Abstract
Yeast cells approach a mating partner by polarizing along a gradient of mating pheromones that are secreted by cells of the opposite mating type. The Bar1 protease is secreted by a-cells and, paradoxically, degrades the α-factor pheromones which are produced by cells of the opposite mating type and trigger mating in a-cells. This degradation may assist in the recovery from pheromone signaling but has also been shown to play a positive role in mating. Previous studies suggested that widely diffusing protease can bias the pheromone gradient towards the closest secreting cell. Here, we show that restricting the Bar1 protease to the secreting cell itself, preventing its wide diffusion, facilitates discrimination between equivalent mating partners. This may be mostly relevant during spore germination, where most mating events occur in nature.
Keywords: Gradient, Yeast, Mating
Introduction
Gradients of signaling peptides play a key role in directing polarized growth or movement of cells. Such gradients are generated by the diffusion of nutrients or toxins from localized sources that can be either external (e.g., a localized source of food that directs chemotaxis [1]) or internal (e.g., cellular signaling centers that guide axon movement during development [2]). The gradient directs cells towards the signaling source, and this is easily achieved when a single signaling source is present. The situation becomes more complex, however, when several secreting sources are present since, in this case, the overall gradient will be summed over multiple contributions and will not necessarily point in the direction of any one specific source [3].
Yeast mating is a case in point. During mating, a pair of haploid cells of different mating type (a and α) fuse to generate a diploid cell [4]. Since yeast cells do not move, the key to successful mating is that the mating cells polarize and grow towards each other. In fact, an efficient and safe fusion requires a precise polarization of both cells [5]. The cells therefore signal their position by secreting the mating-type-specific pheromones a-factor and α-factor [6]. These pheromones bind to cell-type-specific receptors expressed by the opposite cell-type and elicit the mating response [7]. Once mating is initiated, cells begin growing a mating projection (shmoo) and align this projection in the direction of the pheromone gradient, growing toward increasing pheromone levels [8].
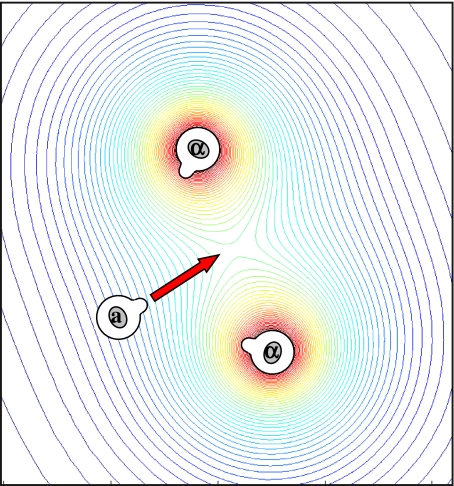
Does the pheromone gradient properly direct polarized growth toward the pheromone-secreting cell? When an a-cell is presented with a single α-factor secreting cell, this is clearly the case. In the wild, however, most mating events occur during spore germination, where the four haploid spores that are tightly packed inside an ascus begin to grow. These four spores consist of two pairs of a- and α-type cells, and consequently, each haploid is now faced with two equivalent partners [9]. Similarly, in mating mixtures, or upon mating-type switching, cells are surrounded by several nearby partners, all of which secrete mating pheromone (Fig. 1). Under these situations, the gradient sums over the contributions of all α-factor secreting cells and need not necessarily point towards a specific mating partner.
Fig. 1.
Mating in the presence of alternative mating partners. α-cell preparing for mating signals its position by secreting mating pheromone (α-factor). a-cells polarize their growth according to the resulting spatial gradient of α-factor. A problem may arise when the a-cell is surrounded by a number of α-factor secreting cells. In this case, the cell needs to polarize towards one particular partner, whereas the pheromone gradient is influenced by all other partners and may point towards some midpoint between the cells
Previous theoretical work had shown that a uniform degradation of the mating pheromone during its diffusion in the medium can screen the pheromone signal coming from distant cells and thus amplify the contribution of nearby cells [3]. It was argued that this screening can improve the orientation of the pheromone gradient by biasing it towards the closest mating partner. Indeed, the mating pheromone α-factor is degraded by the protease Bar1p [10, 11], which is secreted by a-cells and is widely diffusible, as manifested by its ability to affect pheromone sensitivity at large distances from its point of secretion [12, 13]. However, the relevance of this mechanism to realistic mating situations, where equivalent mating partners are present at distances that are about one cell-diameter away, has not been critically addressed.
We re-examined the contribution of Bar1p to the detection of mating partner position and found that uniform degradation can assist in distinguishing between equivalent mating partners only when pheromone is degraded at exceedingly high rates. We argue that under realistic conditions, diffusible Bar1p cannot assist in discriminating between partners but serves primarily to avoid saturating pheromone dosages in semi-closed volumes (e.g., ascus) or in the presence of a large population of pheromone-secreting cells. We also show that in symmetric situations resembling germination, cells can largely improve their ability to orient towards a specific mating partner by localizing the pheromone degradation to the membrane of the stimulated cells. We suggest that a combination of cell-wall-bound Bar1p and α-factor endocytosis significantly enhances α-factor at the surface of a-type cells and in this way facilitates the choice of a specific mating partner from among several potential partners.
Results
Contribution of uniform degradation to the shaping of pheromone gradient
The pheromone profile established when α-factor is subject to uniform degradation can be easily understood by considering a single α-factor secreting cell and calculating the spatial distribution of the pheromone around this cell. In the presence of multiple sources, the gradient at each position will simply be the sum over those single-source contributions.
Consider a single source of radius R which secretes pheromone peptides at some rate η. Assume further that the pheromone is degraded at some rate τ − 1 and diffuses with a characteristic diffusion coefficient D. The pheromone profile around such a source is obtained by solving the reaction-diffusion equation:
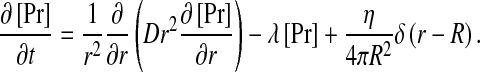 |
1 |
At long times, the pheromone distribution will approach the steady-state profile, given by the well-known solution to (1):
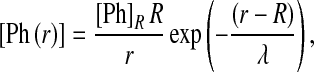 |
2 |
where [Ph]R is the pheromone distribution on the source boundary, given by 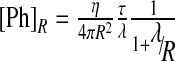 and λ is the decay of the pheromone away from the secreting cell:
and λ is the decay of the pheromone away from the secreting cell:
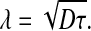 |
3 |
The pheromone concentration thus decays over distances characterized by the decay length λ. Consequently, when multiple secreting cells are present, each source will contribute to cells that are positioned within a distance comparable to this decay length. When considering a specific a-cell that is positioned in a field of many α-factor secreting cells, the screening of distant sources would lead to a significant amplification of the contribution of cells that are within a radius λ of this cell. Thus, as was noted before [3], to effectively bias the pheromone gradient towards a single mating partner, the decay length of the pheromone gradient, λ, needs to be smaller than the distance between cells.
Mating cells are typically found at distances that are less than one cell diameter away. Effective screening would thus require λ < 3 μm. Neither the diffusion coefficient, nor the degradation rate of the mating pheromone, has yet been measured. Using the Stokes–Einstein relation [14], we can estimate α-factor diffusion as D ~ 300 μm2/s ([8], see Section 4). With such a wide diffusion, a decay length relevant for screening would imply rapid degradation, with pheromone half-life shorter than τ ~ 0.03 s.
The catalytic activity of Bar1p toward mating pheromones is not known and the levels of Bar1p in mating mixtures, or in spores, have not yet been measured. Three lines of argument, however, suggest that degradation needs to be significantly slower. First, considering the diffusion-limited upper-bound on Bar1p catalytic activity (1 nM − 1 s − 1), a half-life of τ ~ 0.03 s requires that Bar1p is present at a concentration of at least 30 nM, implying that an isolated a-cell secretes over 3 × 104 Bar1p molecules/s (using (2), η = 4π a Dbar1 c with Dbar1~ 30 μm2/s estimated by the Stokes–Einstein relation [14]). With the estimate of 5 × 107 total proteins in a yeast cell [15], and a division time of 90 min, cells would need to devote over 30% of their total protein synthesis to the production of Bar1p in order to achieve this concentration.
Second, since the pheromone decay length depends on the level of Bar1p, 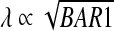 , a decay length that is of the order of the distance between cells would render the concentration of pheromone at the a-cell boundary exponentially-sensitive to the level of Bar1p in the medium and thus to the number of Bar1p-secreting cells. Finally, an upper-bound on Bar1p activity can be obtained from the observation that 1 μM pheromone that is added to 107 Bar1p-secreting a-cells maintains its activity for at least 1 h [16], implying a half-life of over τ > 3,600/ln(104) ~ 400 s if Bar1p functions in the linear regime. We note, however, that if Bar1p is saturated at these high concentrations and degrades pheromone at a fixed levels, the maximal rate of pheromone degradation is limited by 108/3,600 ~ 3 × 104 molecules per second, which does allow for a half-life of 0.2 s at 1 nM, within an order of magnitude of the 0.03 second half-time required for ensuring λ < 3 μm.
, a decay length that is of the order of the distance between cells would render the concentration of pheromone at the a-cell boundary exponentially-sensitive to the level of Bar1p in the medium and thus to the number of Bar1p-secreting cells. Finally, an upper-bound on Bar1p activity can be obtained from the observation that 1 μM pheromone that is added to 107 Bar1p-secreting a-cells maintains its activity for at least 1 h [16], implying a half-life of over τ > 3,600/ln(104) ~ 400 s if Bar1p functions in the linear regime. We note, however, that if Bar1p is saturated at these high concentrations and degrades pheromone at a fixed levels, the maximal rate of pheromone degradation is limited by 108/3,600 ~ 3 × 104 molecules per second, which does allow for a half-life of 0.2 s at 1 nM, within an order of magnitude of the 0.03 second half-time required for ensuring λ < 3 μm.
Taken together, it appears unlikely that uniform degradation by Bar1 protease is sufficiently rapid to reshape the pheromone gradient on the scale of one cell diameter. Thus, although such degradation may assist in reducing the overall magnitude of the signal, and in preventing receptor saturation in a large field of secreting cells, it is unlikely to assist in distinguishing between equivalent mating partners located at similar, biologically relevant distances.
Localized degradation can shape the pheromone gradient
Our analysis above assumed that pheromone is degraded uniformly across the field. Pheromone degradation can shape the pheromone gradient also when localized to a particular position, e.g., the receiving a-cells themselves. Such degradation can result from the endocytosis of the activated receptors [17, 18]. Alternatively, although Bar1p is secreted into the medium, some of the activity of Bar1 protease might be localized to the a-cell itself. In support of that, Bar1p was identified as one of the major disulfide bridged components of the cell wall of a-cells [19].
The impact of localized degradation on the pheromone distribution can be understood by considering a single a-cell that is challenged by a single α-factor secreting cell. The a-cell functions as a sink, effectively drawing the flux lines (the lines that define the net direction of pheromone diffusion) towards its position and thus influencing the global distribution of α-factor. In one dimension, for example, where no steady-state ensues in the absence of degradation, the addition of such a sink leads to a steady-state profile that decays linearly from the secreting to the absorbing cell. Note that in this one-dimension example, the slope of the gradient, and hence the steady-state flux of incoming pheromone molecules, is determined by the rate of pheromone production.
Mating in germinating spores as an “electric quadrupole”
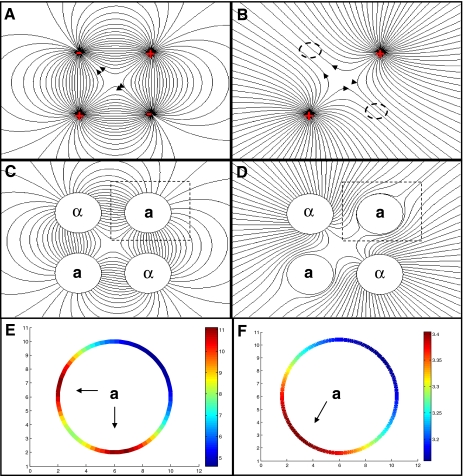
As noted above, localized degradation can draw the pheromone flux toward the receiving a-cell. A previous study has shown that localized degradation facilitates gradient sensing, because each particle is measured only once [20]. We asked whether this shaping of the pheromone flux may assist also in disentangling gradients generated by equivalent partners. In the wild, yeast are found almost exclusively as diploid, suggesting that most mating events occur during germination, where four spores mate in pairs to generate two diploid cells [9]. Our intuition that localized degradation might help in this case is based on the analogy between the diffusion problem and electric field calculations in electrostatics. In this analogy, cells secreting pheromone can be viewed as positive charges, pheromone flux as an electric field and pheromone concentration as an electric potential, with contours joining points with equal pheromone concentrations perpendicular to the pheromone flux lines.
If the receiving a-cells do not perturb the pheromone gradient, we can draw this gradient as the electric field generated by two positive charges (Fig. 2b). In this case, the field lines (pheromone flux) at the a-cell position will be concentrated at the location facing the mid-point between the two secreting cells (Fig. 2d, f).
Fig. 2.
α-factor gradient in germinating spores. As explained in the text, the diffusion of α-factor in the medium can be mapped to the problem of defining an electric field. In this analogy, α-factor secreting cells can be viewed as positive charges, whereas cells that locally degrade α-factor can be viewed as negative changes. The α-factor flux is analogous to the electric field. a Electric field lines of a quadrupole: two positive charges and two negative charges. b Electric field lines due to two positive stationary charges of equal magnitude in a dipole arrangement. c Diffusive field lines for localized degradation model. The system is composed of two α-factor secreting cells and two a-cells which locally degrade a-factor. We assumed that local degradation on the a-cell is strong enough, that the cells can be viewed as perfect sinks with zero concentration on the boundary. D Diffusive field lines for simple diffusion model. The system is composed of two α-factor secreting cells and two responding a-cells. Since in this model a-cells do not degrade α-factor locally, they are assumed to be reflective with a normal flux of zero. Field lines are similar for the uniform degradation model, where degradation is assumed across the field. e The α-factor flux along the perimeter of an a-cell in the presence of localized degradation. Note the two peaks, corresponding to equivalent maxima pointing towards the two alternative partners. f The α-factor concentration in the absence of local degradation. Note that only a single peak is present, pointing midway between the two secreting cells
The situation becomes quite different when the a-cells degrade the impeding α-factor, functioning as effective sinks. Given this boundary condition, the a-cells can be viewed as negative charges, since they draw all incoming field lines. The four germinating cells are thus analogous to an electric quadrupole (Fig. 2a). The resulting field lines are well characterized, showing a bias of the flux lines at each a-cell towards the two secreting α-cells. We reasoned that this symmetric configuration of the flux lines may allow for disentangling the gradients generated by the two equivalent α-cells (Fig. 2c, e).
Localized degradation assists in distinguishing between equivalent mating partners
For simplicity, the discussion above neglected the finite size of the cells, and considered them as point charges. We used numerical simulations to analyze a more precise geometry, where two equivalent pairs of haploid cells are located at distance that is about one cell-diameter away from each other (Fig. 3a). We assumed that the α-cells are secreting pheromone at some constant rate η and calculated the resulting steady-state pheromone distribution assuming 3D diffusion (see Section 4). This solution was used to characterize the level of pheromone, and its flux, along the perimeter defining the a-cell boundaries. The case of uniform degradation was compared to that of a localized degradation over a wide span of parameters.
Fig. 3.
Mating in the presence of alternative partners: a Germination-inspired topology with two a-cells and two α-factor secreting α-cells positioned in close proximity. Simulations are performed in three dimensions (see Section 4 for details). b Illustration of α-factor (concentration of flux) around the perimeter of the a-cell, as shown. For good recognition, the amount of occupied receptors should peak at two angles, corresponding to the direction of the two α-cells. “max” stands for the maximal receptor occupancy, “mid” is the receptor occupancy at the midline between the two expected maxima, and “min” is the minimum receptor occupancy. c Based on the definitions of “max,” “mid,” and “min,” we define three measures for the ability to locate mating partners, sensitivity, relative peak strength, and relative sensitivity, as shown. d Relative peak strength vs. diffusion coefficient for three models: uniform degradation, simple diffusion without degradation, and localized degradation on cell borders. Results are plotted for diffusion coefficient of 1 μm2/s (black) and 300 μm2/s (gray). e Relative peak strength vs. secretion rate for three models as in a. Results are plotted for secretion rate of 1 molecule/second (gray) and 10 molecule/second (black). f Relative sensitivity vs. diffusion coefficient for three models as in a. Results are plotted for diffusion coefficient of 1 μm2/s (black) and 300 μm2/s (gray). g Relative sensitivity vs. secretion rate for three models as in a. Results are plotted for secretion rate of 1 molecule/second (gray) and 10 molecule/second (black)
As expected from the general analysis above, uniform degradation required exceptionally high degradation to properly align the cells toward a specific mating partner. In fact, as long as the degradation rate was lower then 0.01 1/s, the maximum concentration of pheromone along the cell perimeter was found at the midpoint between the two secreting cells. Only for higher degradation rates, did we start to observe two small concentration peaks that point towards the two secreting cells (Figs. 2f and 3d–g). Thus, it appears that for relevant parameters, the distribution of pheromone around the a-cell exhibited a single maximum and did not possess information about the existence of the two alternative partners.
In contrast, in the case of localized degradation, assuming that the a-cells function as a sink for α-factor, the pheromone flux impinging upon the cell perimeter displayed two significant local maxima, pointing in the direction of either mating partners (Fig. 2e). This pattern was observed for a wide range of diffusion coefficients and secretion rates (Fig. 3d–g). Thus, in the presence of localized degradation, the pheromone flux distribution contained the information about the position of both potential mating partners.
When the two secreting cells are equivalent, the two peaks of α-factor flux are of the same height. The decision in which direction to polarize is likely to be stochastic, and possible alternation between the partners may occur. However, any small asymmetry between the two secreting cells breaks this balance, leading to a single global maximum pointing in a direction of a specific partner. For example, if the rate of α-factor secretion differs between the competing partners, the peak facing the rapidly secreting cell will be higher, and this cell is likely to be preferred. Moreover, if the competing cells are at different distances from the a-cell, the peak facing the closer one will be higher and will be established faster.
Compatibility of the “perfect sink” hypothesis
As we have shown above, by biasing the pheromone flux lines towards the cells, localized degradation can assist in discriminating between equivalent mating partners. To be effective, this localized degradation needs to define a perfect, or nearly perfect, sink, such that most molecules which impinge upon the a-cells are absorbed rather than released back into the medium. Cells are triggered to initiate the mating response (as judge by the formation of polarized projections) when subject to uniform concentration of pheromone that is higher than 1 nM. At this concentration, the cell is subject to a flux of η = 4π D a c ~ 0.6 × 104 pheromone molecules per second [21], taking into account a D value of 300 μm2/s [8] and cell radius of 3 μm. Accordingly, to function as a perfect sink in the relevant parameter range, cells need to be capable of absorbing the same order of 104 molecules each second.
Is this high absorbance rate compatible with known biological parameters? The number of pheromone receptors is about 104 [16, 22], but the associated kon for pheromone binding and the endocytosis rates are sufficiently slower (on the order of minutes) and cannot account for such high absorption. A second alternative is that pheromone is degraded by the fraction of the Bar1 protease that is localized to the cell wall. If over 104 molecules of Bar1p are localized to the cell wall, each of which degrades a pheromone molecule at a rate of at least 1 s − 1, effective an sink will be ensured. Considering the known abundance of proteins in the Saccharomyces cerevisiae cell, and the known activities of proteases, these parameters are well within the plausible biological range. Notably, an absorption limit of 104 molecules per second is also compatible with the observation that the addition of 1 μM of pheromone to a-cells present at a density of 107 cells/mL maintains its activity for over an hour (in fact, it predicts that the concentration will be reduced to less than 1 nM at a time t ~ 108/104 s ~ 3 h).
While maintaining maximal absorption rate of 104 pheromone molecules per second appears well within the expected biological regime, it is important to note that the estimated pheromone secretion rates required for efficient activation of the mating response (η ~ 6,000 mol/s) appears exceedingly high. Although direct measurements of α-factor secretion rates are not available, corresponding estimates for protein secretion rate are on the order of magnitude of 10 molecules/second [23, 24]. Moreover, since α-factor mRNA is present at only ~10–50 copies per cell [25], it is unlikely that more than ~100 mature α-factor molecules will be produced per second, considering a translation initiation rate of about ~0.1–1 s − 1 (rate limiting step), even when taking into account the fact that there are six repetitions of the peptide within the genome [26]. Considering the estimated 5 × 107 proteins in a S. cerevisiae cell, and its 90 min doubling time, one would conclude that the production of mating pheromone accounts for ~17% of total protein production in the cell. This result appears to be a direct consequence of the fact α-factor elicits the mating response only at concentrations that are higher than 1 nM and does not require and further assumptions. We do not currently have further explanation for this result.
Notably, the relevant parameter defining the α-factor sensing is the flux of pheromone reaching the cell. Each pheromone molecule reaching the cell is eventually degraded but can potentially bind to receptor and signal before being degraded, either following its release and subsequent cleavage by Bar1p or while endocytosed with the signaling receptor.
Discussion
Yeast cells mate by polarizing their growth toward a cell of the opposite mating type. To signal their position, α-cells secrete a diffusible α-factor pheromone; a-cells, in turn, polarize along gradients of this pheromone. Our study was triggered by the common observation that, in many mating conditions, every a-cell is surrounded by multiple α-factor secreting cells. This raised the question of what ensures the proper polarization of the a-cell towards a specific mating partner, rather than in some average direction reflecting the sum of the pheromone contributions from all nearby α cells. Our central result is that localizing the degradation of the pheromone at the a-cell position can assist in disentangling gradients generated by equivalent sources.
Previous theoretical studies suggested that uniform degradation of α-factor by the secreted protease Bar1 can screen the contribution of distant cells [3]. In mating mixtures, such screening may be essential for reducing the overall level of α-factor and preventing receptor saturation. Indeed, in the absence of degradation, the contribution of cells positioned at a distance R from an a-cell will decrease as 1/R, but if the cells are roughly uniformly distributed, the number of such cells will scale as R2, leading to macroscopic accumulation of α-factor. Screening by Bar1p can prevent this saturation, limiting the location of cells that contribute to the local α-factor concentration to a moderate distance.
In contrast to this global screening function, however, it appears unlikely that diffusible Bar1p can help a-cells discriminate between equivalent partners that are positioned nearby (approximately one cell diameter away). Screening cells at distance of just one cell diameter requires exceedingly fast pheromone degradation, which appears to be inconsistent with experimental evidence, and would have placed a tremendous biosynthesis burden on the a-cell that produces protease.
The results change dramatically if the a-cells consume α-factor on their perimeter. In this case, the capacity to distinguish between equivalent partners is greatly facilitated, in particular under symmetric geometries like the pattern of spores germinating in an ascus, where most mating events appear to take place in the wild. This is achieved when the localized degradation is sufficiently high to function as a “sink” of α-factor, drawing the pheromone flux lines towards the a-cells. In contrast, in the absence of such degradation, α-factor that collides with the a-cells is reflected back into the medium, and diffuses in a random direction, thus diluting the signal generated by the secreting cells.
At least two mechanisms could contribute in principle to a localized degradation of pheromone. First, binding of the α-factor to its receptor triggers a rapid endocytosis and degradation of both the receptor and the bound pheromone [16, 22, 23]. Known binding rates of pheromone to its receptor, however, suggest that these reactions are not rapid enough to define the required sink. Second, although Bar1p is widely diffusible and can function at long distances, a significant fraction of Bar1p remains bound to the cell wall of the secreting a-cells [17]. This bound form is likely to be active, as three independent studies have reported that a-cells exhibit proteolytic activity against α-factor that is cell-wall bound, increases with cell-density, and is inhibited by protease inhibitors [27–29].
In conclusion, we propose that localized degradation can help disentangle signaling gradients generated by equivalent signaling centers. In the case of yeast mating, the need for such discrimination during most mating events in the wild may explain the large amount of the protease Bar1 that is bound to the walls of the a-cells. It will be interesting to examine whether a similar mechanism also functions in other systems which rely on signaling gradients for directing cell fusion, as is the case, for example, in the wiring of axons in the developing neural system.
Methods
Simulations
Simulations were implemented in Matlab 7 (Mathworks) and Femlab 3.1 (Comsol). We considered four cells in a symmetrical geometry, as illustrated in Fig. 3. We consider α-factor secreted by α-cells and impinges on a-cells. We assume that a-cells respond based on the number of occupied receptors on their boundaries. Simulations were done in three or two dimensions, as specified, and were either enclosed in a semi-permeable ascus or within a large, fully absorbing sphere (with radius of 15 times cell radius). Three models were examined. First, the “No degradation” model (1) assumes simple diffusion of α-factor. Second, the “Uniform degradation” model (2) assumes a uniform degradation rate across the domain. Finally, the “Localized degradation” model (3) assumes that the boundaries of the a-cells are absorbing. The model considers the following parameters: α (α-factor), R (receptor), and Rα (α-factor-receptor complex). The modeled equations are:
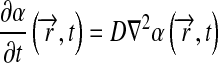 |
4 |
for models 1 and 3, and:
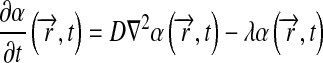 |
5 |
for model 2.
For models 1 and 2, complex level was computed as:
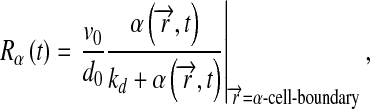 |
6 |
and the boundary condition at the a-cells is:
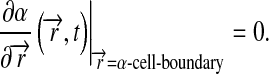 |
7 |
For model 3, complex level was computed as:
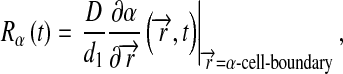 |
8 |
with the following boundary condition at the a-cells:
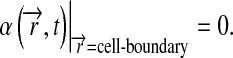 |
9 |
The boundary condition on the α-cells for all models was:
 |
10 |
Rcell is the cell radius, η0 is the α-factor secretion rate, λ is the uniform degradation rate. Boundary conditions at the outer sphere are:
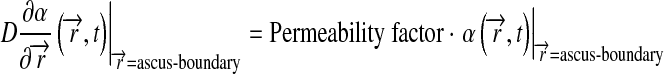 |
11 |
for limited permeability ascus, or:
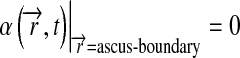 |
12 |
to simulate diffusion in an effective open space, while the ascus radius was 15-fold larger than the cell radius.
Parameters
The following parameters were used (unless stated otherwise): kd—receptor dissociation constant—3.6 molecules/μm3 [22], η0—α-factor secretion rate (secreted mol. per whole cell)—10 molecules/s/μm2 [23], d0—degradation rate of unbound receptor—1/60 1/s, d1—degradation rate of bound receptor—1/60 1/s [16, 22], v0—receptor production rate—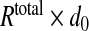 molecules/second, λ—uniform degradation rate, 1/60 1/s, D–α-factor diffusion coefficient—30 μm2/s, taking into account possibility for an order of magnitude over-estimation [8], permeability factor—0.1, Rtotal—8000 molecules [16, 18].
molecules/second, λ—uniform degradation rate, 1/60 1/s, D–α-factor diffusion coefficient—30 μm2/s, taking into account possibility for an order of magnitude over-estimation [8], permeability factor—0.1, Rtotal—8000 molecules [16, 18].
Dimensions: Spore radius—1 μm [30], ascus radius—3 μm [30], distance from adjacent cell—1/2 cell radius.
Acknowledgements
We thank Prof. Andrew Murray for insightful comments and suggestions and members of our group for discussions. This work was supported by the NIH (P50GM068763), the European Research Council, the Israel Science Foundation, Minerva, and the Hellen and Martin Kimmel Award for Innovative Investigation.
References
- 1.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichmann A, Makinen T, Alitalo K. Neural guidance molecules regulate vascular remodeling and vessel navigation. Genes Dev. 2005;19:1013–1021. doi: 10.1101/gad.1305405. [DOI] [PubMed] [Google Scholar]
- 3.Barkai N, Rose MD, Wingreen NS.Protease helps yeast find mating partners Nature 1998396422–423.1998Natur.396..422B 10.1038/24760 [DOI] [PubMed] [Google Scholar]
- 4.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2004;25:1465–1476. doi: 10.1016/j.peptides.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duntze W, MacKay V, Manney TR.Saccharomyces cerevisiae: a diffusible sex factor Science 19701681472–1473.1970Sci...168.1472D 10.1126/science.168.3938.1472 [DOI] [PubMed] [Google Scholar]
- 7.Cross F, Hartwell LH, Jackson C, Konopka JB. Conjugation in Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 1988;4:429–457. doi: 10.1146/annurev.cb.04.110188.002241. [DOI] [PubMed] [Google Scholar]
- 8.Segall JE.Polarization of yeast cells in spatial gradients of alpha mating factor Proc. Natl. Acad. Sci. U. S. A. 1993908332–8336.1993PNAS...90.8332S 10.1073/pnas.90.18.8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taxis C, Keller P, Kavagiou Z, Jensen LJ, Colombelli J, Bork P, Stelzer EH, Knop M. Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J. Cell Biol. 2005;171:627–640. doi: 10.1083/jcb.200507168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballensiefen W, Schmitt HD. Periplasmic Bar1 protease of Saccharomyces cerevisiae is active before reaching its extracellular destination. Eur. J. Biochem. 1997;247:142–147. doi: 10.1111/j.1432-1033.1997.00142.x. [DOI] [PubMed] [Google Scholar]
- 11.Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol. Cell. Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks JB, Herskowitz I.Evidence for a new diffusible element of mating pheromones in yeast Nature 1976260246–248.1976Natur.260..246H 10.1038/260246a0 [DOI] [PubMed] [Google Scholar]
- 13.Manney TR. Expression of the BAR1 gene in Saccharomyces cerevisiae: induction by the alpha mating pheromone of an activity associated with a secreted protein. J. Bacteriol. 1983;155:291–301. doi: 10.1128/jb.155.1.291-301.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crank J. The Mathematics of Diffusion. 1. London: Oxford University Press; 1956. [Google Scholar]
- 15.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS.Global analysis of protein expression in yeast Nature 2003425737–741.2003Natur.425..737G 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 16.Yi TM, Kitano H, Simon MI.A quantitative characterization of the yeast heterotrimeric G protein cycle Proc. Natl. Acad. Sci. U. S. A. 200310010764–10769.2003PNAS..10010764Y 10.1073/pnas.1834247100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajaj A, Celic A, Ding FX, Naider F, Becker JM, Dumont ME. A fluorescent alpha-factor analogue exhibits multiple steps on binding to its G protein coupled receptor in yeast. Biochemistry. 2004;43:13564–13578. doi: 10.1021/bi0494018. [DOI] [PubMed] [Google Scholar]
- 18.Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol. Cell. Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moukadiri I, Jaafar L, Zueco J. Identification of two mannoproteins released from cell walls of a Saccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J. Bacteriol. 1999;181:4741–4745. doi: 10.1128/jb.181.16.4741-4745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endres RG, Wingreen NS.Accuracy of direct gradient sensing by single cells Proc. Natl. Acad. Sci. U. S. A. 200810515749–15754.2008PNAS..10515749E 10.1073/pnas.0804688105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg HC, Purcell EM.Physics of chemoreception Biophys. J. 197720193–219.1977BpJ....20..193B 10.1016/S0006-3495(77)85544-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenness DD, Burkholder AC, Hartwell LH. Binding of alpha-factor pheromone to Saccharomyces cerevisiae a cells: dissociation constant and number of binding sites. Mol. Cell. Biol. 1986;6:318–320. doi: 10.1128/mcb.6.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frykman S, Srienc F. Quantitating secretion rates of individual cells: design of secretion assays. Biotechnol. Bioeng. 1998;59:214–226. doi: 10.1002/(SICI)1097-0290(19980720)59:2<214::AID-BIT9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 24.Frykman S, Srienc F. Cell cycle-dependent protein secretion by Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001;76:259–268. doi: 10.1002/bit.10003. [DOI] [PubMed] [Google Scholar]
- 25.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/S0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 26.Singh A, Chen EY, Lugovoy JM, Chang CN, Hitzeman RA, Seeburg PH. Saccharomyces cerevisiae contains two discrete genes coding for the alpha-factor pheromone. Nucleic Acids Res. 1983;11:4049–4063. doi: 10.1093/nar/11.12.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finkelstein DB, Strausberg S. Metabolism of alpha-factor by a mating type cells of Saccharomyces cerevisiae. J. Biol. Chem. 1979;254:796–803. [PubMed] [Google Scholar]
- 28.Ciejek E, Thorner J. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell. 1979;18:623–635. doi: 10.1016/0092-8674(79)90117-X. [DOI] [PubMed] [Google Scholar]
- 29.Maness PF, Edelman GM.Inactivation and chemical alteration of mating factor a by cells and spheroplasts Proc. Natl. Acad. Sci. U. S. A. 1978751304–1308.1978PNAS...75.1304M 10.1073/pnas.75.3.1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckett A, Illingworth RF, Rose AH. Ascospore wall development in Saccharomyces cerevisiae. J. Bacteriol. 1973;113:1054–1057. doi: 10.1128/jb.113.2.1054-1057.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]