Abstract
Critical to homeostasis of blood cell production by hematopoietic stem/progenitor (HSC/P) cells is the regulation of HSC/P retention within the bone marrow microenvironment and migration between the bone marrow and the blood. Key extracellular regulatory elements for this process have been defined (cell–cell adhesion, growth factors, chemokines), but the mechanism by which HSC/P cells reconcile multiple external signals has not been elucidated. Rac and related small GTPases are candidates for this role and were studied in HSC/P deficient in Rac2, a hematopoietic cell-specific family member. Rac2 appears to be critical for HSC/P adhesion both in vitro and in vivo, whereas a compensatory increase in Cdc42 activation regulates HSC/P migration. This genetic analysis provides physiological evidence of cross-talk between GTPase proteins and suggests that a balance of these two GTPases controls HSC/P adhesion and mobilization in vivo.
Multipotential hematopoietic stem and progenitor (HSC/P) cells reside within the hematopoietic microenvironment (HM) located in the bone medullary cavity in mammals (1). Adhesion and localization of HSC/P in this HM have been shown to play a critical role in the maintenance of stem cell survival, proliferation, and function (2). With the use of a variety of indirect methods, such as stimulation of the entry of HSC/P into the blood from the marrow space (called mobilization) or inhibition of homing into the bone marrow space, several integrin adhesion molecules have been implicated in HSC/P localization in the HM (3–6). Pharmacologically induced mobilization and collection of mobilized HSC/P from the blood have gained wide therapeutic applications in stem cell transplantation protocols. Whether active cell movement (i.e., migration) or change in adhesive interactions or both play a dominant role in mobilization has not been clarified.
Rho-related small GTPases, including Rho, Rac, and Cdc42, are known to regulate cell shape changes, movement and adhesion in multiple mammalian cell types (reviewed in ref. 7). Furthermore, Rho GTPases have been demonstrated to activate a number of signal transduction pathways involved in cell cycle progression, gene expression, cell survival, and Ras transformation (8, 9). Biochemical pathways controlling the adhesion, migration, or mobilization of HSC/P cells have not been completely defined, and to date GTPases have not been implicated in HSC/P function. However, a member of the Rac family, Rac2, has recently been implicated in the migration of other, more differentiated hematopoietic cells in mice (10, 11), and a mutant of Rac2 has been implicated in a human phagocyte immunodeficiency (12, 13). We used mice deficient in Rac2 and retroviral-mediated gene transfer to study the role of Cdc42 and Rac in hematopoietic HSC/P cell adhesion and migration.
Materials and Methods
Hematopoietic Cell Purification.
In all experiments, 6- to 10-week-old age- and gender-matched 129Sv/C57BL6 mice or 129Sv inbred mice were used, and littermates were analyzed in parallel wherever possible. Mature cell lineage antigen-negative (Lin−) cells were enriched by immuno-magnetic negative selection (MACS; Miltenyi Biotech, Auburn, CA), with the use of a mixture of purified rat anti-mouse mAbs specific for the mature cell lineage antigens CD45R (B220, Clone RA3–6B2), Gr-1 (Ly-6G, Clone RB6-8C5), CD4 (L3T4, Clone RM4-5), CD8a (Ly-2, Clone 53-6.7), TER119 (TER119), and Mac-1 (CD11b, Clone M1/70) (all purchased from PharMingen). The nonmagnetic Lin− fraction was collected, washed, and counted. The cells were then incubated with rat anti-mouse CD32/CD16 to avoid nonspecific antibody binding, after which they were stained with fluoresceinated (FITC) rat anti-mouse CD117 (c-Kit) and phycoerythrin-conjugated rat anti-mouse Sca-1 (both from PharMingen). Negative control cells were stained with phycoerythrin-conjugated IgG2a and FITC-conjugated IgG2b. Based on these controls, Lin−c-Kit+Sca-1+ cells were isolated by sorting with a fluorescence-activated cell sorter (FACStar Plus; Becton Dickinson) under sterile conditions. To avoid contamination of different cell types, the bright population was gated. The purity of Lin−c-Kit+Sca-1+ cells thus obtained was >90%. Reverse transcription–PCR was performed with the use of primers and methods described (11).
Real-Time PCR Analysis.
Expression of the receptor for G-CSF was analyzed by real-time PCR (14), with the use of duplicate samples pooled from seven animals. For G-CSF receptor expression, CACCAGCTTCATCCTAAAGAGCTT (forward primer) and TTGCCACACAATCCGGG (reverse primer) were used. The TaqMan probe was 6FAM-CTGACAGTCGGCGCGGCTCCT-TAMRA (6FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine). TaqMan rodent glyceraldehyde-3-phosphate dehydrogenase control reagents were used as the control; all of these were purchased from Applied Biosystems. For CXCR4 receptor expression, GGTGATCCTGGTCATGGGTT (forward primer) and CTGACAGGTGCAGCCGGTA (reverse primer) were used. The TaqMan probe was 6FAM-TGTCCGTCATGCTCCTTAGCTTCTTCTGG-TAMRA. Data are expressed as the cycle threshold (Ct, the cycle at which mRNA is initially detected).
Cell Adhesion, Migration, and Mobilization Assays.
Day 12 colony-forming units-spleen (CFU-S12) and in vitro colony-forming units (CFU-C) in peripheral blood [assays were as described (3)] were assayed 5 days after the beginning of treatment with 250 μg/kg human G-CSF given at 12-h intervals. Animals were bled 12 h after the last injection and either plated (CFU-C) or injected into secondary recipients (CFU-S12). CFU-S12 in peripheral blood after treatment with G-CSF alone or with both G-CSF and anti-α4β1 antibody [purified anti-mouse CD49e antibody (R1–2; PharMingen) at 2 mg/kg/day for 3 days] were counted as described (15). Animals were killed the day after the third dose.
Adhesion of Lin−c-Kit+Sca-1+ bone marrow cells was assayed as described (4). Briefly, nontissue culture plates were coated with fibronectin (FN) fragments (H-296, which contains the VLA-4 binding site; CH-271, which contains the VLA-5 binding site) at 8 μg/cm2 or BSA (as control) overnight at 4°C. The plates were subsequently blocked with 2% BSA for 30 min at room temperature. A total of 1 × 105 wild-type (WT) or Rac2−/− cells in RPMI 1640 medium containing 10% FBS were then allowed to adhere to the coated plates for 1 h at 37°C. After incubation, we collected nonadherent cells by carefully rinsing the plates multiple times with medium. Adherent cells are harvested by vigorously rinsing the plates with PBS. The cells are counted with a hemocytometer and replated in CFU assay.
Migration assays were performed in transwells as described (16). All assays were performed in triplicate. Briefly, 100 μl of serum-free chemotaxis buffer (RPMI 1640 medium, 0.5% crystallized deionized BSA) (Calbiochem) containing 2 × 105 Lin−c-Kit+Sca-1+ cells was added to the upper chamber of a 5-μm-pore filter (Transwell, 24-well cell clusters; Costar), and 0.6 ml of serum-free chemotaxis buffer with various concentrations of stromal-derived factor-1 (SDF-1) was added to the lower chamber. After 4 h of incubation at 37°C in 5% CO2, the upper chamber was carefully removed, and the cells in the bottom chamber were resuspended and divided into aliquots for cell enumeration and CFU assay.
Motility of Lin−c-Kit+Sca-1+ cells was also directly observed by time lapse imaging of cells exposed to a gradient of 0–100 nM SDF-1 in a Dunn chemotaxis chamber (Weber Scientific, Surrey, U.K.) (17) as described (10). Lin−c-Kit+Sca-1+ cells (2–5 × 104 cells in 10 ml of Hanks' balanced salt solution) were applied to glass coverslips coated with fibronectin fragment CH-296 as described above and allowed to adhere for 10 min at 37°C. The coverslips were mounted on the Dunn chamber, the inner well of which was filled with Hanks' balanced salt solution, and the outer well was filled with Hanks' balanced salt solution/SDF-1. The chamber was sealed and mounted on the stage of a Nikon Diaphot 300 inverted microscope equipped with differential interference contrast optics. The chamber temperature was maintained at 37°C with a stage heater (Instec Instruments, Boulder, CO). The chamber was allowed to equilibrate for 20 min to allow a stable gradient to form. Images were recorded digitally at 15-s intervals with a Spot RT cooled charge-coupled device camera. Images were collected for 1 h. The microscope was calibrated with the use of the grating of a hemocytometer. Tracks of the centroids of individual cells were plotted over a 10-min segment of the recording with the use of metamorph software (Universal Imaging, Brandywine, PA). The scalar speed of movement was calculated from the total distance traveled over 10 min. In four experiments >250 cells were analyzed for each genotype. Cells moving at >2 μm/min were considered to show a motile response. The frequency of WT HSC/P cells moving in this assay was much lower than that observed for WT bone marrow neutrophils (35%; see ref. 10) but was comparable to our observations of mast cells with this assay (S.J.A., F.-C.Y., and D.A.W., unpublished observations).
Glutathione S-transferase-p21-Activated Kinase-1 (GST-PAK1) p21-Binding Domain Pull-Down Assay.
PAK1 p21-binding domain (PBD)-GST was expressed in Escherichia coli strain BL21 and purified as described (18). Purified Lin−c-Kit+ bone marrow cells (1 × 106 cells per lane) were treated with 100 ng/ml of SDF-1 for 5 min, mixed with cold PBS, and pelleted. The pellets were resuspended in PBS, lysed, and clarified as described. For in vitro guanine nucleotide binding, cell lysates were incubated for 15 min at 30°C in the presence of 10 mM EDTA and 100 mM GTPγS or 1 mM GDP for nucleotide exchange. The loading was stopped by the addition of MgCl2 to 30 mM (19). The crude or guanine nucleotide-loaded cell lysates (100 μl) were added to 200 μl of binding buffer (25 mM Tris⋅HCl, pH 7.5/1 mM DTT/30 mM MgCl2/40 mM NaCl/0.5% Nonidet P-40) with 10 μg PAK1 PBD-GST recombinant protein and 5 μl of glutathione-Sepharose 4B beads. The binding reaction was incubated for 1 h at 4°C and then washed two times with washing buffer (25 mM Tris⋅HCl, pH 7.5/1 mM DTT/30 mM MgCl2/40 mM NaCl/0.5% Nonidet P-40) and three times with washing buffer without detergent. The bead pellets were finally resuspended in 15 μl of Laemmli sample buffer. Each sample was analyzed on 12% SDS/PAGE and blotted by specific antibodies for Rac1 (1:2,000, clone 23A8; Upstate Biotechnology, Lake Placid, NY) and Cdc42 (1:2,000, sc87G; Santa Cruz Biotechnology). The secondary antibodies were horseradish peroxidase conjugated (1:2,500; New England Biolabs). The immunoblots were detected with a New England Biolabs Luminol kit and Kodak Biomax film.
Retrovirus Transduction.
Transduction of hematopoietic stem/progenitor cells was performed by published methods (20), with the use of vectors described (see references in specific figure legends). Briefly, mice were injected with 5-fluorouracil (150 mg/kg; Sigma). Bone marrow cells were harvested 48 h after injection. Mononuclear cells obtained after density gradient centrifugation were cultured for 2 days in the presence of stem cell factor, G-CSF, and megakaryocyte growth and development factor (all at 100 ng/ml, all from Amgen Biologicals). The cells were infected with MIEG3, MIEG-FR2, and T17NCdc42 virus supernatant on FN CH-296-coated plates. On day 3 after infection, the cells were stained with anti-mouse CD117 (c-Kit) phycoerythrin (PharMingen), and the c-Kit+/green fluorescent protein-positive cells were isolated by fluorescence-activated cell sorter (FACStar Plus; Becton Dickinson) under sterile conditions. Transfection efficiency ranged from 30% to 80%, depending on the vector. The double-positive cells were used for biological assays.
Results
Rac2-Deficient HSC/P Demonstrate Defective Cell Adhesion and Enhanced Mobilization.
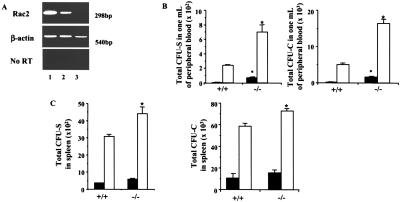
HSC/P cells (c-kit+Sca-1+Lin−, which make up <1% of the bone marrow cells but are highly enriched in HSC/P) were isolated by immunodepletion and fluorescence-activated cell sorting from Rac2−/− mice (10) and analyzed for the expression of Rac2. As demonstrated by reverse transcription–PCR, c-kit+Sca-1+Lin− cells from WT mice express Rac2 mRNA, whereas the 298-bp Rac2 amplicon (11) is absent from cells derived from Rac2−/− mice (Fig. 1A). Compared with WT littermates, the blood of Rac2−/− mice contained significantly higher numbers of these cells, measured either by CFU-C assays or CFU-S12 assays (Fig. 1B). The increased number of HSC/P in the circulation contrasts with the bone marrow contents of HSC/P in Rac2−/− mice, which are identical to WT mice (data not shown). A slight but insignificant increase in the numbers of CFU-S12 was seen in the spleen at baseline (Fig. 1C). Egress of HSC/P from the bone marrow into the blood can be induced by the administration of G-CSF in a process known as mobilization. Rac2−/− mice injected with G-CSF mobilized HSC/P into the blood in 3- to 4-fold greater numbers than similarly treated WT mice (Fig. 1B). The number of CFU-S12 was also significantly increased in the spleen of Rac2−/− mice compared with WT, but this increase was to a lesser extent than in peripheral blood (Fig. 1C). Because Papayannopoulou et al. (21) have also reported differences in the in vivo effects of antibody to integrin α4β1 on splenic vs. bone marrow content of progenitors, these data suggest that a different mechanism may mediate the adhesion or entrapment of primitive cells in the spleen compared with the bone marrow. Real-time PCR confirmed equivalent expression of the receptor for G-CSF, for which no antibody reagents are available to assess protein expression (Ct = 23.2 vs. 23.4, WT vs. Rac2−/−, mean of two determinations).
Figure 1.
Enhanced mobilization of hematopoietic stem and progenitor cells from the bone marrow of Rac2−/− mice. (A) Reverse transcription–PCR analysis for expression of Rac2 mRNA in c-kit+Sca-1+Lin− cells from WT and Rac2−/− mice. Lane 1: WT neutrophils as a positive control. Lane 2: WT HSC/P cells. Lane 3: Rac2−/− HSC/P cells. (B) Enumeration of day CFU-S12 and CFU-C in peripheral blood at baseline or after treatment with 250 μg/kg human G-CSF. Closed bars, baseline; open bars, G-CSF-treated mice. Mean ± SD, *, P < 0.01. CFU colonies contained mixed myeloid, pure myeloid, or megakaryocytic lineages. (C) Enumeration of CFU-S12 and CFU-C in spleen after treatment with G-CSF. Closed bars, baseline; open bars, G-CSF-treated mice. Mean ± SD; *, P < 0.05.
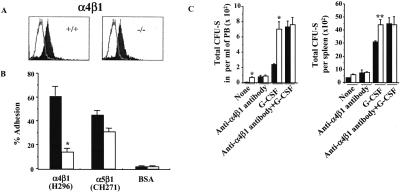
Integrin receptors have been implicated in the adhesion of HSC to proteins in the HM (reviewed in ref. 2). Because migration out of the marrow could reflect diminished adhesion of the cells in the HM, and because the β1 integrins have been demonstrated by our laboratory (3, 4) and by other investigators (5, 6, 22) to mediate adhesion of these cells, further analysis of the expression and function of integrins was carried out. Expression of the integrins α4β1 (Fig. 2A) and α5β1 (not shown), as determined by flow cytometric analysis, was not affected by the lack of Rac2 expression, whereas adhesion of HSC/P cells derived from Rac2−/− mice to the recombinant FN peptide H296 (4), containing specific ligand sequences, CS1, of the integrin α4β1 was significantly reduced compared with WT cells (Fig. 2B). The defect in adhesion via α4β1 appears to be a specific consequence of Rac2 deficiency, inasmuch as adhesion of Rac2−/− cells is restored to normal after expression of the Rac2 cDNA via the retrovirus vector MIEG3-FR2 (13) (WT, 50.0 ± 2% adhesion; Rac2−/−, 17.6 ± 4%; Rac2−/− transduced with MIEG3-FR2, 46.0 ± 7%; P < 0.01, WT vs. Rac2−/−; P > 0.05, Rac2-transduced vs. WT). Adhesion via another β1 integrin expressed in HSC/P cells, α5β1, to an Arg-Gly-Asp-Ser (RGDS) sequence of FN contained in the recombinant peptide CH-271 (4) was lower but not significantly different from WT (Fig. 2B, P > 0.05).
Figure 2.
Effect of Rac2 deficiency on integrin-mediated cell adhesion. (A) Expression of integrin α4β1 (VLA-4) analyzed by flow, one representative experiment of three with identical results, with biotin-conjugated anti-α4 mAb. The solid curve represents expression of α4β1 as detected by mAb; open tracing represents an isotype control. (B) Adhesion of HSC/P cells to FN H296, containing the heparin binding site and the CS-1 binding site for α4β1, or to FN CH271 containing the heparin binding site and the RGDS binding site for α5β1. Closed bars, WT mice; open bars, Rac2−/− mice. Mean ± SD, *, P < 0.01. (C) Enumeration of CFU-S12 in peripheral blood after treatment with G-CSF alone or with both G-CSF and anti-α4β1 antibody. Closed bars, WT mice; open bars, Rac2−/− mice. Mean ± SD; *, P < 0.01.
The defect in adhesion via α4β1 demonstrated in vitro likely explains the increased mobilization seen in vivo, because mobilization into the blood or spleen by the administration of anti-α4 integrin antibody results in equivalent numbers of CFU-S12 in the blood of WT mice as observed at baseline in Rac2−/− mice. Furthermore, no additional increase in the number of circulating CFU-S12 is seen in Rac2−/− mice after treatment with anti-α4 antibody alone (Fig. 2C), and no augmentation of mobilization is seen in Rac2−/− mice with the combined treatment of G-CSF and anti-α4 antibody, as expected in normal mice (15) and seen here in WT mice (Fig. 2C).
Rac2-Deficient HSC/P Cells Are Hypermotile in Vitro.
Chemokine-stimulated migration of primitive hematopoietic cells has also been postulated to be a mechanism of mobilization of hematopoietic progenitor cells (16). Thus, to further explore the relationship between Rac2 deficiency and increased mobilization in vivo of primitive blood cells, purified HSC/P cells were analyzed in vitro for migration in response to SDF-1, a potent chemokine chemoattractant for primitive hematopoietic cells (16, 23). We initially considered enhanced responsiveness to chemokines as an unlikely explanation for the excess of circulating HSC/P, as deficiency of Rac2 function results in decreased migration and F-actin generation in multiple differentiated blood cell types (10–13, 18).
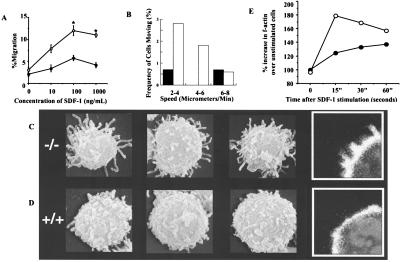
Unexpectedly, despite equivalent expression of the SDF-1 receptor, CXCR4 (Ct = 26.6 vs. 27.5, Rac2WT vs. Rac2−/−, mean of three determinations) as measured by real-time PCR, SDF-1-stimulated primitive cells (c-kit+Sca-1+Lin−) from Rac2−/− mice demonstrated increased migration in transwell chambers (16) across multiple concentrations (Fig. 3A). This increase in cells in the bottom chamber of the transwell was also noted when transwell membranes were coated with FN H296, but not on membranes coated with FN CH296, which contains ligands for both α4β1 and α5β1. In addition, antibody to α4β1 has significantly more effect in diminishing the number of WT vs. Rac2−/− cells in lower chambers coated with FN H296 and in uncoated wells (data not shown), which is similar to the effect seen in in vivo mobilization studies. The difference in apparent migration of FN CH296 in transwells vs. Dunn chambers may relate to the ability of Rac2−/− cells to maintain near-normal adhesion via α5β1 (see Fig. 2B). When the undersides of filters from transwells coated with FN CH296 were examined, nearly equivalent numbers of primitive cells were present in Rac2−/− vs. WT chambers. Thus, with adhesion via α5β1 still effective, fewer cells were detected in the bottom chamber, reducing the apparent migration. Because of the nature of the Dunn analysis, adhesion via α5β1 would still allow the hypermotility phenotype to be seen. Both increased velocity of migration (Fig. 3B) and increased frequency of responding cells (6.7 ± 3.1% vs. 1.1 ± 2.2%, Rac2−/− vs. WT, P = 0.01) were further documented in Rac2-deficient cells by analysis with time-lapsed video microscopy in Dunn chambers.
Figure 3.
Measurement of in vitro migration and F-actin polymerization after stimulation with the chemokine SDF-1. (A) Migration of c-kit+Sca-1+Lin− cells in transwell chamber assay in response to SDF-1. ○, Rac2−/− cells; ●, WT cells. Mean ± SD, *, P < 0.01. (B) Frequency of cells moving as measured with time-lapsed video microscopy in Dunn chambers. Closed bars, WT cells; open bars, Rac2−/− cells. Data show frequency of cells moving at specified speeds and are from one of three representative experiments. See Materials and Methods for details of the assay. (C and D) Scanning electron micrographs and confocal images of cells fixed 10 s after stimulation with 1 μM SDF-1. There was increased length of filopodia on (C) Rac2−/− cells compared with (D) WT stimulated with SDF-1. (Insets) Confocal microscopy performed after staining with 0.1 μg/ml rhodamine phalloidin shows increased F-actin staining and filopodia in SDF-1-stimulated Rac2−/− cells compared with WT. Cells were imaged with a Zeiss LSM 510 Laser scanning confocal microscope at ×100. (E) Increase in F-actin content after stimulation with SDF-1 analyzed by phalloidin staining and flow cytometry (10). ○, Rac2−/− cells; ●, WT cells. One of three experiments showing similar results is presented.
Compensatory Increases in Cdc42 and Rac1 Lead to Increased Cell Migration of HSC/P Cells.
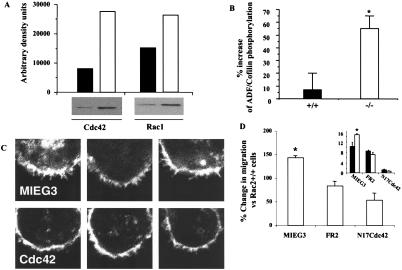
Because previous evidence has suggested that blocking one Rho GTPase protein pathway might result in modulation of other GTPases, affecting cell migration and morphology (24, 25), scanning electron micrographs of purified HSC/P cells were obtained before and after stimulation with SDF-1. Scanning electron micrographs of unstimulated cells showed only subtle differences between genotypes. However, after SDF-1 stimulation many Rac2−/− HSC/P cells demonstrated more uniformly long filopodia (Fig. 3 C vs. D). Confocal images of stimulated cells (Fig. 3 C and D, Inset) revealed pronounced F-actin staining and spike-like projections in Rac2−/− cells. Whereas the content of F-actin seen at baseline was slightly lower in Rac2-deficient cells as measured by quantitative flow analysis (10), there was a significantly larger increase in F-actin content demonstrated in HSC/P from Rac2−/− mice after SDF exposure (Fig. 3E). In fibroblasts, formation of filopodia is commonly associated with activation of the Rho GTPase Cdc42 (26), and with the use of constitutively active mutants, the activity of this Rho GTPase has been shown to positively regulate the activity of Rac (26). Cdc42 has also been implicated in macrophage movement (27). We therefore examined HSC/P cells from Rac2−/− mice for the expression and activity of Cdc42 and for Rac1, the ubiquitously expressed GTPase protein highly homologous to Rac2. Despite comparable levels of the two proteins as measured by immunoblot (data not shown), activated (GTP-bound) Cdc42 measured by GST-PAK1 p21-binding domain pull-down (18) was increased by 3- to 20-fold in Rac2−/− cells after exposure to SDF-1, compared with WT cells (Fig. 4A). Interestingly, activated Rac1 was also increased by about 3-fold (Fig. 4A). In separate experiments, an increase in the percentage of GTP-bound (active) Cdc42 and Rac1 was also demonstrated. Active Cdc42 (GTP-bound) as a percentage of total Cdc42 increased after SDF-1 stimulation from 0.12% to 0.74% (6-fold) in WT cells, but from 3.12% to 49.26% (16-fold) in Rac2−/− cells, showing both some increase in baseline active Cdc42 and a significant apparent compensatory response of Cdc42 activation in the absence of Rac2. Active (GTP-bound) Rac1 increased after stimulation from 4.4% to 28.6% (6.5-fold) in WT cells, but from 21.4% to 49.3% (2.3-fold) in Rac2−/− cells.
Figure 4.
Biochemical analysis of activation of Cdc42 and Rac1 and downstream targets in hematopoietic stem and progenitor cells from Rac2−/− mice. (A) Increased Cdc42 and Rac1 activation in Rac2−/− cells as analyzed by GST-PAK1 p21-binding domain pull-down (18), one of three experiments showing similar results. (Upper) Densitometric determination of immunoblot bands shown below. Closed bars, WT cells; open bars, Rac2−/− cells. (B) Increased inhibitory phosphorylation of ADF/cofilin as measured by immunoblot with phosphorylation-specific polyclonal antibody (anti-pADF, 1:100 dilution) (34). Closed bars, WT cells; open bars, Rac2−/− cells. Data are mean ± SD of densitometric determination of four independent experiments. *, P < 0.01. (C) Reversal of filopodia as analyzed by confocal microscopy performed after staining with 0.1 μg/ml rhodamine phalloidin after expression of dominant negative Cdc42 (Lower) or empty vector MIEG3 (Upper). The T17NCdc42 mutant and WT Rac2 (not shown) were introduced into the cells via the retrovirus vectors pMX-IRES (35, 36) and MIEG3-FR2, respectively, with published methods (20). (D) Reversal of increased migration of Rac2−/− vs. Rac2+/+ HSC/P cells after stimulation with SDF-1 analyzed in a transwell chamber assay after expression of empty vector (MIEG3), WT Rac2 (FR2), or N17Cdc42. The data are expressed as percentage change vs. Rac2WT (+/+) cells (*, P = 0.02, n = 3). (Inset) Migration data expressed as percentage of cells migrating, showing increased migration of Rac2−/− cells (open bars) expressing empty vector (MIEG3), which is reduced by expression of Rac2 (FR2). Expression of N17Cdc42 further reduces migration, but to a larger degree in Rac2−/− cells.
This increased activity appears to be important in the observed motility of Rac2−/− HSC/P cells, inasmuch as actin-depolymerizing factor (ADF)/cofilin phosphorylation, which reflects LIM kinase activities and is a known downstream target of Cdc42 and Rac (28), is significantly elevated after stimulation in Rac2−/− cells (Fig. 4B), whereas expression of the dominant negative Cdc42 bearing Thr→Asp at position 17 is associated with reversal of the enhanced filopodia in Rac2−/− cells (Fig. 4C; MIEG3 represents empty vector, Cdc42 represents N17Cdc42) and a reversal of the increased migration (Fig. 4D) after SDF-1 stimulation, but has no effect on the adhesion of Rac2−/− cells (data not shown). At baseline, the phosphorylation of ADF/cofilin is also increased in Rac2−/− cells compared with WT cells (data not shown). Because ADF/cofilin phosphorylation inhibits the F-actin depolymerization activity of cofilin proteins (28), increased Lim kinase activity and increased phosphorylation of ADF/cofilin could explain the increased F-actin content in Rac2−/− cells.
Discussion
The consequence of Rac2 deficiency in HSC/progenitor cells includes decreased integrin-mediated adhesion and increased HSC/P cells in the blood, with enhanced migration of these cells in response to stimulation. Increased numbers of HSC/progenitor cells in the blood are not likely a consequence of increased numbers of these cells, because the number of CFUs is equivalent in the bone marrow of Rac2−/− and WT mice. These adhesion and migration abnormalities are associated with increased Cdc42 and Rac1 activation, actin polymerization, and increased mobilization of HSC/progenitor cells out of the marrow cavity. The adhesion defects are directly related to Rac2 deficiency and cannot be compensated for by increased Rac1 activity. In contrast, the increased migration of Rac2−/− HSC/P cells reflects Cdc42 activation and implies that normal migration requires the coordinated regulation of Rac and Cdc42 GTPase activities. Data presented here also suggest a feedback mechanism leading to increased activation of Cdc42 and Rac1 in the absence of Rac2. Compensatory changes in GTPase activity have been noted in cell lines after overexpression of activated or dominant negative mutants of Rac, Cdc42, or Rho (24, 25), but have not been demonstrated in primary cells under physiological conditions. More recent experiments have also demonstrated induction of Rac1 protein during ex vivo expansion of Rac2−/− primitive hematopoietic cells (37). These data substantiate the assumption that in primary cells there is considerable cross-talk between GTPases (9).
Because increased activation of other GTPases, in particular of Rac1, is ineffective in reversing the adhesion defects consequent to Rac2 deficiency, these data also imply that Rac2 plays a critical and specific role in the adhesion of these primitive cells. The basis for this potential specificity of Rac function is unknown, but subcellular localization may be important. Both Rac1 and Rac2 contain conserved CAAX motifs in the C-terminal tail, which mediate attachment of farnesyl moieties and therefore may mediate protein/protein or membrane targeting (29). Differences in Rac1 and Rac2 sequences reside primarily in the area immediately preceding this motif, where Rac1 contains a stretch of basic amino acids, whereas nonbasic residues in the Rac2 protein interrupt this region. Signaling proteins with polybasic regions may colocalize within regions of the membranes enriched in acidic phospholipids, and thus these sequences may enhance membrane targeting or colocalization of relevant interacting proteins (29, 30). In addition, polylysines in this region may be involved in specifying effector recognition, as recently shown for Cdc42 interaction with an effector, the γ subunit of coatomer complex (31). In addition, this region of Rac2 may be involved in homodimer formation (32). Additional studies have demonstrated that this motif, TRQQKRP, is essential for the biological function and intracellular localization of Rac2 (W. Tao, J. R. Bailey, S.J.A., B. Connors, A. Evan, and D.A.W., unpublished observations).
Finally, as the increase in cell migration seen in Rac2-deficient HSC/P is in absolute contrast to the marked diminution in movement observed in their mature cell progeny, the consequences of Rac activation clearly depend on cell type, maturation stage, and, potentially, activation signals. These data suggest that receptor pathways, lineage-specific signals (either guanine exchange factors or downstream effectors), and/or the levels of other GTPases differ in progeny of HSC/P cells. In this regard, because adhesion of HSC/P ex vivo appears to be important in stem cell survival and function and because Rac2-deficient mast cells show both decreased adhesion to FN and increased apoptosis (11), an intriguing possibility yet to be explored is that Rac-dependent signaling, specifically via kinase cascades (33), also links adhesion and growth factor stimulation to the survival of HSC/P.
Acknowledgments
We thank Drs. Willem Fibbe, Stuart Orkin, and Anne Ridley and members of the Wells Center for Pediatric Research for reviewing the initial version of the manuscript. We also thank Eva Meunier and Sharon Smoot for assistance in manuscript preparation. We thank Dr. James Bamburg, Colorado State University, for the antibody to ADF/cofilin, and Robert Breese for maintaining Rac2 murine stocks. We are grateful to Drs. Andrew Evan and Brett Connors for assistance with scanning electron microscopy and for use of facilities in the Department of Anatomy, Dr. Jun Chen for assistance in real-time PCR analysis, and Chad Harris for technical assistance. The Wells Center for Pediatric Research is a Core Centers of Excellence in Molecular Hematology (P30 DK49218). F.-C.Y., A.W.R., and Y.G. are Research Associates, and D.A.W. is an Associate Investigator with the Howard Hughes Medical Institute. A.W.R. was also supported by a Neil Hamilton Fairley Fellowship (977211) from the National Health and Medical Research Council of Australia.
Abbreviations
- HSC/P cells
hematopoietic stem/progenitor cells
- HM
hematopoietic microenvironment
- G-CSF
granulocyte colony-stimulating factor
- CFU-S12
day 12 colony-forming units-spleen
- CFU-C
in vitro colony-forming units
- FN
fibronectin
- WT
wild type
- ADF
actin-depolymerizing factor
- SDF-1
stromal-derived factor-1
- PAK1
p21-activated kinase-1
- GST
glutathione-S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Greenberger J S. Crit Rev Oncol/Hematol. 1991;11:65–84. doi: 10.1016/1040-8428(91)90018-8. [DOI] [PubMed] [Google Scholar]
- 2.Papayannopoulou T, Craddock C. Acta Haematol. 1997;97:97–104. doi: 10.1159/000203665. [DOI] [PubMed] [Google Scholar]
- 3.Williams D A, Rios M, Stephens C, Patel V. Nature (London) 1991;352:438–441. doi: 10.1038/352438a0. [DOI] [PubMed] [Google Scholar]
- 4.van der Loo J C, Xiao X, McMillin D, Hashino K, Kato I, Williams D A. J Clin Invest. 1998;102:1051–1061. doi: 10.1172/JCI3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verfaillie C M, McCarthy J B, McGlave P B. J Exp Med. 1991;174:693–703. doi: 10.1084/jem.174.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papayannopoulou T, Nakamoto B. Proc Natl Acad Sci USA. 1993;90:9374–9378. doi: 10.1073/pnas.90.20.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 8.Vojtek A B, Der C J. J Biol Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 9.Bar-Sagi D, Hall A. Cell. 2000;103:227–238. doi: 10.1016/s0092-8674(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 10.Roberts A W, Kim C, Zhen L, Lowe J B, Kapur R, Petryniak B, Spaetti A, Pollock J D, Borneo J B, Bradford G B, et al. Immunity. 1999;10:183–196. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- 11.Yang F C, Kapur R, King A J, Tao W, Kim C, Borneo J, Breese R, Marshall M, Dinauer M C, Williams D A. Immunity. 2000;12:557–568. doi: 10.1016/s1074-7613(00)80207-1. [DOI] [PubMed] [Google Scholar]
- 12.Ambruso D R, Knall C, Abell A N, Panepinto J, Kurkchubasche A, Thurman G, Gonzalez-Aller C, Hiester A, deBoer M, Harbeck R J, et al. Proc Natl Acad Sci USA. 2000;97:4654–4659. doi: 10.1073/pnas.080074897. . (First published April 11, 2000; 10.1073/pnas.080074897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams D A, Tao W, Yang F C, Kim C, Gu Y, Mansfield P, Levine J E, Petryniak B, Derrow C W, Harris C, et al. Blood. 2000;96:1646–1654. [PubMed] [Google Scholar]
- 14.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 15.Craddock C F, Nakamoto B, Andrews R G, Priestley G V, Papayannopoulou T. Blood. 1997;90:4779–4788. [PubMed] [Google Scholar]
- 16.Aiuti A, Webb I J, Bleul C, Springer T, Gutierrez-Ramos J C. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zicha D, Dunn G A, Brown A F. J Cell Sci. 1991;99:769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 18.Benard V, Bohl B P, Bokoch G M. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 19.Knaus U G, Heyworth P G, Kinsella B T, Curnutte J T, Bokoch G M. J Biol Chem. 1992;267:23575–23582. [PubMed] [Google Scholar]
- 20.Hanenberg H, Hashino K, Konishi H, Hock R A, Kato I, Williams D A. Hum Gene Ther. 1997;8:2193–2206. doi: 10.1089/hum.1997.8.18-2193. [DOI] [PubMed] [Google Scholar]
- 21.Papayannopoulou T, Craddock C, Nakamoto B, Priestley G V, Wolf N S. Proc Natl Acad Sci USA. 1995;92:9647–9651. doi: 10.1073/pnas.92.21.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch E, Iglesias A, Potocnik A J, Hartmann U, Fassler R. Nature (London) 1996;380:171–175. doi: 10.1038/380171a0. [DOI] [PubMed] [Google Scholar]
- 23.Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, et al. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 24.Moorman J P, Luu D, Wickham J, Bobak D A, Hahn C S. Oncogene. 1999;18:47–57. doi: 10.1038/sj.onc.1202262. [DOI] [PubMed] [Google Scholar]
- 25.Sander E E, ten Klooster J P, van Delft S, van der Kammen R A, Collard J G. J Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobes C D, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 27.Allen W E, Zicha D, Ridley A J, Jones G E. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumi T, Matsumoto K, Takai Y, Nakamura T. J Cell Biol. 1999;147:1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resh M D. Cell Signal. 1996;8:403–412. doi: 10.1016/s0898-6568(96)00088-5. [DOI] [PubMed] [Google Scholar]
- 30.Choy E, Chiu V K, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov I E, Philips M R. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 31.Wu W J, Erickson J W, Lin R, Cerione R A. Nature (London) 2000;405:800–804. doi: 10.1038/35015585. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Zheng Y. J Biol Chem. 1998;273:25728–25733. doi: 10.1074/jbc.273.40.25728. [DOI] [PubMed] [Google Scholar]
- 33.Vojtek A B, Cooper J A. Cell. 1995;82:527–529. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 34.Meberg P J, Ono S, Minamide L S, Takahashi M, Bamburg J R. Cell Motil Cytoskeleton. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 35.Kinsella T M, Nolan G P. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 36.Liliental J, Moon S Y, Lesche R, Mamillapalli R, Li D, Zheng Y, Sun H, Wu H. Curr Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- 37.Gu, Y., Jia, B., Yang, F.-C., D'Sovzza, M., Harris, C. E., Derrow, C. W., Zheng, Y. & Williams, D. A. (2001) J. Biol. Chem. 276, in press. [DOI] [PubMed]