubule (Caki-1), placenta (BeWo) and colon carcinoma (Caco-2) cells65–69. These are in addition to various human embryonic kidney (HEK293) cell lines expressing recombinant OCTs68,73,83. Functional assessment of OATs generally uses heterologous expression in cultured cells or X. laevis oocytes, and such systems have been used to develop structure–activity relationships for the interaction of OATs70,71. Studies performed in Oct−/− knockout mice have demonstrated the pharmacokinetic, pharmacological and physiological relevance of OCTs in organic cation disposition and activity8,72–74. Studies using Oat1−/− and Oat3−/− knockout mice have also allowed an initial assessment of the influence of each transporter on the renal handling of selected compounds, and have led to the identification of endogenous OAT substrates75,76.
Clinical significance
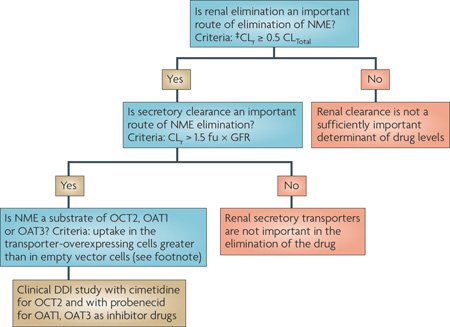
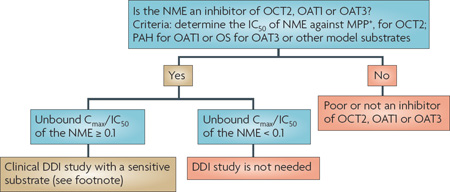
Recent studies suggest that genetic variation in OCT1 may be a significant determinant of inter-individual variability in the disposition and response to cationic drug substrates, particularly metformin73,77. However, such studies have been controversial78. Recent studies suggest that the activity of OCT1 is positively associated with the degree to which patients with chronic myeloid leukaemia respond to imatinib79,80. In Chinese and Korean populations, a common OCT2 variant (A270S) has been associated with a significant reduction in renal clearance of metformin81,82, but the effect of this variant may differ depending on the ethnic group83. By contrast, genetic variants in the OATs have not been associated with changes in drug disposition. DDIs with the OATs and the OCTs can occur, and may result in reduced renal clearance (TABLE 3). Creatinine, which is often used to assess filtration clearance, undergoes renal tubular secretion, which could be inhibited by inhibitors of OCT2. Therefore, other methods should be used to assess filtration clearance of NMEs in DDI studies involving OCT2 inhibitors. DDIs involving the OATs and the OCTs are discussed in more detail in Section 3.
OATPs
General description
The OATPs (SLCOs) represent a superfamily of important membrane transport proteins that mediate the sodium-independent transport of a diverse range of amphiphilic organic compounds. These include bile acids, steroid conjugates, thyroid hormones, anionic peptides, numerous drugs and other xenobiotic substances84 (TABLE 1). The general predicted OATP structure consists of proteins with 12 transmembrane domains85. The mechanism of transport appears to consist of anion exchange by coupling the cellular uptake of substrate with the efflux of endogenous intracellular substances such as bicarbonate in a process that seems to be electroneutral85–87. OATP1B1 was cloned by several groups88–91 and its localization along with other OATPs is shown in FIG. 1. Interestingly, OATP family members are poorly conserved evolutionarily and orthologues for human oATPs may not exist in rodents.
Substrate and inhibitor selectivity
OATP1B1 transports a broad range of compounds such as bile acids; sulphate and glucuronate conjugates; thyroid hormones; peptides; and drugs such as methotrexate and HMG-CoA reductase inhibitors (TABLE 1). Similar to OATP1B1, OATP1B3 also transports bile acids; monoglucuronosyl bilirubin; bromosulphophthalein; steroid conjugates; peptide deltorphin II92; the hepatotoxic cyclic peptide the thyroid hormones T3 and T4; leukotriene C4; and amanitin; and the cardiac glycosides digoxin and ouadrugs such as methotrexate and rifampicin86–91. However, bain92. It is also involved in the uptake of the angiotensin OATP1B3 also exhibits unique transport properties in II receptor antagonist telmisartan and its glucuronide that it is able to mediate the cellular uptake of the opioid conjugate91, and the selective uptake of the intestinal peptide cholecystokinin 8 (refs 93,94). TABLE 1 lists additional details on the substrate specificity of OATP1B3, and the specificities of OATP1A2 and OATP2B1 are also noted.
Methodology for evaluating function
In vitro assessment of OATP transporter function has relied on a number of transient and stable heterologous expression systems84. These include X. laevis oocytes, recombinant virus, or stable cell lines expressing an individual or multiple transporters95. In addition, stable expression of OATP transporters in polarized cells such as MDCK-II has been reported, typically in combination with efflux transporters such as MRP2 (refs 96,97). Isolated hepatocytes with OATP inhibitors can also be used to study OATP transport98,99. More recently, in vivo drug disposition profiles from Oatp1b2−/− knockout mouse models have been reported100,101 and may reflect, in part, the activity of both human OATP1B1 and OATP1B3. Additional studies are needed to more fully validate the utility of rodent models to predict human OATP-mediated drug disposition.
Clinical significance
The clinical relevance and DDIs related to OATPs (TABLe 3) have been noted only for certain OATPs; for example, OATP1B1 and OATP1B3 expressed primarily in the liver. DDIs involving the OATPs have focused primarily on OATP1B1. Inhibition of OATP1B1-mediated hepatic uptake appears to contribute to the significant increase in statin (for example, rosuvastatin) concentrations in blood after cyclosporine administration102,103 (TABLE 3). Because cyclosporine is an inhibitor of multiple transporters, the specific transporter involved in the clinical DDI cannot be ascertained; although OATP1B1 is a likely candidate. In addition, a series of functional polymorphisms of OATP1B1 have been characterized88. The frequencies of the SLCO1B1 388G allele in Caucasians, African Americans, and Asians is approximately 40%, 75% and 60%, respectively88,104–106. Another common single nucleotide polymorphism is 521T>C in codon 174, which has frequencies of approximately 15%, 2% and 15% in Caucasians, African Americans, and Asians, respectively88,104–106. The c.388G allele often occurs in a haplotype with the c.521C allele. These haplotypes — c.388G-c521T (*1B), c.388A-c.521C (*5) and c.388G-c.521C (*15) — have different activities and also occur at different frequencies in various ethnic groups. In addition, the promoter variants (−11187G>A and −10499A>C) may be in linkage disequilibrium with common coding region single nucleotide polymorphisms104.
Pharmacokinetic studies indicate that individuals with the SLCO1B1*5 or *15 haplotypes have increased exposure to statin drugs such as pravastatin106, pitavastatin107, simvastatin acid108, atorvastatin109 and rosuvastatin110. They also have increased exposure to other drugs such as repaglinide111, atrasentan112, irinotecan113 and ezetimibe (Zetia; Merck/Schering–Plough)114. The *5 haplotype is rare (with a frequency of 2% in Caucasians and is absent in individuals of other ancestries), whereas the *15 haplotype is more common (16% in Caucasians, 2% in sub-Saharan Africans and 9–12% in Asians)104. However, because of the large number of patients on statins, even less common variants may have an effect in many individuals. Compelling clinical evidence supporting an important role for SLCO1B1 polymorphisms has come from a genome-wide association study of simvastatin-induced myopathy115.
Section 2: Methods for studying transporters
The sections below provide, in order of increasing complexity, assay systems that are currently available for measuring transporter activity. Although application of these systems is less labor intensive when radiolabelled or fluorescent substrates are available, the use of sensitive liquid chromatography–mass spectrometry methodology has provided alternative high-throughput detection methods116.
Membrane-based assay systems
ATPase assay
Substrate-dependent ATP hydrolysis has been used to evaluate the interactions of substrates and inhibitors with some ABC transporters by colorimetric analysis of inorganic phosphate release during the transport process. The simplicity of this assay makes it a practical technique, which can be used in high-throughput assays to screen for compounds that interact with some ABC transporters117. Drawbacks of this technique include inconsistency between ATPase activity and the transport rate of some substrates and inhibitors; a high incidence of false positives and negatives; and the requirement of high substrate concentrations.
Membrane vesicle transport assay
Inverted plasma membrane vesicles have been used primarily to study efflux transporter activity, in particular for ABC transporters118. A wide variety of cell lines have been used to prepare membrane vesicles, such as drug-selected cells, transfected cells and baculovirus-infected insect cells. Assay mixtures should contain an ATP-regenerating system to maintain constant ATP concentrations during the assay period; a blank in which ATP is replaced by 5′-AMP is recommended. A major advantage of this methodology is that drugs are directly applied to the cytoplasmic compartment and influx, rather than efflux, is measured. This enables detailed kinetic and QSAR analyses for substrate or inhibitor interaction with the target transporter40. Determining the uptake or efflux of the hydrophobic compounds may be problematic because of the high degree of background binding to cell membranes in vesicle or cellular systems. Vesicle transport assays with hydrophobic compounds may require different size-exclusion techniques116–118.
Cell-based assay systems
Cell-based assay systems can be used in drug discovery to identify substrates and inhibitors for individual transporters and for developing QSAR models. In addition, these systems can be used for mechanistic studies to assess transport mechanisms, the rate-limiting step in trans-epithelial transport, and transporter-based DDIs.
Data from multiple experimental systems are often useful in confirming the involvement of a transporter with a potential substrate. Various cell-based assay systems are described below.
Polarized cell lines without recombinant transporters
Vectorial transport systems in which flux can be measured in two directions (namely, apical-to-basolateral and vice versa) are standard methods for evaluating drug transport across the small intestine and the blood–brain barrier. However these systems have some limitations. For example, Caco-2 cells can vary in their transporter expression profiles compared with the small intestine; although some data support strong correlation with human jejunum119. Although endogenous expression levels of transporters may be low in MDCK and LLC-PK1 cell lines, these have similar limitations. Immortalized human kidney cells (for example, Caki, IHKE-1 and NRK52E) can be used to understand regulation of key drug transport processes; although limitations include their inability to replicate transport processes present in the intact kidney120. Recently, chemically synthesized RNA interference molecules have been used to silence expression of various genes in mammalian cell lines. By selectively knocking down endogenous transporters, the new cell lines could be used to study the role of a particular transporter in the transport of drugs121–123.
Single- and double-transfected cell lines
Recombinant transporters that are stably or transiently expressed in various cell lines can be used to characterize drug transporter interactions. Cell-based assay systems for quantitative drug transport studies include cell lines expressing uptake or efflux transporters, or both123,124. Cultured cell lines used to study drug transporter interactions include cell monolayers such as MDCK, LLC-PK1, hEK293 or Cho cells grown on solid support. Such monolayers can be used to assess uptake and efflux by single recombinant transporters, or by multiple recombinant transporters, and kinetic measurements can be obtained. Various ABC transporters have been analysed in monolayers of polarized cells, such as MDR1-transfected MDCK II cells, by measuring B-A/A-B flux ratios of compounds. Apart from single-transfected cell lines, polarized cells that stably express multiple transporters — for example, a recombinant uptake transporter in their basolateral membrane and an efflux transporter in their apical membrane — have been developed97,124–127. These cell lines may overcome the limitation of certain single-transfected cells that often lack the endogenous uptake or efflux transporters to provide a complete mechanism for trans-cellular transport of a molecular entity. It is noted that compounds may undergo uptake or efflux by multiple transporters in intact organs, and therefore even double-transfected cell lines may not predict the true in vivo situation128.
Primary cells
Primary-cell-based assays are derived from intact tissue, which, at the time of isolation, express the full complement of drug transporters present in a given tissue or cell. Thus, human primary cells may be used to study drug disposition and clinically relevant drug interactions. However, these cells can adapt to culture conditions over time, requiring strict definition of culture methods to ensure proper polarization, expression and localization of drug transporters. For example, brain microvessel endothelial cells require monitoring of tight junction integrity, and renal proximal tubule cell usage is limited by tubule collapse in culture. In addition, investigation of intestinal drug transport in isolated intestinal cells or shed enterocytes is limited by their inability to form well-defined monolayers in culture.
Sandwich-cultured primary hepatocytes
Suspended primary hepatocytes have been shown to reproduce hepatic uptake function129. Unfortunately, the in vivo polarity of hepatocytes is rapidly lost upon isolation, leading to the inability to assess the potential for canalicular efflux. This polarity can be regenerated when hepatocytes are cultured in a sandwich configuration between two layers of gelled collagen130,131. The drug-metabolizing capabilities and the regulatory machinery of hepatocytes are retained, as well as expression, localization and function of various hepatic transport proteins in the sinusoidal and canalicular membranes132. Utilizing sandwich-cultured rat and human hepatocytes, good correlations have been demonstrated between in vitro and in vivo biliary clearance for some compounds133. This system can provide an estimate of intracellular drug and/or metabolite concentrations. A caveat of the sandwich-cultured hepatocyte model is that it is not as simple to establish as primary cells, and the nature of the culture system does not allow for considerations of other factors that would influence uptake and efflux of compounds (for example, loss of sinusoidal flow or lack of exteriorized bile flow).
Considerations for use of in vitro systems in evaluating transporter interactions
In vitro technologies for determining the interaction of drugs with transporter proteins are described here and in the literature (see sections above). These techniques and tools continually evolve, and so prescriptive approaches in an article such as this are at risk of being out of date in the near future. The ideal experimental approach considers many factors such as stage of development (discovery versus clinical phases); physicochemical properties of the NME; and the required outputs from an investigation (for example, efflux ratio, kinetic parameters Km and Vmax, and uptake clearance). Whatever the selected experimental approach, it is essential that the system be well characterized with known substrates or inhibitors, and that appropriate controls are incorporated to verify test results. Due consideration should be given to the choice of concentrations; time points (for example, within linear ranges, giving appropriate signal-to-noise ratio); calculation methods appropriate to the test system; and justification that the probe substrates or inhibitors allow tangible extrapolation to the clinical situation.
Intact organ/in vivo models
Numerous transporter-gene knockout and naturally-occurring transporter-deficient animal models have been characterized in recent years, and their commercial availability has been increasing steadily134. Knockout models have illustrated the role of transporters in physiology, protection of major blood–tissue barriers, and the absorption and excretion of xenobiotics and endogenous compounds. For instance, although the localization of P-gp in the blood–brain barrier has been known, its profound role in limiting brain exposure of some xenobiotics became apparent only after demonstrating that Mdr1a−/− mice displayed a 100-fold higher sensitivity towards ivermectin135. And although a key role for BCRP in protecting the blood–brain barrier remains to be demonstrated, recent evidence obtained in Bcrp−/−/Mdr1a−/−/Mdr1b−/− triple-knockout mice treated with lapatinib (Tykerb/Tyverb; GlaxoSmithKline) suggested that Bcrp and P-gp may have a combined effect at the blood–brain barrier136.
Numerous transporters have been implicated in the clearance of various drugs and metabolites. The importance of a transporter-based elimination pathway is determined by the fraction of total clearance mediated by the transporter. Appreciable alterations (greater than twofold) in exposure occur only when a major elimination transport pathway is inactivated or extensively inhibited (that is, unbound inhibitor concentration [I]/Ki ≫ 1, in which [I]/Ki is the concentration of inhibitor relative to its inhibition constant)137. Transporter knockout models are useful in establishing the extent of the contribution of the ablated pathway to overall clearance in that species.
Species, strain, sex, diet and housing condition differences, as well as compensatory mechanisms, are a limitation of knockout and mutant models. All these variables should be considered carefully in the interpretation of data, and in attempts to extrapolate findings across species. For example, hepatic Mrp2 expression in rats is approximately tenfold higher than in humans138, and species differences in substrate specificities have been observed139. Therefore, the relevance of biliary excretion via Mrp2 in rats compared to other species should be confirmed further with, for example, knockout mice and in vitro systems138,140,141. Gender differences in the expression of certain transporters may exist; higher hepatic BCRP expression in males has been established in both mice and humans142. Strain differences have also been reported. For example, hepatic Mrp4 is upregulated only in certain Mrp2−/− mouse strains143,144. Direct translation of preclinical transport findings to the clinic is challenging because of species differences in transporter expression, substrate affinity, physiological function and interplay between transporters and enzymes. humanized transporter animal models145 and stem cells are promising tools that may prove to be useful in the near future to study the absorption, distribution, metabolism and excretion (ADME) properties of compounds.
Contribution of transporters in vivo
Estimating the contribution of transporters to total tissue uptake and excretion is necessary for understanding their importance in drug disposition. This estimate allows the prediction of the extent to which inhibition of, or a genetic polymorphism in, a particular transporter, will affect drug concentrations in plasma and tissues. Unfortunately, unlike cytochrome P450 (CYP) enzymes, selective inhibitors or antibodies for most drug transporters have not been identified; although some progress has been made. For instance, potent inhibitors for BCRP and P-gp have been described2. The relative contribution of OATP1B1 and OATP1B3 can be estimated in human hepatocytes by measuring the proportion of uptake of test compounds that is inhibited by oestrone-3-sulphate, which is an OATP1B1 inhibitor146. A method using reference compounds (substrates) for specific liver uptake transporters also has been proposed (for example, cholecystokinin 8 for OATP1B3)147,148. The reference compound should be a specific substrate for a particular transporter. The contribution of a specific transporter to the uptake of the test compound in human hepatocytes can be calculated by taking the ratio of the uptake of the reference compound in human hepatocytes to the uptake in cells transfected with the specific transporter (relative activity factor) and multiplying that ratio by the uptake of the test compound in the transporter-expressing cells. Unfortunately, for most transporters, reference compounds are not yet available. Instead of relative activity factor values, the ratio of the expression level of a specific transporter in human hepatocytes to that in transporter-expressing cells estimated by the band density of Western blot analysis of cell-surface protein can also be used148.
The relative importance of uptake and efflux transporters in the pharmacokinetics of drugs in vivo in humans can be quantitatively estimated by using physiologically-based pharmacokinetic (PBPK) modeling. For example, the effects of changes in OATP1B1 and MRP2 activity on systemic and hepatic exposure of pravastatin were simulated using a PBPK model incorporating blood, liver (the clearance and pharmacological target organ), and peripheral organs149. Application of this modeling approach to other drugs has demonstrated its broader validity150,151.
Imaging techniques can qualitatively and quantitatively elucidate the role of transport proteins in drug disposition (for example, whole body autoradiography and positron emission tomography (PET)). PET is a noninvasive imaging method that is useful for accurately measuring pharmacodynamic end points in humans and preclinical species. Examples of end points include the level of dopamine D2 receptor occupancy of antipsychotics in the brain152, or the amount of tracer accumulation (for example,11C,13N or18F) in an organ over time to quantify transporter activity. [11C]-N-Acetyl-leukotriene E4 has been used to study hepatobiliary elimination in normal animals, including monkeys, and in Mrp2-deficient mutant rats153. [11C]-verapamil was used to measure the effect of inhibition of P-gp activity in the blood–brain barrier28,152,154. PET probes are currently being developed for defined transporters; however, to use these probes a cyclotron must be in close proximity owing to the short half-lives of some PET tracers, such as [11C]-labelled compounds.
Gamma scintigraphy is a non-invasive imaging method utilizing short-lived gamma-emitting radioisotopes (for example, 99mTc) that can be used to quantify transporter activity and modulation. However, available probes have not been characterized completely for specific transporters. 99mTc-sestamibi has been used to assess MDR1 P-gp activity155,156, while 99mTc-mebrofenin and analogues have been used to measure MRP2 function157–159. Gamma scintigraphy has been used to directly quantify biliary and intestinal drug clearance in humans160,161. Use of 99mTc probes to assess hepatic transport function in patients receiving irinotecan162 and vinorelbine163 provides interesting translational applications for gamma scintigraphy and the possibility to individualize drug dosage regimens. But, practical reasons may limit the application of imaging methods in the later phases of drug development.
Interplay of efflux transporters and enzymes
The considerable overlap in the substrate specificity and tissue localization of CYP3A and P-gp has led to the hypothesis that this enzyme and transporter pair act as a coordinated absorption barrier against xenobiotics164,165. Clinical studies investigating the importance of intestinal CYP3A and P-gp through inhibition or induction of these proteins have demonstrated that the role of P-gp in the intestine extends beyond simply limiting absorption of the parent drug164,166,167. P-gp also increases the access of drug for metabolism by CYP3A through repeated cycles of absorption and efflux. That is, after penetration into enterocytes, molecules that escape metabolism are eliminated from the cells via P-gp or other apical ABC transporters but then re-enter the enterocytes168,169. It has been shown that the residence time of the drug in the intestine is prolonged with the aid of P-gp, thereby increasing the chance of local metabolic conversion by CYP3A4 (refs 168,169).
Transporters and enzymes in drug clearance
Drug uptake followed by metabolism and excretion in hepatocytes and kidney proximal tubule epithelia is a major determinant of the systemic clearance and exposure of many drugs. Vectorial transport of anionic drugs (for example, statins and angiotensin-converting enzyme inhibitors) from the blood to the bile or urine via an uptake and efflux transporter is important for determining drug exposure in the circulating blood and in organs. Sugiyama and co-workers170,171 have characterized the interplay of enzymes and transporters to understand the importance of parameters that determine the intrinsic unbound drug clearance in the intestine, liver and kidney. For highly permeable drugs172–174 in which neither active uptake nor efflux is rate-limiting, the traditional organ clearance models incorporating blood flow, extent of protein binding, and intrinsic metabolic and excretion clearance may hold. However, most anionic drugs and some hydrophilic organic cationic and zwitterionic drugs exhibit poor membrane permeability172–174. These types of drugs require transporters for their efficient penetration into or out of the liver or kidney. In such cases, the intrinsic organ clearance is a hybrid of three parameters: the intrinsic clearances for cellular uptake and efflux into the systemic circulation; the traditional organ intrinsic clearance, representing metabolism; and biliary and renal excretion. Each of these uptake, metabolic or efflux clearances can be rate-determining for certain drugs175. Therefore, the effects of changes in metabolism and transport activity resulting from interactions with co-administered drugs, or polymorphisms, should be considered in predicting potential changes in the systemic exposure of drugs. Modeling systemic exposure changes for some drugs can be done with PBPK modelling as described in the section above, thus providing initial predictions of drug interaction potential. The role of transporters, including transporter–enzyme interplay, in the pharmacokinetics of orally administered drugs has been reviewed recently176.
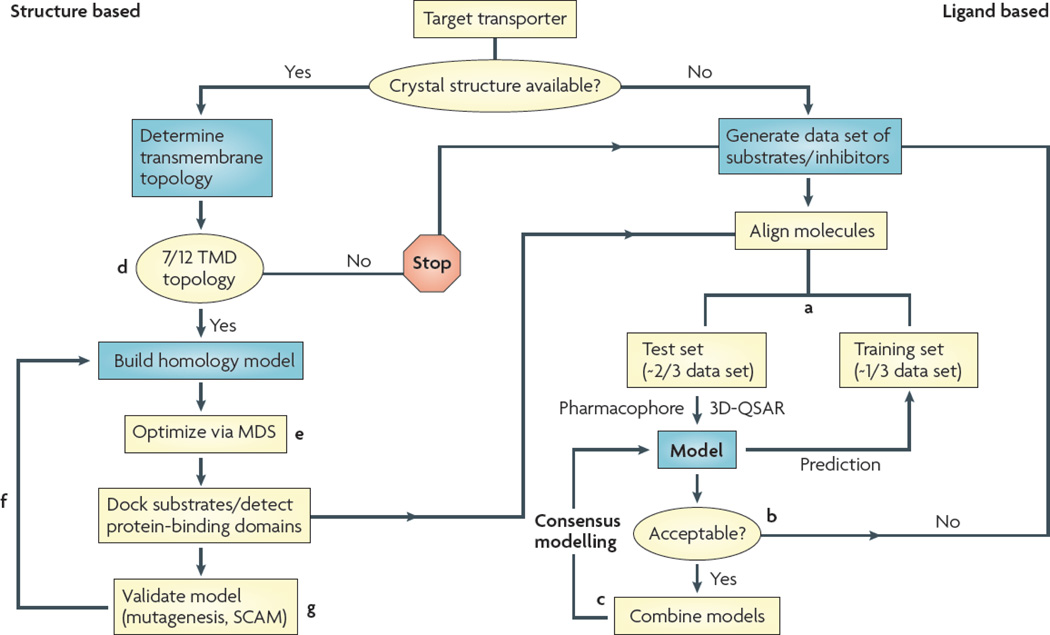
Computational models
The application of computational modeling algorithms to gain insight into transporter–substrate interactions has met with increasing success by the availability of high-quality data sets and atomic resolution structures of several major facilitator proteins. This burgeoning field can be divided broadly into indirect ligand-based techniques, such as pharmacophore and 3D-QSAR modeling, and direct structure-based approaches, such as homology or comparative modeling based on available crystallographic data177 (FIG. 2). However, synergistic models fusing both techniques are becoming increasingly important178. Ligand-based models describe a protein’s structural requirements for substrate or inhibitor interaction by correlating the molecular features of validated substrates or inhibitors with their biological activity38–40,64,178–180. Most models are limited to predicting putative inhibitors; although some models predict substrates. However, such models generally do not robustly predict the rate of substrate transport177,178,181. Pharmacophore models can be used successfully to screen large databases and identify novel transporter ligands. Structure-based approaches generate three-dimensional models of a target transporter based on the structure of an appropriate scaffold protein; currently available structures include bacterial solute transporters and an ABC transporter (Sav1866). A unique challenge in modeling membrane transporters concerns the generation of robust models using low levels of sequence identity and divergent membrane topologies. Ultimately, the docking of ligands may guide ligand-based and rational drug design as well as allow for screening of large databases.
Figure 2. Decision tree for computer modelling of transporter proteins.
Ligand-based methods such as pharmacophore and 3D-quantitative structure–activity relationship modelling (3D-QSAR) will be required when crystal structure templates are not available or can be used to complement existing homology models. Generally, a high-throughput in vitro assay is used to generate a data set of transporter ligands (substrates or inhibitors) and their corresponding activity values (Km/Jmax, Ki, or percentage inhibition). a | Data sets are split into a training set used to construct a model and a test set to validate the model. b | Criteria for model acceptance depend on its ability to successfully predict biological activity of test set molecules. c | Acceptable models may be combined to generate synergistic consensus models. d | To generate homology models with high confidence, the membrane topology of both the target and scaffold proteins must match. Modelling of transporters with divergent topology should be attempted with great caution as it is currently unclear how the arbitrary deletion or insertion of a membrane helix may distort the overall helical packing of membrane proteins. e | To further increase fidelity, models should be optimized using molecular dynamics simulations (MDS), preferably embedded in a lipid bilayer surrounded by an aqueous environment. f | When all criteria are satisfied, homology models may be used to determine regions of substrate binding or interaction and mapping of possible permeation pathways. g | Biochemical validation — for example, substituted cysteine accessibility method (SCAM) — will therefore be needed. TMD, transmembrane domain.
Section 3: Drug development issues
The identification of membrane transporters that influence the disposition and safety of drugs is a new challenge for drug development programmes, as well as for regulatory agencies worldwide. Drug transporter information is becoming common in drug labels, and provides important mechanistic ADME information that is useful for patients, physicians, regulatory agencies and research scientists. For instance, the label for mycophenolate mofetil highlights the role of MRP2 in the hepatic disposition of mycophenolic acid; the atorvastatin label indicates the role of oATP1B1; and the varenicline label discusses the involvement of oCT2 in its interaction with other drugs. other examples include the sitagliptin (Januvia; Merck) label, which has information about the role of P-gp and oAT3 in the drug’s clearance; the cidofovir label, which includes information on the potential modulation of renal toxicity by coadministration of an OAT inhibitor, probenecid; and the lapatinib label, which includes information on P-gp, BCRP and OATPs182. Transporter substrate or inhibition studies should be undertaken specifically to clarify the impact transporters have on a drug’s disposition, efficacy and safety, including their importance in DDIs. Thus, the ability to move a test compound rapidly into the clinic and to plan effectively for its clinical development will, in part, depend on preclinical assessments of the interaction of drug candidates with transporters.
The ITC discussed the characteristics of an NME that would trigger an in vitro interaction study with a specific transporter. An understanding of the physicochemical properties of the drug (for example, molecular mass, charge and lipophilicity) and the organ(s) involved in clearance of the NME will help determine which transporter–NME interactions should be studied in vitro. For example, an NME that is cleared exclusively by the kidneys should not (initially) be evaluated as a substrate for the hepatic transporter OATP1B1. Furthermore, consideration of the physicochemical and pharmacokinetic properties of an NME is also important in assessing the likelihood of a clinical DDI. For example, the solubility, permeability and degree of metabolism of an NME will affect its likelihood for a DDI with a P-gp or BCRP inhibitor (BOX 2).
Of the numerous drug transporters identified so far, P-gp, BCRP, OATPs, OCTs and OATs have been implicated (and studied) most frequently in clinical practice. As outlined in Section 2, various in silico, in vitro and in vivo tools are available to evaluate the role of transporters in the disposition of a drug. Examples of potential decision trees for these clinically relevant transporters are included here (BOXES 2–6), and are intended to provide guidance to sponsors and regulatory agencies as to when to conduct clinical studies to investigate the importance of transporters in a drug’s disposition. The recommendations proposed herein are generally intended to support clinical development and filing of a new drug application. Case studies of decision analyses are provided in BOX 7. The timing of transporter investigations should be driven by efficacy; safety; clinical trial enrolment questions (for example, exclusion and inclusion criteria); the need to further understand the ADME properties of the drug molecule; and transporter information needed for drug labeling. The proposed decision trees represent starting points for discussion. The US Food and Drug Administration draft Drug Interaction Guidance14,15,183is recommended for use in conjunction with this report as a starting point to develop a strategy for addressing transporter issues during drug development.
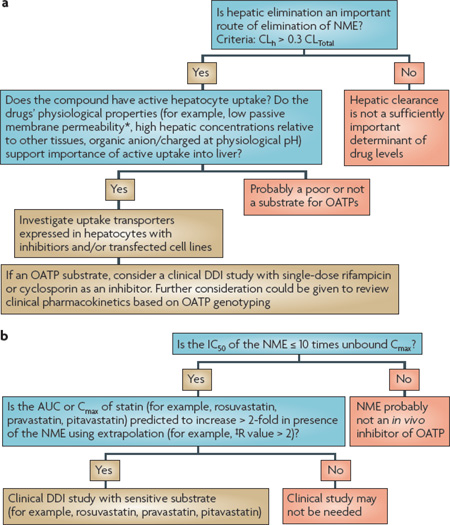
Box 6 | Decision trees for OATP interactions.
organic anion transporting polypeptide (oATP) substrates
Generally, investigations involving OATPs are focused on the OATP1B family in the liver. Most OATP1B substrates are large (350–1,000 daltons) organic anions that are relatively hydrophilic. In addition to parent drugs, a number of Phase II drug conjugates are OATP1B substrates. For the identification of substrates, heterologous expression systems including OATP1B1, OATP1B3 and possibly other hepatic OATPs such as OATP2B1, should be considered. Criteria for establishing whether an NME is an in vitro substrate of an OATP are discussed in the footnote of BOX 4. If a new molecular entity (NME) is a substrate of an OATP, then a detailed assessment of non-clinical (for instance, permeability, metabolism, tissue-to-plasma ratio in liver and other organs) and clinical data (for instance, dose linearity) should be completed to determine whether a human pharmacokinetic study is warranted (a).
If a clinical study is necessary, the NME could be given together with a single dose of an OATP inhibitor such as rifampicin or cyclosporine, as both are broad OATP inhibitors. However, some care should be taken in the interpretation of the clinical data with respect to the interaction because these agents can affect many other transporters as well as absorption, distribution, metabolism and excretion processes. In addition, the pharmacokinetics of the NME could be studied in individuals with OATP1B1 single nucleotide polymorphisms (SNPs) associated with lower OATP activity (as outlined in Section 1). Functional SNPs in OATP1B3 have been described197, and appear to be associated with alterations in bilirubin levels198; however, to date, no phenotypic consequence related to pharmacokinetics has been observed. Therefore, at this time, clinical studies in individuals with OATP1B3 SNPs are not recommended.
oATP inhibitors
To assess the potential of an NME to inhibit OATPs, the uptake of a prototypical substrate such as oestradiol-17-β- glucuronide, a statin or bromosulphophthalein in a heterologous expression system for OATP1B1 or OAT1B3, should be measured in the presence of the NME, and the IC50 value of the NME should be determined (b). A known inhibitor such as rifampicin or cyclosporine should be included as a positive control127,199. The predicted or actual plasma concentration in vivo should be considered as a starting point for interpreting the observed IC50 value. If the IC50 value is ≤ 10 times the unbound Cmax (for example, unbound Cmax/IC50 ≥ 0.1 or [I]/IC50 ≥ 0.1), then the NME may be an in vivo OATP inhibitor. If this criterion is met, it is recommended that an in vitro–in vivo extrapolation approach (R value) be completed192,193, which takes the maximal unbound drug concentration at the inlet of the liver (Iin, max) into account (see figure (b) and BOX 7). If the extrapolation suggests a > 2-fold change in exposure, consider proceeding to a clinical drug–drug interaction (DDI) study using atorvastatin, pravastatin, pitavastatin or rosuvastatin as OATP1B1 and OATP1B3 non-selective in vivo probe substrates192,193. Note that calculation of an R value is only one method of many that are available for extrapolation. In vitro confirmation that the NME inhibits the uptake of the statin, which is proposed as an in vivo probe substrate, should be obtained before proceeding to the clinical DDI study.
To estimate the magnitude of the expected clinical effect of the NME on the statin, calibration of the R value should be performed using OATP inhibitors such as rifampicin or cyclosporine as positive controls127,199. It has been noted that telmisartan may prove to be useful as an OATP1B3 selective substrate; thus telmisartan can be considered if a potential drug interaction specific to OATP1B3 needs to be assessed in vivo98. Examples of how the R value is calculated and used are described in BOX 7.
*Low permeability needs to be defined by each laboratory based on standards, such as atenolol (a Biopharmaceutics Classification System reference drug). A general guide would be 1 × 10−6 cm per sec (10 nm per sec) or lower is classed as low permeability. ‡R value is defined as 1+(fu × Iin, max/IC50), in which Iin, max is the estimated maximum inhibitor concentration at the inlet to the liver and is equal to: Imax + (Fa × Dose × ka/Qh). Imax is the maximum systemic plasma concentration of the inhibitor; Fa is the fraction of the dose of the inhibitor, Dose, which is absorbed; ka is the absorption rate constant of the inhibitor; and Qh is the hepatic blood flow (for example, 1,500 ml per min).
Box 7 | OATP1B1 decision analysis: case studies.
Current information suggests that HIV protease inhibitors are organic anion transporting polypeptide (OATP) inhibitors. Lopinavir is administered at high doses (400 mg) yielding total Cmax values of approximately 15–20 µM, representing approximately 0.23–0.3 µM free drug (fraction unbound (fu) = 0.015)200. In vitro inhibition assays demonstrated that lopinavir inhibited OATP1B1 with an IC50 value of 0.1 µM. As the IC50 value is less than ten times the unbound Cmax, completion of an extrapolation is recommended for OATP1B1 (see BOX 6 for OATP inhibition decision tree). An R value for lopinavir of 9.2 is calculated201 using the following equation and parameters: R = 1+(fu × Iin, max/IC50); Iin, max = 54.8 µM200 (see BOX 6 footnote for further explanation of calculation of the R value and of definition of Iin, max); the fraction of drug absorbed, Fa = 1.0; the absorption rate constant, ka = 0.03 per min202; hepatic blood flow, Qh = 1,500 ml per min. These estimates assume that the fraction of the substrate transported by OATP1B1 is 100%. However, the magnitude of clinical interaction will be attenuated if the fraction transported by a single pathway is less than 100% (for example, a second route of transport). For rosuvastatin, the contribution of OATP1B1 to the hepatic uptake is estimated to be 54%. As shown in the drug label, the clinical drug–drug interaction (DDI) with lopinavir and ritonavir combination (400 mg and 100 mg) twice a day for 10 days yields a two fold change in AUC and five fold change in Cmax of rosuvastatin. This is probably due to lopinavir inhibiting OATP1B1 (ritonavir’s effect on OATP1B1 is discussed below).
A similar analysis can be completed for two other HIV protease inhibitors: for ritonavir — 100 mg dose; fu = 0.02198; Cmax = 1.73 µM203; Iin, max = 4.51 µM, (using Fa = 1.0, Qh = 1,500 ml per min and ka = 0.03 per min202) — the unbound concentration is < 1 µM, when used as a boosting agent only. However, the IC50 value (0.3 µM) is less than ten times the unbound ritonavir concentration (0.05 µM). The extrapolation yields an R value of 1.3, suggesting that the drug interaction potential of ritonavir with OATP substrates is minimal based solely on plasma concentrations, and that a DDI study to evaluate the clinical significance of OATP inhibition is not needed.
For amprenavir (600 mg dose; fu = 0.1; Cmax = 12.0 µM; Iin, max = 35.8 µM)203, unbound concentrations are > 1 µM and the IC50 value (5.5 µM) for OATP1B1 is five fold higher than the unbound concentration (1.2 µM). Extrapolation provides an R value of 1.7, suggesting that a DDI study may not be required. Indeed, the lack of an interaction between amprenavir (when administered as fosamprenavir) and rosuvastatin was confirmed in a clinical study201. Furthermore, in this study, ritonavir was present as a boosting agent, suggesting that this low-dose ritonavir does not inhibit OATP in vivo.
The lack of specific inhibitors and substrates for transporters is well recognized, and represents an area of research that warrants further research. However, this should not be seen as a reason not to move forward with a clinical (or preclinical) drug interaction study, as the primary reason for conducting a drug interaction study is to provide information for dose adjustments. Even though it is desirable to understand the mechanisms responsible for DDIs, elucidating the mechanisms is not the primary objective of clinical DDI studies; understanding the required dose adjustments for patients is the single most important objective. The supporting preclinical and in vitro transporter data are essential in the design of the clinical DDI study.
Summary and conclusions
The focus of the ITC has been to provide, where possible, consensus on the current knowledge of transporters in drug development. The ITC identified areas in which there was agreement on the current status of the field, existing challenges and future directions. Importantly, the ITC has provided some guidelines that industry and regulatory agencies can apply to assist in the development of safe and effective medications (see decision trees in Section 3). Although this topic has been addressed to some extent previously184, the field of drug transporters is rapidly advancing. Consideration must be given to the role of transporters in the absorption (for example, intestinal P-gp and BCRP); distribution (for example, P-gp at the blood–brain barrier, and OATP1B1, OATP1B3 and OATP2B1 for hepatocyte uptake); and excretion (for example, OATs and OCTs for renal elimination) of NMEs in development. One could also argue that metabolism should be included to complete the ADME paradigm, as intracellular drug concentrations can be affected by an interplay between transporters and DMEs, as summarized in Section 2 (refs 166,169).
There are significant challenges that result from these rapid developments. Our applied knowledge of DMEs progressed through the 1990s and was successfully practiced to address specific issues in the pharmaceutical industry. A comparison between the experiences with DMEs and the current challenges with transporters is inevitable, and can also be informative. Will successes in the transporter field parallel the successes in the field of DMEs? And will the fields be integrated to provide a more holistic approach to clinical trials of NMEs? Overall, from a mechanistic perspective, transporters offer complexities that are distinct from those of DMEs. Typically, tissue-specific drug concentrations are determined by both uptake and efflux transporters within a tissue. Drug concentrations measured in plasma, therefore, may not reflect levels in organs such as liver and brain; this important concept has been illustrated in studies with transporter-knockout mice. Furthermore, in contrast to DMEs, which are largely concentrated in the liver and intestine, transporters are present in varying abundance in all tissues in the body, and have important roles in drug distribution and tissue-specific drug targeting, as well as in drug absorption and elimination. Another complexity of drug transporters, which is also seen for DMEs, is the issue of redundant specificities among transporters within a particular tissue. For example, in a given tissue, there may be redundant influx and efflux transporters such that multiple transporters with overlapping substrate specificities will determine tissue-specific drug concentrations. Standardized, well-characterized, generally accessible in vitro assay systems are needed to define the roles of the different transporters in drug absorption, distribution and elimination. These are in addition to clinically relevant, selective probe substrates, inhibitors and tracers suitable for imaging studies. The Critical Path Initiative has highlighted the enormous need for modern tools for use in drug development185. In this manuscript, we have suggested models for decision trees that can be used in the development of drugs. Much time was spent by the ITC in discussing whether to include such decision trees here. Many expressed concerns that decision trees could and would be rigidly interpreted as the sole path for in vitro and in vivo testing of transporter–drug interactions. Importantly, there was concern that the field was not sufficiently advanced, and that there are gaps in our knowledge of the in vivo role that transporters play in drug absorption and disposition in humans. Moreover, tools such as selective probes and inhibitors were not available at this time. However, others felt that the field was evolving and that decision trees, interpreted with flexibility, might be useful in the development of drugs and the collection of data to refine the decision trees. All felt that Considerable caution must be exercised in the use and interpretation of the decision trees presented here, and all acknowledged that the decision trees will need to be modified regularly as the field of transporter pharmacology grows. At some point in the future, integrated transporter and enzyme decision trees should be considered. The proposed decision trees attempt to define what can be extrapolated from current knowledge to aid in guiding the need, or lack thereof, for clinical studies. The decision trees that were constructed included conservative criteria for each decision so as to reduce false negatives. However, concern was expressed that an abundance of false positives will have a detrimental effect on the development of new drugs. Thus, a balance in the decision trees is clearly needed. The evolution and appropriate application of these decision trees will require constant monitoring. How can this be achieved with an assured and encompassing measure of success? Constant questioning of current practices with ongoing dialogue between academic institutions, regulatory agencies and industry will ensure the development of safe drugs.
Box 3 | Decision trees for P-glycoprotein or BCRP inhibitor interactions.
A new molecular entity (NME) is considered to be a potential P-glycoprotein (P-gp; also known as MDR1, ABCB1) and/or breast cancer resistance protein (BCRP; also known as ABCG2) inhibitor if the net flux ratio of a P-gp (or BCRP) probe substrate is decreased in the presence of the NME using a bi-directional transport assay (see footnote 1 below). Generally, the potency of inhibition is reported only as an IC50 value, which is dependent on the test system used. There are several practical challenges in evaluating whether NMEs that inhibit efflux transporters in vitro are also inhibitors in vivo. Orally administered drugs that are classified as in vitro efflux inhibitors provide the dual challenge of considering both intestinal efflux inhibition, in which intestinal drug concentrations will be the highest, and tissue efflux inhibition, in which circulating concentrations will be more relevant. Zhang et al.15 proposed that drugs exhibiting an [I]1/IC50 ≥ 0.1 or [I]2/IC50 ≥10 should be evaluated to determine whether inhibition occurs in vivo. In that publication, [I]1 is the mean NME steady-state total Cmax at the highest clinical dose, and [I]2 is the theoretical maximal gastrointestinal NME concentration after oral administration calculated as the highest clinical dose (mg) in a volume of 250 ml15. The [I]1/IC50 ≥ 0.1 calculation is familiar to those working on metabolic inhibition, as this approach has been used for many years.
In this paper, to be consistent with decision trees for other transporters (for example, organic cation transporter 2 (OCT2) and organic anion transporting polypeptide (OATP)) and the notion that unbound drug is the pharmacologically active species, mean steady-state Cmax values for unbound drug following administration of the highest proposed clinical dose is used for [I]1 in the decision tree (see footnote 2 for further discussion). Using this approach, a drug (molecular mass of 500 daltons) with an in vitro IC50 value of 10 µM will exceed an [I]1/IC50 value > 0.1 at an unbound Cmax > 1 µM (assumes fraction unbound (fu) = 0.1 and Cmax = 10 µM). This suggests that there is limited drug interaction potential with P-gp or BCRP. By contrast, for the same drug, an [I]2/IC50 value > 10 will be exceeded at a dose of approximately 12 mg or greater. Thus, a drug with an inhibition potency of approximately 10 µM or less in vitro is likely to exceed [I]2/IC50 value > 10 as many drugs require oral doses greater than 12 mg.
If an NME meets either [I]1/IC50 ≥ 0.1 (for example, IC50 is ≤ 10-fold the unbound Cmax value) or [I]2/IC50 ≥ 10, an in vivo drug interaction study with digoxin is recommended. Digoxin is a unique drug (see footnote 3), and may not be the best reference for decisions regarding interaction studies with other P-gp substrate drugs. Fenner et al.188 analysed drug interactions with digoxin and suggested that interactions of digoxin with P-gp inhibitors were limited (for example, most inhibitors caused < 2-fold changes in plasma concentrations of digoxin). However, owing to its narrow therapeutic index, interactions that alter the digoxin AUC > 1.25-fold are clinically important for digoxin. Therefore, the interpretation of the significance of P-gp for drug–drug interactions should not be over-extrapolated based on the importance for digoxin safety, as most other drugs have a much larger therapeutic index. In addition, if the NME inhibits a cytochrome P450 enzyme as well as P-gp, selection of a drug that is a dual substrate of cytochrome P450 and P-gp may be deemed appropriate. If the NME is a pure P-gp inhibitor, other than for clarifying a digoxin dose adjustment requirement, no drug–drug interaction studies may be indicated depending on the therapeutic area of the NME and its drug labeling considerations.
For an NME that is a BCRP inhibitor, possible candidate probe substrates include sulphasalazine189, rosuvastatin190, pitavastatin41, ciprofloxacin47 and dipyridamole191. However, many of these remain to be tested as selective BCRP probe substrates in clinical studies.
Note 1: A unidirectional assay based on the probe substrate can also be considered188. Inside-out vesicles expressing P-gp or BCRP, or transfected cell monolayers grown on solid support with appropriate controls (for example, inhibitors and positive controls, as described in Section 2), can also be used for inhibition assays. Note 2: If fu cannot be accurately determined owing to high protein binding, then assume fu = 0.01, to err on the conservative side. Alternatively, unbound portal vein concentrations may be used and might be more applicable to liver uptake192,193. Note 3: Digoxin has an absolute bioavailability of approximately 70% from immediate release tablets. By contrast, the absolute bioavailability from a liquid-filled formulation (for example, Lanoxicaps) is approximately 90%. Therefore, consideration should also be given to the dosage form of digoxin used in an interaction study. In addition, digoxin may be a substrate for other transporters15,92,188,194. Hence, caution is advised when extrapolating results from digoxin to other P-gp substrates.
Box 4 | Decision trees for OCT or OAT substrate interactions.
Generally, most organic cation transporter (OCT) and organic anion transporter (OAT) substrates are small organic ions (less than 400 daltons) that are relatively hydrophilic. OCT substrates are typically cations and OAT substrates anions. Of note, OAT3 can also transport weakly basic compounds such as cimetidine and sitagliptin (Januvia; Merck). The key clinical renal transporters in the basolateral membrane of the proximal tubule are OCT2, OAT1 and OAT3. Transporters in the apical membrane of the proximal tubule that are important, but not a focus of this manuscript, include the multidrug and toxin extrusion transporters (MATE1 and MATE2-K) and the organic anion transporter (OAT4), as well as some ATP-binding cassette transporters (for example, MRP2 and MRP4).
In evaluating whether a new molecular entity (NME) is an OCT or OAT substrate, the first step to consider is whether the renal clearance (CLr) of the NME is an important determinant of the total clearance (CLTotal). That is, if CLr ≥ 0.5 × CLTotal (0.5 was selected because 50% was considered to represent a substantial and clinically important percentage of the CLTotal). When CLr is a substantial component of CLTotal (or when the fraction of CLr to CLTotal cannot be estimated because oral bioavailability cannot be determined), the second condition to establish is whether renal clearance involves secretion. That is, if CLr > 1.5 fraction unbound (fu) × glomerular filtration rate (GFR). If fu cannot be accurately determined, for example, plasma protein binding is greater than 99.9%, or is not known, then assume fu = 0.01, to err on the conservative side. Greater than 1.5 implies that a substantial fraction of the CLr is due to renal tubular secretion.
If both conditions are met, that is, CLr ≥ 0.5 CLTotal and CLr > 1.5 fu × GFR, then in vitro uptake into a recombinant cell line expressing renal transporters should be conducted. These assays should include control cells without recombinant transporters, or transfected cells with known OCT or OAT inhibitors, or both. To determine whether an NME is a substrate of an OCT or OAT (or OATP as discussed in BOX 6), the ratio of NME uptake in the transporter-expressing cells versus the control (or empty vector) cells should be statistically greater than 1 (see footnote). Michaelis–Menten studies can be used in the transfected cells to determine the kinetic parameters of the NME.
For confirmed in vitro substrates of OCT2 or OAT1/3, a clinical drug–drug interaction (DDI) study could be performed with cimetidine as an inhibitor of organic cation transport, or probenecid as a broad non-specific OAT inhibitor. Cimetidine and probenecid are model inhibitors; however, these drugs may not be prescribed commonly. Therefore, sponsors should consider other more clinically relevant renal transporter inhibitors, if available, for use in DDI studies. Examples of labels that describe DDIs mediated by OCTs or OATs include metformin, pramipexole and varenicline.
The ratio of NME uptake in the cells expressing the transporter versus the control (or empty vector) cells should be statistically greater than 1. No agreement was reached by the International Transporter Consortium regarding the magnitude of the ratio. However, it is important that uptake into the transfected cells be significantly greater than background in a control cell line and be inhibited by a known inhibitor of the transporter. A positive control should be included. ‡If the CLr/CLTotal is unknown, go to ‘Yes’.
Box 5 | Decision trees for OCT or OAT inhibitor interactions.
Evaluation of organic cation transporter (OCT) or organic anion transporter (OAT) inhibitors requires determination of an IC50 value in an in vitro study. If the IC50 value is ≤ 10-fold the unbound Cmax value in cells expressing OCT2 (or OAT1/3), a clinical drug–drug interaction (DDI) study is recommended. Ten-fold was selected to err on the conservative side and to be consistent with the P-glycoprotein (P-gp; also known as MDR1, ABCB1) and breast cancer resistance protein (BCRP; also known as ABCG2) decision trees. For inhibitors of OCT2, the new molecular entity (NME) can be clinically evaluated for its potential to inhibit the renal clearance of metformin, a commonly used anti-diabetic drug. However, OCT2 inhibitors may also be multidrug and toxin extrusion transporter (MATE) inhibitors, and clinical inhibition interactions with metformin may involve MATE1 or MATE2-K in addition to, or even instead of, OCT2. Therefore, some caution in the interpretation of in vivo inhibition studies for OCT2 should be exercised. For OAT1 and OAT3, several potential substrates may be used as ‘victim’ drugs (see footnote). Some inhibitors of OAT1/3 may also inhibit apical transporters (for example, ATP-binding cassette transporters); therefore, clinical inhibition data should be interpreted cautiously.
Selection of an appropriate probe substrate victim drug for the clinical DDI study should be based upon therapeutic considerations; for example, a victim drug should be selected that is likely to be co-administered with the NME in a therapeutic setting. Other considerations should include the therapeutic window of the victim drug and the maximum effect that would be expected if the secretory clearance of the victim drug was totally inhibited.
For NMEs that are OAT inhibitors, multiple candidate probe substrates could be used in clinical DDI studies, including zidovudine, lamivudine, zalcitabine, acyclovir, ciprofloxacin, tenofovir and methotrexate. Some of these substrates may be transported by multiple transporters. For example, some data suggest that methotrexate renal elimination may be affected by co-administration of non-steroidal anti-inflammatory drugs; however, the mechanisms may include other transporters in addition to OATs195,196. MPP+, 1-methyl-4-phenylpyridinium; OS, oestrone-3-sulphate; PAH, para-aminohippuric acid.
Acknowledgements
J.M.S’s insight and research career focused on understanding the determinants of drug–drug interactions, clinical pharmacology, drug-induced liver injury, and the translation of transporter biology to the drug safety evaluation paradigm. We are grateful for the able assistance of R. Bogenrief and C. Weiss in preparing this manuscript. We would like to acknowledge partial support from a National Institutes of Health grant, GM61390 and funds from the FDA Critical Path for the workshop leading to this report.
Glossary
- Absorption, Distribution, Metabolism and Excretion (ADME)
The acronym ADMe describes the factors that determine the overall exposure to an administered drug. Absorption describes uptake from the site of administration. Although usually referring to oral route, other routes can apply, for example, subcutaneous or buccal. As drug levels are typically measured systemically, oral absorption can be a function of movement across the mucosal surface of the intestinal tract, through the liver and into the systemic circulation via the vena cava. Distribution describes the extent of partitioning into tissues. Metabolism refers to a chemical change usually catalysed by enzymes such as the cytochrome P450s. The drug and its metabolites are excreted or eliminated usually by the kidney (in urine) or from the liver (via bile) or intestines into the faeces
- Blood brain barrier (BBB)
The BBB consists of endothelial cells connected by tight junctions. The endothelial cells, which are surrounded by astrocytes, separate the circulating blood and the brain interstitial space. The role of the BBB is to protect the central nervous system by restricting and preventing the entry of toxic substances including drug molecules and bacteria into the brain. Included in this protective barrier are transporters such as P-gp
- Drug Clearance
The clearance of a drug is defined as the proportionality constant between rate of elimination and plasma concentration. The total clearance (CLTotal) reflects the rate of removal from the systemic circulation (the site of monitoring concentrations). Clearance by an organ can also be defined (e.g. renal clearance (CLr) via excretion and hepatic clearance (CLh) via metabolism and/or biliary excretion). Renal clearance of a drug is determined by glomerular filtration and tubular secretion and reabsorption, both of which may be transporter-mediated.
- Drug-drug interaction (DDI)
Concomitant administration of multiple drugs can result in altered levels of the drugs, compared to administration of the drugs alone. DDI can result in higher (inhibition) or lower (induction) levels of the drug. For example, if drug A (“perpetrator” drug) inhibits a membrane transporter that drug B (“victim” drug) uses to enter into the cell, administration of drug A can lower the level of drug B in the cell and also potentially increase the level of drug B in the systemic circulation
- Drug-metabolizing enzymes (DMEs)
These are enzymes responsible for chemical modification of drugs usually to increase rate of elimination. The activity of these enzymes can be altered through inhibition or induction (up-regulation) thus affecting the rate of metabolism
- Fraction unbound, fu
This fraction represents the ratio of unbound drug concentration to total drug concentration in the plasma. The fu is determined by the affinity of drugs to the binding proteins (e.g., albumin and α1-acid glycoprotein) in plasma and the total number of binding sites on the proteins
- IC50 and Ki
IC50 value, as used in this paper, is the concentration of an inhibitor needed to inhibit one-half of the transport rate of the substrate measured in the absence of the inhibitor. The IC50 of a drug is determined by a concentration - transport rate curve. To determine the IC50 value of an inhibitor of a drug transporter, the effect of increasing concentrations of the inhibitor on the transport rate of a substrate is determined.
- Ki
Ki is the inhibition constant of the drug inhibitor, and for competitive inhibition is determined as, Ki = IC50/[1+([s]/Km)], in which [s] is the concentration of the drug substrate and Km is the affinity of the drug substrate for the drug transporter.
- Membrane transporters
Membrane-associated proteins that govern the transport of solutes (e.g., drugs and other xenobiotics) into and out of cells. Transporters can play a critical role in determining drug concentrations in the systemic circulation and in cells. The two major superfamilies of membrane transporters are the ABC (ATP-binding cassette) and SLC (Solute Carrier) superfamilies
- Pharmacodynamics
Pharmacodynamics describe the extent and time course of effects of drugs on physiologic and pathophysiologic processes. The relationship between drug concentration and drug effect is important in determining pharmacodynamics
- Pharmacokinetics
Pharmacokinetics describe what the body does to a drug. Pharmacokinetic parameters result from the ADME properties of a drug, and provide information about the rates of elimination and systemic levels of drugs. The functional activity and level of a drug transporter can determine the amount of drug molecule transported into or out of specific tissues, which will determine the level of drugs in that tissue and also the systemic circulation
- Substrate or Inhibitor
A substrate of a drug transporter is translocated across a membrane by the transporter. An inhibitor of a drug transporter can impair the uptake and/or efflux of another drug. Many inhibitors are not substrates, i.e., are not transported by the transporter. Substrates can competitively inhibit other substrates that interact at the same site(s) on the transporter
- Unbound inhibitor concentration [I]
The unbound drug is the pharmacologically active species and is available for inhibition of transporters. Typically, mean unbound steady-state Cmax values following administration of the highest proposed clinical dose is used for [I]1 in the decision tree. Note that the definition of [I]1 used in this report differs from that described by Zhang et al. (ref. 15) in the original published P-glycoprotein decision tree. Defining [I] as the unbound steady-state maximum concentration was intentionally done to provide consistency across the all the decision trees included in this report
Footnotes
Competing interests statement
The authors declare competing financial interests: see web version for details.
DATABASES
UniProtKB: http://www.uniprot.org
BCRP | MRP2 | OCT1 | OCT2 | OAT1 | OAT3 | OAT4 | OATP1A2 | OATP1A2 | OATP1B1 | OATP1B3 | P-gp
FURTHER INFORMATION
US FDA Guidance, Compliance and Regulatory Information — Clinical Pharmacology: http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm064982.htm
US FDA Drug Development and Approval Processes —
Drug Development and Drug Interactions: http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm080499.htm
All links are active in the online PDF
References
- 1. Giacomini KM, Sugiyama Y. In: Gilman’s The Pharmacological Basis of Therapeutics. Brunton LL, Lazo JS, Parker RL, editors. New York: McGraw-Hill; 2006. pp. 41–70. This chapter in a textbook provides an excellent overview of transporters.
- 2. Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv. Drug Deliv. Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. This manuscript provides an excellent review of ABC transporters that are important in drug response.
- 3.Sai Y. Biochemical and molecular pharmacological aspects of transporters as determinants of drug disposition. Drug Metab. Pharmacokinet. 2005;20:91–99. doi: 10.2133/dmpk.20.91. [DOI] [PubMed] [Google Scholar]
- 4.Cascorbi I. Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol. Ther. 2006;112:457–473. doi: 10.1016/j.pharmthera.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Choudhuri S, Klaassen CD. Structure, function, expression, genomic organization, and single nucleotide polymorphisms of human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) efflux transporters. Int. J. Toxicol. 2006;25:231–259. doi: 10.1080/10915810600746023. [DOI] [PubMed] [Google Scholar]
- 6.Hediger MA, et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 7. Koepsell H, Lips K, Volk C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm. Res. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. This manuscript provides an excellent review of various transporters for organic cations (OCTs, OCTNs and MATEs).
- 8.Jonker JW, Wagenaar E, Van Eijl S, Schinkel AH. Deficiency in the organic cation transporters 1 and 2 (Oct1/Oct2 [Slc22a1/Slc22a2]) in mice abolishes renal secretion of organic cations. Mol. Cell Biol. 2003;23:7902–7908. doi: 10.1128/MCB.23.21.7902-7908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schinkel AH, et al. Multidrug resistance and the role of P-glycoprotein knockout mice. Eur. J. Cancer. 1995;31A:1295–1298. doi: 10.1016/0959-8049(95)00130-b. [DOI] [PubMed] [Google Scholar]
- 10.Maeda K, Sugiyama Y. Impact of genetic polymorphisms of transporters on the pharmacokinetic, pharmacodynamic and toxicological properties of anionic drugs. Drug Metab. Pharmacokinet. 2008;23:223–235. doi: 10.2133/dmpk.23.223. [DOI] [PubMed] [Google Scholar]
- 11.Sissung TM, Gardner ER, Gao R, Figg WD. Pharmacogenetics of membrane transporters: a review of current approaches. Methods Mol. Biol. 2008;448:41–62. doi: 10.1007/978-1-59745-205-2_4. [DOI] [PubMed] [Google Scholar]
- 12.Huang SM, Temple R, Throckmorton DC, Lesko LJ. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin. Pharmacol. Ther. 2007;81:298–304. doi: 10.1038/sj.clpt.6100054. [DOI] [PubMed] [Google Scholar]
- 13.Huang SM, et al. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J. Clin. Pharmacol. 2008;48:662–670. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- 14. US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Biologics Evaluation and Research (CBER) Guidance for Industry. Drug Interaction Studies — Study Design, Data Analysis, and Implications for Dosing and Labeling. US FDA website. 2006 [online], http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm072101.pdf. This website provides current FDA guidances for DDI studies. For updated guidances see the websites in Further information.
- 15. Zhang L, Zhang YD, Strong JM, Reynolds KS, Huang SM. A regulatory viewpoint on transporter based drug interactions. Xenobiotica. 2008;38:709–724. doi: 10.1080/00498250802017715. This article provides an FDA perspective about the role of transporters in DDIs.
- 16.Zhang L, Strong JM, Qiu W, Lesko LJ, Huang SM. Scientific perspectives on drug transporters and their role in drug interactionst. Mol. Pharm. 2006;3:62–69. doi: 10.1021/mp050095h. [DOI] [PubMed] [Google Scholar]
- 17.Raub TJ. P-glycoprotein recognition of substrates and circumvention through rational drug design. Mol. Pharm. 2006;3:3–25. doi: 10.1021/mp0500871. [DOI] [PubMed] [Google Scholar]
- 18.Miller DS, Bauer B, Hartz AM. Modulation of P-glycoprotein at the blood–brain barrier: opportunities to improve central nervous system pharmacotherapy. Pharmacol. Rev. 2008;60:196–209. doi: 10.1124/pr.107.07109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura Y, Morita SY, Matsuo M, Ueda K. Mechanism of multidrug recognition by MDR1/ ABCB1. Cancer Sci. 2007;98:1303–1310. doi: 10.1111/j.1349-7006.2007.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clin. Pharmacol. Ther. 2007;81:265–269. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- 21.Zhou SF. Structure, function and regulation of P-glycoprotein and its clinical relevance in drug disposition. Xenobiotica. 2008;38:802–832. doi: 10.1080/00498250701867889. [DOI] [PubMed] [Google Scholar]
- 22. Aller SG, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. This manuscript describes the first molecular structure of mouse P-gp.
- 23.Zhou Y, Gottesman MM, Pastan I. Studies of human MDR1–MDR2 chimeras demonstrate the functional exchangeability of a major transmembrane segment of the multidrug transporter and phosphatidylcholine flippase. Mol. Cell Biol. 1999;19:1450–1459. doi: 10.1128/mcb.19.2.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrier I, Julien M, Gros P. Analysis of catalytic carboxylate mutants E552Q and E1197Q suggests asymmetric ATP hydrolysis by the two nucleotide binding domains of P-glycoprotein. Biochemistry. 2003;42:12875–12885. doi: 10.1021/bi034257w. [DOI] [PubMed] [Google Scholar]
- 25.Loo TW, Bartlett MC, Clarke DM. Val133 and Cys137 in transmembrane segment 2 are close to Arg935 and Gly939 in transmembrane segment 11 of human P-glycoprotein. J. Biol. Chem. 2004;279:18232–18238. doi: 10.1074/jbc.M400229200. [DOI] [PubMed] [Google Scholar]
- 26.Feng B, et al. In vitro P-glycoprotein assays to predict the in vivo interactions of P-glycoprotein with drugs in the central nervous system. Drug Metab. Dispos. 2008;36:268–275. doi: 10.1124/dmd.107.017434. [DOI] [PubMed] [Google Scholar]
- 27.Yamazaki M, et al. In vitro substrate identification studies for P-glycoprotein-mediated transport: species difference and predictability of in vivo results. J. Pharmacol. Exp. Ther. 2001;296:723–735. [PubMed] [Google Scholar]
- 28.Sasongko L, et al. Imaging P-glycoprotein transport activity at the human blood–brain barrier with positron emission tomography. Clin. Pharmacol. Ther. 2005;77:503–514. doi: 10.1016/j.clpt.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 29.Kurnik D, et al. Tariquidar, a selective P-glycoprotein inhibitor, does not potentiate loperamide’ opioid brain effects in humans despite full inhibition of lymphocyte P-glycoprotein. Anesthesiology. 2008;109:1092–1099. doi: 10.1097/ALN.0b013e31818d8f28. [DOI] [PubMed] [Google Scholar]
- 30.Sadeque AJ, Wandel C, He H, Shah S, Wood AJ. Increased drug delivery to the brain by P-glycoprotein inhibition. Clin. Pharmacol. Ther. 2000;68:231–237. doi: 10.1067/mcp.2000.109156. [DOI] [PubMed] [Google Scholar]
- 31.Eyal S, Hsiao P, Unadkat JD. Drug interactions at the blood–brain barrier: fact or fantasy? Pharmacol. Ther. 2009;123:80–104. doi: 10.1016/j.pharmthera.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams JA, et al. PhRMA white paper on ADME pharmacogenomics. J. Clin. Pharmacol. 2008;48:849–889. doi: 10.1177/0091270008319329. [DOI] [PubMed] [Google Scholar]
- 33.Wakabayashi K, Tamura A, Saito H, Onishi Y, Ishikawa T. Human ABC transporter ABCG2 in xenobiotic protection and redox biology. Drug Metab. Rev. 2006;38:371–391. doi: 10.1080/03602530600727947. [DOI] [PubMed] [Google Scholar]
- 34.Robey RW, et al. ABCG2: a perspective. Adv. Drug Deliv. Rev. 2009;61:3–13. doi: 10.1016/j.addr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle LA, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl Acad. Sci. USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Herwaarden AE, Schinkel AH. The function of breast cancer resistance protein in epithelial barriers, stem cells and milk secretion of drugs and xenotoxins. Trends Pharmacol. Sci. 2006;27:10–16. doi: 10.1016/j.tips.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Vlaming ML, Lagas JS, Schinkel AH. Physiological and pharmacological roles of ABCG2 (BCRP): recent findings in Abcg2 knockout mice. Adv. Drug Deliv. Rev. 2009;61:14–25. doi: 10.1016/j.addr.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Matsson P, et al. A global drug inhibition pattern for the human ATP-binding cassette transporter breast cancer resistance protein (ABCG2) J. Pharmacol. Exp. Ther. 2007;323:19–30. doi: 10.1124/jpet.107.124768. [DOI] [PubMed] [Google Scholar]
- 39.Nicolle E, et al. QSAR analysis and molecular modeling of ABCG2-specific inhibitors. Adv. Drug Deliv. Rev. 2009;61:34–46. doi: 10.1016/j.addr.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Saito H, et al. A new strategy of high-speed screening and quantitative structure–activity relationship analysis to evaluate human ATP-binding cassette transporter ABCG2-drug interactions. J. Pharmacol. Exp. Ther. 2006;317:1114–1124. doi: 10.1124/jpet.105.099036. [DOI] [PubMed] [Google Scholar]
- 41.Hirano M, et al. Involvement of BCRP (ABCG2) in the biliary excretion of pitavastatin. Mol. Pharmacol. 2005;68:800–807. doi: 10.1124/mol.105.014019. [DOI] [PubMed] [Google Scholar]
- 42.Enokizono J, Kusuhara H, Sugiyama Y. Effect of breast cancer resistance protein (Bcrp/Abcg2) on the disposition of phytoestrogens. Mol. Pharmacol. 2007;72:967–975. doi: 10.1124/mol.107.034751. [DOI] [PubMed] [Google Scholar]
- 43.Hegedus C, et al. Ins and outs of the ABCG2 multidrug transporter: an update on in vitro functional assays. Adv. Drug Deliv. Rev. 2009;61:47–56. doi: 10.1016/j.addr.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 44.de Vries NA, et al. P-glycoprotein and breast cancer resistance protein: two dominant transporters working together in limiting the brain penetration of topotecan. Clin. Cancer Res. 2007;13:6440–6449. doi: 10.1158/1078-0432.CCR-07-1335. [DOI] [PubMed] [Google Scholar]
- 45.Zaher H, et al. Breast cancer resistance protein (Bcrp/abcg2) is a major determinant of sulfasalazine absorption and elimination in the mouse. Mol. Pharm. 2006;3:55–61. doi: 10.1021/mp050113v. [DOI] [PubMed] [Google Scholar]
- 46.Merino G, Jonker JW, Wagenaar E, van Herwaarden AE, Schinkel AH. The breast cancer resistance protein (BCRP/ABCG2) affects pharmacokinetics, hepatobiliary excretion, and milk secretion of the antibiotic nitrofurantoin. Mol. Pharmacol. 2005;67:1758–1764. doi: 10.1124/mol.104.010439. [DOI] [PubMed] [Google Scholar]
- 47.Merino G, et al. Breast cancer resistance protein (BCRP/ABCG2) transports fluoroquinolone antibiotics and affects their oral availability, pharmacokinetics, and milk secretion. Drug Metab. Dispos. 2006;34:690–695. doi: 10.1124/dmd.105.008219. [DOI] [PubMed] [Google Scholar]
- 48.Oostendorp RL, Buckle T, Beijnen JH, van Tellingen O, Schellens JH. The effect of P-gp (Mdr1a/1b), BCRP (Bcrp1) and P-gp/BCRP inhibitors on the in vivo absorption, distribution, metabolism and excretion of imatinib. Invest. New Drugs. 2009;27:31–40. doi: 10.1007/s10637-008-9138-z. [DOI] [PubMed] [Google Scholar]
- 49.Yamasaki Y, et al. Pharmacogenetic characterization of sulfasalazine disposition based on NAT2 and ABCG2 (BCRP) gene polymorphisms in humans. Clin. Pharmacol. Ther. 2008;84:95–103. doi: 10.1038/sj.clpt.6100459. [DOI] [PubMed] [Google Scholar]
- 50.Cusatis G, Sparreboom A. Pharmacogenomic importance of ABCG2. Pharmacogenomics. 2008;9:1005–1009. doi: 10.2217/14622416.9.8.1005. [DOI] [PubMed] [Google Scholar]
- 51.Cusatis G, et al. Pharmacogenetics of ABCG2 and adverse reactions to gefitinib. J. Natl Cancer Inst. 2006;98:1739–1742. doi: 10.1093/jnci/djj469. [DOI] [PubMed] [Google Scholar]
- 52.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin. Drug Metab. Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 53.Keskitalo JE, et al. ABCG2 polymorphism markedly affects the pharmacokinetics of atorvastatin and rosuvastatin. Clin. Pharmacol. Ther. 2009;86:197–203. doi: 10.1038/clpt.2009.79. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, et al. Role of BCRP 421C>A polymorphism on rosuvastatin pharmacokinetics in healthy Chinese males. Clin. Chim. Acta. 2006;373:99–103. doi: 10.1016/j.cca.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Morisaki K, et al. Single nucleotide polymorphisms modify the transporter activity of ABCG2. Cancer Chemother. Pharmacol. 2005;56:161–172. doi: 10.1007/s00280-004-0931-x. [DOI] [PubMed] [Google Scholar]
- 56.Aoki M, et al. Kidney-specific expression of human organic cation transporter 2 (OCT2/SLC22A2) is regulated by DNA methylation. Am. J. Physiol. Renal Physiol. 2008;295:F165–F170. doi: 10.1152/ajprenal.90257.2008. [DOI] [PubMed] [Google Scholar]
- 57.Koepsell H, Endou H. The SLC22 drug transporter family. Pflugers Arch. 2004;447:666–676. doi: 10.1007/s00424-003-1089-9. [DOI] [PubMed] [Google Scholar]
- 58.Wright SH. Role of organic cation transporters in the renal handling of therapeutic agents and xenobiotics. Toxicol. Appl. Pharmacol. 2005;204:309–319. doi: 10.1016/j.taap.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 59.Urban TJ, Giacomini KM. In: Drug Transporters. You G, Morris ME, editors. New York: John Wiley & Sons; 2007. pp. 11–33. [Google Scholar]
- 60.Ciarimboli G. Organic cation transporters. Xenobiotica. 2008;38:936–971. doi: 10.1080/00498250701882482. [DOI] [PubMed] [Google Scholar]
- 61. Nigam SK, Bush KT, Bhatnagar V. Drug and toxicant handling by the OAT organic anion transporters in the kidney and other tissues. Nature Clin. Pract. Nephrol. 2007;3:443–448. doi: 10.1038/ncpneph0558. This manuscript provides an excellent review of OATs.
- 62.Rizwan AN, Burckhardt G. Organic anion transporters of the SLC22 family: biopharmaceutical, physiological, and pathological roles. Pharm. Res. 2007;24:450–470. doi: 10.1007/s11095-006-9181-4. [DOI] [PubMed] [Google Scholar]
- 63.Srimaroeng C, Perry JL, Pritchard JB. Physiology, structure, and regulation of the cloned organic anion transporters. Xenobiotica. 2008;38:889–935. doi: 10.1080/00498250801927435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahlin G, et al. Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J. Med. Chem. 2008;51:5932–5942. doi: 10.1021/jm8003152. [DOI] [PubMed] [Google Scholar]
- 65.Okura T, Ito R, Ishiguro N, Tamai I, Deguchi Y. Blood–brain barrier transport of pramipexole, a dopamine D2 agonist. Life Sci. 2007;80:1564–1571. doi: 10.1016/j.lfs.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 66.Glube N, Langguth P. Caki-1 cells as a model system for the interaction of renally secreted drugs with OCT3. Nephron Physiol. 2008;108:18–28. doi: 10.1159/000115040. [DOI] [PubMed] [Google Scholar]
- 67.Glube N, Giessl A, Wolfrum U, Langguth P. Caki-1 cells represent an in vitro model system for studying the human proximal tubule epithelium. Nephron Exp. Nephrol. 2007;107:e47–e56. doi: 10.1159/000107804. [DOI] [PubMed] [Google Scholar]
- 68.Muller J, et al. Drug specificity and intestinal membrane localization of human organic cation transporters (OCT) Biochem. Pharmacol. 2005;70:1851–1860. doi: 10.1016/j.bcp.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 69.Rytting E, Bryan J, Southard M, Audus KL. Low-affinity uptake of the fluorescent organic cation 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide (4-Di-1-ASP) in BeWo cells. Biochem. Pharmacol. 2007;73:891–900. doi: 10.1016/j.bcp.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaler G, et al. Structural variation governs substrate specificity for organic anion transporter (OAT) homologs. Potential remote sensing by OAT family members. J. Biol. Chem. 2007;282:23841–23853. doi: 10.1074/jbc.M703467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Truong DM, Kaler G, Khandelwal A, Swaan PW, Nigam SK. Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J. Biol. Chem. 2008;283:8654–8663. doi: 10.1074/jbc.M708615200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jonker JW, et al. Reduced hepatic uptake and intestinal excretion of organic cations in mice with a targeted disruption of the organic cation transporter 1 (Oct1 [Slc22a1]) gene. Mol. Cell Biol. 2001;21:5471–5477. doi: 10.1128/MCB.21.16.5471-5477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang DS, et al. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 2002;302:510–515. doi: 10.1124/jpet.102.034140. [DOI] [PubMed] [Google Scholar]
- 75.Eraly SA, et al. Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem. 2006;281:5072–5083. doi: 10.1074/jbc.M508050200. [DOI] [PubMed] [Google Scholar]
- 76.Vallon V, et al. Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am. J. Physiol. Renal Physiol. 2008;294:F867–F873. doi: 10.1152/ajprenal.00528.2007. [DOI] [PubMed] [Google Scholar]
- 77.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou K, et al. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes. 2009;58:1434–1439. doi: 10.2337/db08-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.White DL, et al. OCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood. 2006;108:697–704. doi: 10.1182/blood-2005-11-4687. [DOI] [PubMed] [Google Scholar]
- 80.White DL, et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood. 2007;110:4064–4072. doi: 10.1182/blood-2007-06-093617. [DOI] [PubMed] [Google Scholar]
- 81.Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet. Genomics. 2008;18:637–645. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
- 82.Song IS, et al. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin. Pharmacol. Ther. 2008;84:559–562. doi: 10.1038/clpt.2008.61. [DOI] [PubMed] [Google Scholar]
- 83.Chen Y, et al. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet. Genomics. 2009;19:497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tirona RG, Kim RB. In: Drug Transporters. You G, Morris ME, editors. New York: John Wiley & Sons; 2007. pp. 75–104. [Google Scholar]
- 85. Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. This manuscript provides an excellent review of OATPs.
- 86.Leuthold S, et al. Mechanisms of pH-gradient driven transport mediated by organic anion polypeptide transporters. Am. J. Physiol. Cell Physiol. 2009;296:C570–C582. doi: 10.1152/ajpcell.00436.2008. [DOI] [PubMed] [Google Scholar]
- 87.Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim. Biophys. Acta. 2003;1609:1–18. doi: 10.1016/s0005-2736(02)00633-8. [DOI] [PubMed] [Google Scholar]
- 88.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem. 2001;276:35669–35675. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- 89.Hsiang B, et al. A novel human hepatic organic anion transporting polypeptide (OATP2). Identification of a liver-specific human organic anion transporting polypeptide and identification of rat and human hydroxymethylglutaryl-CoA reductase inhibitor transporters. J. Biol. Chem. 1999;274:37161–37168. doi: 10.1074/jbc.274.52.37161. [DOI] [PubMed] [Google Scholar]
- 90.Konig J, Cui Y, Nies AT, Keppler D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;278:G156–G164. doi: 10.1152/ajpgi.2000.278.1.G156. [DOI] [PubMed] [Google Scholar]
- 91.Abe T, et al. Identification of a novel gene family encoding human liver-specific organic anion transporter LST-1. J. Biol. Chem. 1999;274:17159–17163. doi: 10.1074/jbc.274.24.17159. [DOI] [PubMed] [Google Scholar]
- 92.Kullak-Ublick GA, et al. Organic anion-transporting polypeptide B (OATP-B) and its functional comparison with three other OATPs of human liver. Gastroenterology. 2001;120:525–533. doi: 10.1053/gast.2001.21176. [DOI] [PubMed] [Google Scholar]
- 93.Gui C, Hagenbuch B. Amino acid residues in transmembrane domain 10 of organic anion transporting polypeptide 1B3 are critical for cholecystokinin octapeptide transport. Biochemistry. 2008;47:9090–9097. doi: 10.1021/bi8008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ismair MG, et al. Hepatic uptake of cholecystokinin octapeptide by organic anion-transporting polypeptides OATP4 and OATP8 of rat and human liver. Gastroenterology. 2001;121:1185–1190. doi: 10.1053/gast.2001.28704. [DOI] [PubMed] [Google Scholar]
- 95.Shitara Y, Sato H, Sugiyama Y. Evaluation of drug–drug interaction in the hepatobiliary and renal transport of drugs. Annu. Rev. Pharmacol. Toxicol. 2005;45:689–723. doi: 10.1146/annurev.pharmtox.44.101802.121444. [DOI] [PubMed] [Google Scholar]
- 96.Letschert K, Komatsu M, Hummel-Eisenbeiss J, Keppler D. Vectorial transport of the peptide CCK-8 by double-transfected MDCKII cells stably expressing the organic anion transporter OATP1B3 (OATP8) and the export pump ABCC2. J. Pharmacol. Exp. Ther. 2005;313:549–556. doi: 10.1124/jpet.104.081224. [DOI] [PubMed] [Google Scholar]
- 97.Matsushima S, et al. Identification of the hepatic efflux transporters of organic anions using doubletransfected Madin-Darby canine kidney II cells expressing human organic anion-transporting polypeptide 1B1 (OATP1B1)/multidrug resistance associated protein 2, OATP1B1/multidrug resistance 1, and OATP1B1/breast cancer resistance protein. J. Pharmacol. Exp. Ther. 2005;314:1059–1067. doi: 10.1124/jpet.105.085589. [DOI] [PubMed] [Google Scholar]
- 98.Ishiguro N, et al. Predominant contribution of OATP1B3 to the hepatic uptake of telmisartan, an angiotensin II receptor antagonist, in humans. Drug Metab. Dispos. 2006;34:1109–1115. doi: 10.1124/dmd.105.009175. [DOI] [PubMed] [Google Scholar]
- 99.Noe J, Portmann R, Brun ME, Funk C. Substrate-dependent drug–drug interactions between gemfibrozil, fluvastatin and other organic anion transporting peptide (OATP) substrates on OATP1B1, OATP2B1, and OATP1B3. Drug Metab. Dispos. 2007;35:1308–1314. doi: 10.1124/dmd.106.012930. [DOI] [PubMed] [Google Scholar]
- 100.Zaher H, et al. Targeted disruption of murine organic anion-transporting polypeptide 1b2 (Oatp1b2/ Slco1b2) significantly alters disposition of prototypical drug substrates pravastatin and rifampin. Mol. Pharmacol. 2008;74:320–329. doi: 10.1124/mol.108.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu H, et al. Characterization of organic anion transporting polypeptide 1b2-null mice: essential role in hepatic uptake/toxicity of phalloidin and microcystin-LR. Toxicol. Sci. 2008;103:35–45. doi: 10.1093/toxsci/kfn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neuvonen PJ, Niemi M, Backman JT. Drug interactions with lipid-lowering drugs: mechanisms and clinical relevance. Clin. Pharmacol. Ther. 2006;80:565–581. doi: 10.1016/j.clpt.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 103.Shitara Y, Itoh T, Sato H, Li AP, Sugiyama Y. Inhibition of transporter-mediated hepatic uptake as a mechanism for drug–drug interaction between cerivastatin and cyclosporin A. J. Pharmacol. Exp. Ther. 2003;304:610–616. doi: 10.1124/jpet.102.041921. [DOI] [PubMed] [Google Scholar]
- 104.Pasanen MK, Neuvonen PJ, Niemi M. Global analysis of genetic variation in SLCO1B1. Pharmacogenomics. 2008;9:19–33. doi: 10.2217/14622416.9.1.19. [DOI] [PubMed] [Google Scholar]
- 105.Mwinyi J, Johne A, Bauer S, Roots I, Gerloff T. Evidence for inverse effects of OATP-C (SLC21A6) 5 and 1b haplotypes on pravastatin kinetics. Clin. Pharmacol. Ther. 2004;75:415–421. doi: 10.1016/j.clpt.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 106.Nishizato Y, et al. Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes: consequences for pravastatin pharmacokinetics. Clin. Pharmacol. Ther. 2003;73:554–565. doi: 10.1016/S0009-9236(03)00060-2. [DOI] [PubMed] [Google Scholar]
- 107.Chung JY, et al. Effect of OATP1B1 (SLCO1B1) variant alleles on the pharmacokinetics of pitavastatin in healthy volunteers. Clin. Pharmacol. Ther. 2005;78:342–350. doi: 10.1016/j.clpt.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 108.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet. Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 109.Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M. Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin. Pharmacol. Ther. 2007;82:726–733. doi: 10.1038/sj.clpt.6100220. [DOI] [PubMed] [Google Scholar]
- 110.Lee E, et al. Rosuvastatin pharmacokinetics and pharmacogenetics in white and Asian subjects residing in the same environment. Clin. Pharmacol. Ther. 2005;78:330–341. doi: 10.1016/j.clpt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 111.Niemi M, et al. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin. Pharmacol. Ther. 2005;77:468–478. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 112.Katz DA, et al. Organic anion transporting polypeptide 1B1 activity classified by SLCO1B1 genotype influences atrasentan pharmacokinetics. Clin. Pharmacol. Ther. 2006;79:186–196. doi: 10.1016/j.clpt.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 113.Xiang X, et al. Pharmacogenetics of SLCO1B1 gene and the impact of *1b and *15 haplotypes on irinotecan disposition in Asian cancer patients. Pharmacogenet. Genomics. 2006;16:683–691. doi: 10.1097/01.fpc.0000230420.05221.71. [DOI] [PubMed] [Google Scholar]
- 114.Oswald S, Scheuch E, Cascorbi I, Siegmund W. A LC-MS/MS method to quantify the novel cholesterol lowering drug ezetimibe in human serum, urine and feces in healthy subjects genotyped for SLCO1B1. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006;830:143–150. doi: 10.1016/j.jchromb.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 115. Link E, et al. SLCO1B1 variants and statin-induced myopathy — a genome wide study. N. Engl. J. Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. This manuscript represents the first genome-wide association study in the pharmacogenomics of, highlights the important role of, drug transporters in drug safety.
- 116.Uchida Y, Kamiie J, Ohtsuki S, Terasaki T. Multichannel liquid chromatography-tandem mass spectrometry cocktail method for comprehensive substrate characterization of multidrug resistance associated protein 4 transporter. Pharm. Res. 2007;24:2281–2296. doi: 10.1007/s11095-007-9453-7. [DOI] [PubMed] [Google Scholar]
- 117.Ishikawa T, et al. High-speed screening of human ATP-binding cassette transporter function and genetic polymorphisms: new strategies in pharmacogenomics. Methods Enzymol. 2005;400:485–510. doi: 10.1016/S0076-6879(05)00027-3. [DOI] [PubMed] [Google Scholar]
- 118.Keppler D, Jedlitschky G, Leier I. Transport function and substrate specificity of multidrug resistance protein. Methods Enzymol. 1998;292:607–616. doi: 10.1016/s0076-6879(98)92047-x. [DOI] [PubMed] [Google Scholar]
- 119.Hilgendorf C, et al. Expression of thirty-six drug transporter genes in human intestine, liver, kidney, and organotypic cell lines. Drug Metab. Dispos. 2007;35:1333–1340. doi: 10.1124/dmd.107.014902. [DOI] [PubMed] [Google Scholar]
- 120.Sauvant C, et al. Action of EGF and PGE2 on basolateral organic anion uptake in rabbit proximal renal tubules and hOAT1 expressed in human kidney epithelial cells. Am. J. Physiol. Renal Physiol. 2004;286:F774–F783. doi: 10.1152/ajprenal.00326.2003. [DOI] [PubMed] [Google Scholar]
- 121.Lee SH, Sinko PJ. siRNA — getting the message out. Eur. J. Pharm. Sci. 2006;27:401–410. doi: 10.1016/j.ejps.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 122.Yue W, Abe K, Brouwer KL. Knocking down breast cancer resistance protein (Bcrp) by adenoviral vector-mediated RNA interference (RNAi) in sandwich cultured rat hepatocytes: a novel tool to assess the contribution of Bcrp to drug biliary excretion. Mol. Pharm. 2009;6:134–143. doi: 10.1021/mp800100e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang W, et al. Silencing the breast cancer resistance protein expression and function in caco-2 cells using lentiviral vector-based short hairpin RNA. Drug Metab. Dispos. 2009;37:737–744. doi: 10.1124/dmd.108.023309. [DOI] [PubMed] [Google Scholar]
- 124.Keppler D. Uptake and efflux transporters for conjugates in human hepatocytes. Methods Enzymol. 2005;400:531–542. doi: 10.1016/S0076-6879(05)00029-7. [DOI] [PubMed] [Google Scholar]
- 125.Sasaki M, et al. Prediction of in vivo biliary clearance from the in vitro transcellular transport of organic anions across a double-transfected Madin-Darby canine kidney II monolayer expressing both rat organic anion transporting polypeptide 4 and multidrug resistance associated protein 2. Mol. Pharmacol. 2004;66:450–459. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 126.Sasaki M, Suzuki H, Ito K, Abe T, Sugiyama Y. Transcellular transport of organic anions across a double-transfected Madin-Darby canine kidney II cell monolayer expressing both human organic anion transporting polypeptide (OATP2/SLC21A6) and multidrug resistance-associated protein 2 (MRP2/ ABCC2) J. Biol. Chem. 2002;277:6497–6503. doi: 10.1074/jbc.M109081200. [DOI] [PubMed] [Google Scholar]
- 127.Cui Y, Konig J, Keppler D. Vectorial transport by double-transfected cells expressing the human uptake transporter SLC21A8 and the apical export pump ABCC2. Mol. Pharmacol. 2001;60:934–943. doi: 10.1124/mol.60.5.934. [DOI] [PubMed] [Google Scholar]
- 128.Bartholome K, et al. Data-based mathematical modeling of vectorial transport across doubletransfected polarized cells. Drug Metab. Dispos. 2007;35:1476–1481. doi: 10.1124/dmd.107.015636. [DOI] [PubMed] [Google Scholar]
- 129.Marion TL, Leslie EM, Brouwer KL. Use of sandwich-cultured hepatocytes to evaluate impaired bile acid transport as a mechanism of drug-induced hepatotoxicity. Mol. Pharm. 2007;4:911–918. doi: 10.1021/mp0700357. [DOI] [PubMed] [Google Scholar]
- 130.Liu X, et al. Biliary excretion in primary rat hepatocytes cultured in a collagen-sandwich configuration. Am. J. Physiol. 1999;277:G12–G21. doi: 10.1152/ajpgi.1999.277.1.G12. [DOI] [PubMed] [Google Scholar]
- 131.LeCluyse EL, Audus KL, Hochman JH. Formation of extensive canalicular networks by rat hepatocytes cultured in collagen-sandwich configuration. Am. J. Physiol. 1994;266:C1764–C1774. doi: 10.1152/ajpcell.1994.266.6.C1764. [DOI] [PubMed] [Google Scholar]
- 132.Hoffmaster KA, et al. P-glycoprotein expression, localization, and function in sandwich-cultured primary rat and human hepatocytes: relevance to the hepatobiliary disposition of a model opioid peptide. Pharm. Res. 2004;21:1294–1302. doi: 10.1023/b:pham.0000033018.97745.0d. [DOI] [PubMed] [Google Scholar]
- 133.Abe K, Bridges AS, Brouwer KL. Use of sandwich-cultured human hepatocytes to predict biliary clearance of angiotensin II receptor blockers and HMG-CoA reductase inhibitors. Drug Metab. Dispos. 2009;37:447–452. doi: 10.1124/dmd.108.023465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Klaassen CD, Lu H. Xenobiotic transporters: ascribing function from gene knockout and mutation studies. Toxicol. Sci. 2008;101:186–196. doi: 10.1093/toxsci/kfm214. [DOI] [PubMed] [Google Scholar]
- 135. Schinkel AH, et al. Disruption of the mouse Mdr1a P-glycoprotein gene leads to a deficiency in the blood–brain barrier and to increased sensitivity to drugs. Cell. 1994;77:491–502. doi: 10.1016/0092-8674(94)90212-7. This manuscript was the first study that demonstrated the important role of P-gp in the blood–brain barrier in the mouse and its role in determining drug sensitivity.
- 136.Polli JW, et al. An unexpected synergist role of P-glycoprotein and breast cancer resistance protein on the central nervous system penetration of the tyrosine kinase inhibitor lapatinib (N-{3-chloro4-[(3- fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl) ethy l]amino}methyl)-2-furyl]-4-quinazolinamine, GW572016) Drug Metab. Dispos. 2009;37:439–442. doi: 10.1124/dmd.108.024646. [DOI] [PubMed] [Google Scholar]
- 137.Zamek-Gliszczynski MJ, Kalvass JC, Pollack GM, Brouwer KL. Relationship between drug/ metabolite exposure and impairment of excretory transport function. Drug Metab. Dispos. 2009;37:386–390. doi: 10.1124/dmd.108.023648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li M, et al. Identification of interspecies difference in efflux transporters of hepatocytes from dog, rat, monkey and human. Eur. J. Pharm. Sci. 2008;35:114–126. doi: 10.1016/j.ejps.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 139.Takekuma Y, et al. Difference between pharmacokinetics of mycophenolic acid (MPA) in rats and that in humans is caused by different affinities of MRP2 to a glucuronized form. J. Pharm. Pharm. Sci. 2007;10:71–85. [PubMed] [Google Scholar]
- 140.Zamek-Gliszczynski MJ, et al. Differential involvement of Mrp2 (Abcc2) and Bcrp (Abcg2) in biliary excretion of 4-methylumbelliferyl glucuronide and sulfate in the rat. J. Pharmacol. Exp. Ther. 2006;319:459–467. doi: 10.1124/jpet.106.101840. [DOI] [PubMed] [Google Scholar]
- 141.Zamek-Gliszczynski MJ, et al. The important role of Bcrp (Abcg2) in the biliary excretion of sulfate and glucuronide metabolites of acetaminophen, 4-methylumbelliferone, and harmol in mice. Mol. Pharmacol. 2006;70:2127–2133. doi: 10.1124/mol.106.026955. [DOI] [PubMed] [Google Scholar]
- 142.Merino G, van Herwaarden AE, Wagenaar E, Jonker JW, Schinkel AH. Sex-dependent expression and activity of the ATP-binding cassette transporter breast cancer resistance protein (BCRP/ ABCG2) in liver. Mol. Pharmacol. 2005;67:1765–1771. doi: 10.1124/mol.105.011080. [DOI] [PubMed] [Google Scholar]
- 143.Vlaming ML, et al. Carcinogen and anticancer drug transport by Mrp2 in vivo: studies using Mrp2 (Abcc2) knockout mice. J. Pharmacol. Exp. Ther. 2006;318:319–327. doi: 10.1124/jpet.106.101774. [DOI] [PubMed] [Google Scholar]
- 144.Chu XY, et al. Characterization of mice lacking the multidrug resistance protein MRP2 (ABCC2) J. Pharmacol. Exp. Ther. 2006;317:579–589. doi: 10.1124/jpet.105.098665. [DOI] [PubMed] [Google Scholar]
- 145.van de Steeg E, et al. Methotrexate pharmacokinetics in transgenic mice with liver-specific expression of human organic anion-transporting polypeptide 1B1 (SLCO1B1) Drug Metab. Dispos. 2009;37:277–281. doi: 10.1124/dmd.108.024315. [DOI] [PubMed] [Google Scholar]
- 146.Hirano M, Maeda K, Shitara Y, Sugiyama Y. Drug–drug interaction between pitavastatin and various drugs via OATP1B1. Drug Metab. Dispos. 2006;34:1229–1236. doi: 10.1124/dmd.106.009290. [DOI] [PubMed] [Google Scholar]
- 147.Maeda K, Sugiyama Y. In: Drug Transporters. You G, Morris ME, editors. Hoboken, New York: John Wiley & Sons; 2007. pp. 557–588. [Google Scholar]
- 148.Hirano M, Maeda K, Shitara Y, Sugiyama Y. Contribution of OATP2 (OATP1B1) and OATP8 (OATP1B3) to the hepatic uptake of pitavastatin in humans. J. Pharmacol. Exp. Ther. 2004;311:139–146. doi: 10.1124/jpet.104.068056. [DOI] [PubMed] [Google Scholar]
- 149.Watanabe T, Kusuhara H, Maeda K, Shitara Y, Sugiyama Y. Physiologically based pharmacokinetic modeling to predict transporter-mediated clearance and distribution of pravastatin in humans. J. Pharmacol. Exp. Ther. 2009;328:652–662. doi: 10.1124/jpet.108.146647. [DOI] [PubMed] [Google Scholar]
- 150.Paine SW, Parker AJ, Gardiner P, Webborn PJ, Riley RJ. Prediction of the pharmacokinetics of atorvastatin, cerivastatin, and indomethacin using kinetic models applied to isolated rat hepatocytes. Drug Metab. Dispos. 2008;36:1365–1374. doi: 10.1124/dmd.107.019455. [DOI] [PubMed] [Google Scholar]
- 151.Poirier A, Funk C, Scherrmann JM, Lave T. Mechanistic modeling of hepatic transport from cells to whole body: application to napsagatran and fexofenadine. Mol. Pharm. 2009;6:1716–1733. doi: 10.1021/mp8002495. [DOI] [PubMed] [Google Scholar]
- 152.Takano A, et al. Evaluation of in vivo P-glycoprotein function at the blood–brain barrier among MDR1 gene polymorphisms by using 11C-verapamil. J. Nucl. Med. 2006;47:1427–1433. [PubMed] [Google Scholar]
- 153.Guhlmann A, et al. Noninvasive assessment of hepatobiliary and renal elimination of cysteinyl leukotrienes by positron emission tomography. Hepatology. 1995;21:1568–1575. [PubMed] [Google Scholar]
- 154.de Vries EF, et al. Can celecoxib affect P-glycoprotein-mediated drug efflux? A microPET study. Nucl. Med. Biol. 2008;35:459–466. doi: 10.1016/j.nucmedbio.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 155.Piwnica-Worms D, et al. Functional imaging of multidrug-resistant P-glycoprotein with an organotechnetium complex. Cancer Res. 1993;53:977–984. [PubMed] [Google Scholar]
- 156.Hendrikse NH, et al. In vivo imaging of hepatobiliary transport function mediated by multidrug resistance associated protein and P-glycoprotein. Cancer Chemother. Pharmacol. 2004;54:131–138. doi: 10.1007/s00280-004-0793-2. [DOI] [PubMed] [Google Scholar]
- 157.Cebecauerova D, et al. Dual hereditary jaundice: simultaneous occurrence of mutations causing Gilbert’s and Dubin–Johnson syndrome. Gastroenterology. 2005;129:315–320. doi: 10.1053/j.gastro.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 158.Bujanover Y, Bar-Meir S, Hayman I, Baron J. 99mTc-HIDA cholescintigraphy in children with Dubin–Johnson syndrome. J. Pediatr. Gastroenterol. Nutr. 1983;2:311–312. [PubMed] [Google Scholar]
- 159.Ghibellini G, Leslie EM, Pollack GM, Brouwer KL. Use of Tc-99m mebrofenin as a clinical probe to assess altered hepatobiliary transport: integration of in vitro, pharmacokinetic modeling, and simulation studies. Pharm. Res. 2008;25:1851–1860. doi: 10.1007/s11095-008-9597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ghibellini G, et al. In vitro–in vivo correlation of hepatobiliary drug clearance in humans. Clin. Pharmacol. Ther. 2007;81:406–413. doi: 10.1038/sj.clpt.6100059. [DOI] [PubMed] [Google Scholar]
- 161.Ghibellini G, Johnson BM, Kowalsky RJ, Heizer WD, Brouwer KL. A novel method for the determination of biliary clearance in humans. AAPS J. 2004;6:e33. doi: 10.1208/aapsj060433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Michael M, et al. Relationship of hepatic functional imaging to irinotecan pharmacokinetics and genetic parameters of drug elimination. J. Clin. Oncol. 2006;24:4228–4235. doi: 10.1200/JCO.2005.04.8496. [DOI] [PubMed] [Google Scholar]
- 163.Wong M, et al. Predictors of vinorelbine pharmacokinetics and pharmacodynamics in patients with cancer. J. Clin. Oncol. 2006;24:2448–2455. doi: 10.1200/JCO.2005.02.1295. [DOI] [PubMed] [Google Scholar]
- 164.Zhang Y, Benet LZ. The gut as a barrier to drug absorption: combined role of cytochrome P450 3A and P-glycoprotein. Clin. Pharmacokinet. 2001;40:159–168. doi: 10.2165/00003088-200140030-00002. [DOI] [PubMed] [Google Scholar]
- 165.Wacher VJ, Wu CY, Benet LZ. Overlapping substrate specificities and tissue distribution of cytochrome P450 3A and P-glycoprotein: implications for drug delivery and activity in cancer chemotherapy. Mol. Carcinog. 1995;13:129–134. doi: 10.1002/mc.2940130302. [DOI] [PubMed] [Google Scholar]
- 166.Benet LZ, Cummins CL. The drug efflux– metabolism alliance: biochemical aspects. Adv. Drug Deliv. Rev. 2001;50 Suppl. 1:S3–S11. doi: 10.1016/s0169-409x(01)00178-8. [DOI] [PubMed] [Google Scholar]
- 167.Wacher VJ, Salphati L, Benet LZ. Active secretion and enterocytic drug metabolism barriers to drug absorption. Adv. Drug Deliv. Rev. 2001;46:89–102. doi: 10.1016/s0169-409x(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 168.Lown KS, et al. Role of intestinal P-glycoprotein (mdr1) in interpatient variation in the oral bioavailability of cyclosporine. Clin. Pharmacol. Ther. 1997;62:248–260. doi: 10.1016/S0009-9236(97)90027-8. [DOI] [PubMed] [Google Scholar]
- 169.Gomez DY, Wacher VJ, Tomlanovich SJ, Hebert MF, Benet LZ. The effects of ketoconazole on the intestinal metabolism and bioavailability of cyclosporine. Clin. Pharmacol. Ther. 1995;58:15–19. doi: 10.1016/0009-9236(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 170.Shitara Y, Horie T, Sugiyama Y. Transporters as a determinant of drug clearance and tissue distribution. Eur. J. Pharm. Sci. 2006;27:425–446. doi: 10.1016/j.ejps.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 171.Kusuhara H, Sugiyama Y. In vitro–in vivo extrapolation of transporter-mediated clearance in the liver and kidney. Drug Metab. Pharmacokinet. 2009;24:37–52. doi: 10.2133/dmpk.24.37. [DOI] [PubMed] [Google Scholar]
- 172.Amidon GL, Lennernas H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm. Res. 1995;12:413–420. doi: 10.1023/a:1016212804288. [DOI] [PubMed] [Google Scholar]
- 173.Wu CY, Benet LZ. Predicting drug disposition via application of BCS: transport/absorption/ elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm. Res. 2005;22:11–23. doi: 10.1007/s11095-004-9004-4. [DOI] [PubMed] [Google Scholar]
- 174.US Department of Health and Human Services Food, Food and Drug Administration & Center for Drug Evaluation and Research (CDER) Guidance for Industry. Waiver of In Vivo Bioavailability and Bioequivalence Studies for Immediate-Release Solid Oral Dosage Forms Based on a Biopharmaceutics Classification System. 2000 US FDA website [online], http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM070246.pdf.
- 175.Watanabe T, et al. Prediction of the hepatic and renal clearance of transporter substrates in rats using in vitro uptake experiments. Drug Metab. Dispos. 2009;37:1471–1479. doi: 10.1124/dmd.108.026062. [DOI] [PubMed] [Google Scholar]
- 176.Shugarts S, Benet LZ. The role of transporters in the pharmacokinetics of orally administered drugs. Pharm. Res. 2009;26:2039–2054. doi: 10.1007/s11095-009-9924-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chang C, Ekins S, Bahadduri P, Swaan PW. Pharmacophore-based discovery of ligands for drug transporters. Adv. Drug Deliv. Rev. 2006;58:1431–1450. doi: 10.1016/j.addr.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gombar VK, Polli JW, Humphreys JE, Wring SA, Serabjit-Singh CS. Predicting P-glycoprotein substrates by a quantitative structure–activity relationship model. J. Pharm. Sci. 2004;93:957–968. doi: 10.1002/jps.20035. [DOI] [PubMed] [Google Scholar]
- 179.Ha SN, Hochman J, Sheridan RP. Mini review on molecular modeling of P-glycoprotein (Pgp) Curr. Top. Med. Chem. 2007;7:1525–1529. doi: 10.2174/156802607782194806. [DOI] [PubMed] [Google Scholar]
- 180.Ekins S, et al. In vitro and pharmacophore-based discovery of novel hPEPT1 inhibitors. Pharm. Res. 2005;22:512–517. doi: 10.1007/s11095-005-2505-y. [DOI] [PubMed] [Google Scholar]
- 181.Ekins S, Ecker GF, Chiba P, Swaan PW. Future directions for drug transporter modelling. Xenobiotica. 2007;37:1152–1170. doi: 10.1080/00498250701646341. [DOI] [PubMed] [Google Scholar]
- 182.US Food and Drug Administration. Highlights of Prescribing Information Tykerb (Lapatinib) 2007 US FDA website [online], http://www.accessdata.fda.gov/drugsatfda_docs/label/2010/022059s3s6lbl.pdf.
- 183.US Food and Drug Administration. Drug Development and Drug Interactions. 2009 US FDA website [online], http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm080499.htm.
- 184.Tucker GT, Houston JB, Huang SM. EUFEPS conference report. Optimising drug development: strategies to assess drug metabolism/transporter interaction potential — towards a consensus. European Federation of Pharmaceutical Sciences. Eur. J. Pharm. Sci. 2001;13:417–428. doi: 10.1016/s0928-0987(01)00148-8. [DOI] [PubMed] [Google Scholar]
- 185.Buckman S, Huang SM, Murphy S. Medical product development and regulatory science for the 21st century: the critical path vision and its impact on health care. Clin. Pharmacol. Ther. 2007;81:141–144. doi: 10.1038/sj.clpt.6100085. [DOI] [PubMed] [Google Scholar]
- 186.US Department of Health and Human Services; Food and Drug Administration; Center for Drug Evaluation and Research (CDER); Center for Biologics Evaluation and Research (CBER) Guidance for Industry. Exposure–Response Relationships — Study Design, Data Analysis and Regulatory Applications. 2003 US FDA website [online], http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatory Information/Guidances/UCM072109.pdf.
- 187.Huang SM, Temple R. Is this the drug or dose for you? Impact and consideration of ethnic factors in global drug development, regulatory review, and clinical practice. Clin. Pharmacol. Ther. 2008;84:287–294. doi: 10.1038/clpt.2008.144. [DOI] [PubMed] [Google Scholar]
- 188.Fenner KS, et al. Drug–drug interactions mediated through P-glycoprotein: clinical relevance and in vitro–in vivo correlation using digoxin as a probe drug. Clin. Pharmacol. Ther. 2009;85:173–181. doi: 10.1038/clpt.2008.195. [DOI] [PubMed] [Google Scholar]
- 189.Urquhart BL, et al. Breast cancer resistance protein (ABCG2) and drug disposition: intestinal expression, polymorphisms and sulfasalazine as an in vivo probe. Pharmacogenet. Genomics. 2008;18:439–448. doi: 10.1097/FPC.0b013e3282f974dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kitamura S, Maeda K, Wang Y, Sugiyama Y. Involvement of multiple transporters in the hepatobiliary transport of rosuvastatin. Drug Metab. Dispos. 2008;36:2014–2023. doi: 10.1124/dmd.108.021410. [DOI] [PubMed] [Google Scholar]
- 191.Zhang Y, et al. BCRP transports dipyridamole and is inhibited by calcium channel blockers. Pharm. Res. 2005;22:2023–2034. doi: 10.1007/s11095-005-8384-4. [DOI] [PubMed] [Google Scholar]
- 192.Ito K, et al. Prediction of pharmacokinetic alterations caused by drug–drug interactions: metabolic interaction in the liver. Pharmacol. Rev. 1998;50:387–412. [PubMed] [Google Scholar]
- 193. Kanamitsu S, Ito K, Sugiyama Y. Quantitative prediction of in vivo drug–drug interactions from in vitro data based on physiological pharmacokinetics: use of maximum unbound concentration of inhibitor at the inlet to the liver. Pharm. Res. 2000;17:336–343. doi: 10.1023/a:1007509324428. This manuscript provides methodologies for the quantitative prediction of in vivo DDIs from in vitro studies of drug–transporter interactions.
- 194.Mikkaichi T, et al. Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney. Proc. Natl Acad. Sci. USA. 2004;101:3569–3574. doi: 10.1073/pnas.0304987101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nozaki Y, et al. Species difference in the inhibitory effect of nonsteroidal anti-inflammatory drugs on the uptake of methotrexate by human kidney slices. J. Pharmacol. Exp. Ther. 2007;322:1162–1170. doi: 10.1124/jpet.107.121491. [DOI] [PubMed] [Google Scholar]
- 196.Maeda A, et al. Evaluation of the interaction between nonsteroidal anti-inflammatory drugs and methotrexate using human organic anion transporter 3-transfected cells. Eur. J. Pharmacol. 2008;596:166–172. doi: 10.1016/j.ejphar.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 197.Letschert K, Keppler D, Konig J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8) Pharmacogenetics. 2004;14:441–452. doi: 10.1097/01.fpc.0000114744.08559.92. [DOI] [PubMed] [Google Scholar]
- 198.Sanna S, et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum. Mol. Genet. 2009;18:2711–2718. doi: 10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Treiber A, Schneiter R, Hausler S, Stieger B. Bosentan is a substrate of human OATP1B1 and OATP1B3: inhibition of hepatic uptake as the common mechanism of its interactions with cyclosporin A, rifampicin, and sildenafil. Drug Metab. Dispos. 2007;35:1400–1407. doi: 10.1124/dmd.106.013615. [DOI] [PubMed] [Google Scholar]
- 200.Matsushima S, Maeda K, Ishiguro N, Igarashi T, Sugiyama Y. Investigation of the inhibitory effects of various drugs on the hepatic uptake of fexofenadine in humans. Drug Metab. Dispos. 2008;36:663–669. doi: 10.1124/dmd.107.017814. [DOI] [PubMed] [Google Scholar]
- 201.Busti AJ, et al. Effects of atazanavir/ritonavir or fosamprenavir/ritonavir on the pharmacokinetics of rosuvastatin. J. Cardiovasc. Pharmacol. 2008;51:605–610. doi: 10.1097/FJC.0b013e31817b5b5a. [DOI] [PubMed] [Google Scholar]
- 202.Obach RS, et al. The utility of in vitro cytochrome P450 inhibition data in the prediction of drug–drug interactions. J. Pharmacol. Exp. Ther. 2006;316:336–348. doi: 10.1124/jpet.105.093229. [DOI] [PubMed] [Google Scholar]
- 203.Wire MB, Shelton MJ, Studenberg S. Fosamprenavir: clinical pharmacokinetics and drug interactions of the amprenavir prodrug. Clin. Pharmacokinet. 2006;45:137–168. doi: 10.2165/00003088-200645020-00002. [DOI] [PubMed] [Google Scholar]
- 204.Hedman M, Neuvonen PJ, Neuvonen M, Holmberg C, Antikainen M. Pharmacokinetics and pharmacodynamics of pravastatin in pediatric and adolescent cardiac transplant recipients on a regimen of triple immunosuppression. Clin. Pharmacol. Ther. 2004;75:101–109. doi: 10.1016/j.clpt.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 205.Simonson SG, et al. Rosuvastatin pharmacokinetics in heart transplant recipients administered an anti rejection regimen including cyclosporine. Clin. Pharmacol. Ther. 2004;76:167–177. doi: 10.1016/j.clpt.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 206.US Food and Drug Administration. Highlights of Prescribing Information Livalo (Pitavastatin) 2009 US FDA website [online], http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/022363s000lbl.pdf.
- 207.Zheng HX, Huang Y, Frassetto LA, Benet LZ. Elucidating rifampin’s inducing and inhibiting effects on glyburide pharmacokinetics and blood glucose in healthy volunteers: unmasking the differential effects of enzyme induction and transporter inhibition for a drug and its primary metabolite. Clin. Pharmacol. Ther. 2009;85:78–85. doi: 10.1038/clpt.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.US Food and Drug Administration. Highlights of Prescribing Information: Tracleer (Bosentan) 2001 US FDA website [online], http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021290s012lbl.pdf.
- 209.Kiser JJ, et al. Drug/Drug interaction between lopinavir/ritonavir and rosuvastatin in healthy volunteers. J. Acquir. Immune Defic. Syndr. 2008;47:570–578. doi: 10.1097/QAI.0b013e318160a542. [DOI] [PubMed] [Google Scholar]
- 210.Li M, Anderson GD, Wang J. Drug–drug interactions involving membrane transporters in the human kidney. Expert Opin. Drug Metab. Toxicol. 2006;2:505–532. doi: 10.1517/17425255.2.4.505. [DOI] [PubMed] [Google Scholar]
- 211. Cundy KC. Clinical pharmacokinetics of the antiviral nucleotide analogues cidofovir and adefovir. Clin. Pharmacokinet. 1999;36:127–143. doi: 10.2165/00003088-199936020-00004. This article provides an excellent overview of clinical DDIs in the kidney.
- 212.Laskin OL, et al. Effects of probenecid on the pharmacokinetics and elimination of acyclovir in humans. Antimicrob. Agents Chemother. 1982;21:804–807. doi: 10.1128/aac.21.5.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Somogyi A, Stockley C, Keal J, Rolan P, Bochner F. Reduction of metformin renal tubular secretion by cimetidine in man. Br. J. Clin. Pharmacol. 1987;23:545–551. doi: 10.1111/j.1365-2125.1987.tb03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Somogyi A, Muirhead M. Pharmacokinetic interactions of cimetidine 1987. Clin. Pharmacokinet. 1987;12:321–366. doi: 10.2165/00003088-198712050-00002. [DOI] [PubMed] [Google Scholar]
- 215.Somogyi AA, Bochner F, Sallustio BC. Stereoselective inhibition of pindolol renal clearance by cimetidine in humans. Clin. Pharmacol. Ther. 1992;51:379–387. doi: 10.1038/clpt.1992.37. [DOI] [PubMed] [Google Scholar]
- 216.Feng B, et al. Effect of human renal cationic transporter inhibition on the pharmacokinetics of varenicline, a new therapy for smoking cessation: an in vitro–in vivo study. Clin. Pharmacol. Ther. 2008;83:567–576. doi: 10.1038/sj.clpt.6100405. [DOI] [PubMed] [Google Scholar]
- 217.Shiga T, Hashiguchi M, Urae A, Kasanuki H, Rikihisa T. Effect of cimetidine and probenecid on pilsicainide renal clearance in humans. Clin. Pharmacol. Ther. 2000;67:222–228. doi: 10.1067/mcp.2000.104018. [DOI] [PubMed] [Google Scholar]
- 218.Tsuruoka S, et al. Severe arrhythmia as a result of the interaction of cetirizine and pilsicainide in a patient with renal insufficiency: first case presentation showing competition for excretion via renal multidrug resistance protein 1 and organic cation transporter 2. Clin. Pharmacol. Ther. 2006;79:389–396. doi: 10.1016/j.clpt.2005.12.302. [DOI] [PubMed] [Google Scholar]
- 219.Abel S, Nichols DJ, Brearley CJ, Eve MD. Effect of cimetidine and ranitidine on pharmacokinetics and pharmacodynamics of a single dose of dofetilide. Br. J. Clin. Pharmacol. 2000;49:64–71. doi: 10.1046/j.1365-2125.2000.00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rameis H. Quinidine–digoxin interaction: are the pharmacokinetics of both drugs altered? Int. J. Clin. Pharmacol. Ther. Toxicol. 1985;23:145–153. [PubMed] [Google Scholar]
- 221.Hager WD, et al. Digoxin–quinidine interaction pharmacokinetic evaluation. N. Engl. J. Med. 1979;300:1238–1241. doi: 10.1056/NEJM197905313002202. [DOI] [PubMed] [Google Scholar]
- 222.Ding R, et al. Substantial pharmacokinetic interaction between digoxin and ritonavir in healthy volunteers. Clin. Pharmacol. Ther. 2004;76:73–84. doi: 10.1016/j.clpt.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 223.US Food and Drug Administration. NDA 21–913 Dronedarone HCl. 2006 US FDA website [online], http://www.accessdata.fda.gov/drugsatfda_docs/nda/2009/022425s000_ClinPharm_P2.pdf.
- 224.Jerling M. Clinical pharmacokinetics of ranolazine. Clin. Pharmacokinet. 2006;45:469–491. doi: 10.2165/00003088-200645050-00003. [DOI] [PubMed] [Google Scholar]
- 225.Kruijtzer CM, et al. Increased oral bioavailability of topotecan in combination with the breast cancer resistance protein and P-glycoprotein inhibitor GF120918. J. Clin. Oncol. 2002;20:2943–2950. doi: 10.1200/JCO.2002.12.116. [DOI] [PubMed] [Google Scholar]