Abstract
Unlike small molecule drugs, therapeutic protein pharmaceuticals must not only have the correct amino acid sequence and modifications, but also the correct conformation to ensure safety and efficacy. Here, we describe a method for comparison of therapeutic protein conformations by hydroxyl radical protein footprinting using liquid chromatography-mass spectrometry (LC-MS) as an analytical platform. Hydroxyl radical protein footprinting allows for rapid analysis of the conformation of therapeutic proteins based on the apparent rate of oxidation of various amino acids by hydroxyl radicals generated in situ. Conformations of Neupogen®, a patented granulocyte colony-stimulating factor (GCSF), were compared to several expired samples of recombinant GCSF, as well as heat-treated Neupogen®. Conformations of different samples of the therapeutic proteins interferon α-2A and erythropoietin were also compared. Differences in the hydroxyl radical footprint were measured between Neupogen® and the expired or mishandled GCSF samples, and confirmed by circular dichroism spectroscopy. Samples that had identical circular dichroism spectra were also found to be indistinguishable by hydroxyl radical footprinting. The method is applicable to a wide variety of therapeutic proteins and formulations through the use of separations techniques to clean up the protein samples after radical oxidation. The reaction products are stable, allowing for flexibility in sample handling, as well as archiving and reanalysis of samples. Initial screening can be performed on small amounts of therapeutic protein with minimal training in LC-MS, but samples with structural differences from the reference can be more carefully analyzed by LC-MS/MS to attain higher spatial resolution, which can aid in engineering and troubleshooting.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-012-9336-7) contains supplementary material, which is available to authorized users.
Key words: biosimilars, hydroxyl radical protein footprinting, mass spectrometry, protein conformation, therapeutic proteins
INTRODUCTION
Since the introduction of recombinant human insulin in 1982, therapeutic protein pharmaceuticals have grown into an estimated $102.4 billion industry in 2011, with more than 120 drugs on the market. Assays for protein pharmaceuticals have not kept pace because of the inherent complexity and variability of the proteins and their biological manufacturing processes. Unlike small molecules, therapeutic protein pharmaceuticals must have the proper primary structure of amino acid sequence, post-translational modification(s), disulfide bond formation, and also maintain the proper three-dimensional conformation. The conformation of a therapeutic protein is often sensitive to small changes of many factors in its production, formulation, and handling, including temperature, pH, and buffer composition. Therapeutic proteins with improper conformation can lose efficacy, or even become highly toxic. For example, subcutaneously injected Eprex®, a recombinant human erythropoietin pharmaceutical manufactured by Johnson and Johnson, was linked to an increase in incidents of pure red-cell aplasia. A change in Eprex® formulation resulted in a decrease in the conformational stability of the protein, making the protein more sensitive to disruptions in the cold chain (1,2).
Analysis of protein conformation is necessary during development of expression and purification methods for the therapeutic protein, development of deliverable formulations of the therapeutic protein, tests of drug stability and shelf-life, and quality control during the manufacturing process. Given the number of therapeutic protein patents that are either expired or will be expiring shortly, follow-on formulations of the therapeutic protein must demonstrate equivalency of both the primary protein structure and the protein conformation to ensure equivalency with the therapeutic protein approved by regulatory agencies to avoid separate clinical trials. Conformational analysis is often cumbersome, requiring large amounts of sample, labor, and expertise, making routine conformational screening of drug lots difficult to achieve. High-resolution analysis of most protein therapeutics is possible by nuclear magnetic resonance (NMR) spectroscopy and/or X-ray crystallography, but these analyses are very laborious, require large amounts of sample, and have strict limitations on the formulation components that can be tolerated. Of particular note is the fact that neither X-ray crystallography nor NMR spectroscopy can readily analyze protein aggregation or highly heterogeneous mixtures of protein conformations, which are often problems with therapeutic protein formulations and can play a large role in the inducement of immune responses (3). Spectroscopic techniques are also available for conformational analysis, and are often quite useful for detecting differences in conformation and stability. However, they typically yield little information about the regions of the therapeutic proteins that have been altered in conformation or stability, making troubleshooting more difficult, and usually require relatively large amounts of protein.
Mass spectrometry is the method of choice for analysis of the primary structure of therapeutic proteins due to its versatility and sensitivity (4). Development of mass spectrometry (MS)-based methods for conformation analysis would allow for a rapid and sensitive analysis of both protein primary structure and conformation in one set of experiments using a single analytical platform. Several methods for probing the solution-phase conformation of proteins have been developed previously, such as native spray charge state distribution analysis (5). Perhaps the most widely used method is amide hydrogen-deuterium exchange (HDX) analysis coupled with MS (6). In HDX-MS, the protein of interest is diluted into deuterium oxide. The amide proton found in the backbone of every amino acid except proline undergoes exchange with water on a time scale that is amenable to analysis. In HDX-MS, the protein is allowed to exchange for differing periods of time, after which the rate of exchange is drastically reduced by lowering the pH and cooling the sample to near freezing. The protein is usually rapidly digested with a nonspecific protease, and the peptides are analyzed by rapid liquid chromatography (LC-MS) to determine the shift in mass caused by exchange of a hydrogen for a deuterium in order to determine the kinetics of exchange between the amide hydrogen and the deuterated solvent. The rate of exchange is a function of the amino acid sequence, as well as the stability of any hydrogen bonds that the amide hydrogen is involved with (the solvent accessibility of the amide hydrogen may also play a role). By comparing two or more samples of the same amino acid sequence, changes in the kinetics of HDX can be interpreted as changes in the conformation of the protein (7). HDX is a powerful technique for examining changes in protein secondary structure and dynamics in therapeutic proteins (8); however, it is technically a very challenging technique. Challenges that arise are largely due to the reversible nature of the HDX process and difficulties with reproducibility due to small changes in pH, temperature, analysis time, protease digestion efficiency, or chromatography. Even after the exchange is “quenched” by acidification of the solution, exchange still occurs and the continued exchange results in loss of information because incorporated deuteriums back exchange with water. The back-exchange problem also limits the post-analysis sample handling that can be done, posing problems for therapeutic protein formulations that contain compounds that make mass spectrometric analysis more difficult.
To avoid the back-exchange problems of HDX, a group of methods uses differences in the apparent rate of covalent modification of a protein in two or more different conformations to probe the structure of the protein. In these methods, a protein of interest is mixed with a reactive agent and allowed to react under controlled conditions. This process is then repeated for the same amino acid sequence thought to be in a different conformation. The protein is then enzymatically digested into peptides, and the peptides are subjected to LC to clean up samples and separate peptides, coupled to MS to measure the mass and relative abundance of each modified and unmodified peptide. Tandem mass spectrometry (MS/MS) is used to identify the site(s) of modification of each peptide. Differences in the apparent rate of reaction as measured by changes in the relative abundance(s) of the modified form(s) of the peptide compared to the relative abundance of the unmodified form are then interpreted as differences in either the accessibility of the reactive protein functional group to the reagent, or differences in the local environment of the functional group that alter its reactivity (9). Covalent modification has fewer technical challenges than HDX due to the usually irreversible nature of the modification; however, most covalent modification techniques have their own set of difficulties. Most notable is the fact that covalent modification itself is known to alter the conformation of the protein. This caveat is a central problem in covalent modification techniques. Usually, other biophysical techniques or functional analyses must be carried out to ensure that the modification is controlled to an extent that does not compromise the conformational integrity of the protein analyte. In addition, most covalent labeling techniques target specific functional groups of protein side chains (e.g., primary amines, free thiols, etc.). This specificity limits the amount of information that can be gathered for the protein, because only a strictly limited subset of amino acids is probed.
One covalent modification technique has been quite successful at overcoming the aforementioned limitations. Hydroxyl radical protein footprinting is a technique wherein freely diffusing hydroxyl radicals are generated in situ with the protein of interest (10). Hydroxyl radicals are highly reactive, and will oxidize any amino acid side chain, albeit with different inherent rate constants. The rate of reaction depends primarily on two factors: the inherent reactivity of the amino acids—with aromatic and sulfur-containing amino acids being the most reactive, followed by aliphatic amino acids and arginine, followed by the other charged and hydrophilic amino acids (11, 12)—and the time-averaged solvent accessible surface area of the side chain (13). Like other covalent modification technique, hydroxyl radical protein footprinting has been shown to alter the conformation of the protein if not sufficiently controlled. However, recent work has demonstrated that techniques that complete the oxidation process in less than 1 μs are capable of heavily oxidizing protein analytes faster than they can change their conformation due to the modifications. While the protein still changes conformation after the labeling process, the information from the original conformation is frozen in a chemical “snapshot” of the oxidation rates of the amino acid side chains, completed before the protein can change conformation (14). Sub-microsecond oxidation has been reported to be performed by either radiolysis of water by brief electron pulses from a Van de Graaff accelerator (15) or by nanosecond laser photolysis of hydrogen peroxide by a KrF excimer laser (16). Either method is capable of generating short bursts of very high concentrations of hydroxyl radicals, and through the proper use of scavengers and quenchers, the labeling chemistry can be controlled to the sub-microsecond regime (14). After the oxidation chemistry is completed, the protein is digested with a protease and subjected to LC-MS/MS to identify oxidized and unoxidized peptides, quantitate the amount of oxidation of each peptide based on the relative abundance of the oxidized form(s) of the peptide to the relative abundance of the unoxidized form, and finally to identify the major sites of oxidation of the peptide. The “snapshot” nature of hydroxyl radical footprinting (HRF) also decouples the conformational probe from the electrospray ionization process, allowing for post-oxidation clean-up of samples containing components that interfere with the electrospray ionization of proteins and peptides and giving the technique more flexibility than native spray charge distribution analysis (5). While hydroxyl radical protein footprinting manages to overcome many of the problems with traditional covalent-labeling techniques, data interpretation remains a daunting challenge. Due to the promiscuity of the hydroxyl radical, the MS/MS spectra to determine the major site(s) of oxidation of each peptide is challenging, and often requires manual interpretation from a mass spectrometrist experienced in analyzing oxidized peptides.
Here, we describe the application of an abbreviated hydroxyl radical protein footprinting protocol towards the conformational analysis of therapeutic protein formulations. We take samples of recombinant erythropoietin (EPO), interferon α-2A (IFN), and granulocyte colony-stimulating factor (GCSF), including samples of a Food and Drug Administration-approved formulation of GCSF (Neupogen®) and subject them to hydroxyl radical protein footprinting using fast photolysis of peroxide (FPOP) to generate the radical in situ. The samples are then digested and subjected to LC-MS in order to quantitate the amount of oxidation of each peptide. As MS/MS analysis is the most laborious step in the process, and the only step requiring a substantial amount of highly specialized expertise, we do not perform MS/MS analysis of oxidized peptides. Even without the MS/MS analysis, we demonstrate that we can successfully compare different samples of therapeutic proteins. We confirm all of our analyses with circular dichroism spectroscopy in order to validate the data from the abbreviated hydroxyl radical footprinting technique. We propose abbreviated hydroxyl radical protein footprinting as a rapid, flexible, and sensitive technique for conformational comparison of therapeutic protein samples (consuming ~300 pmol of sample per triplicate analysis) that can be performed by any laboratory with typical expertise in protein LC-MS. The technique is amenable to any typical protein LC-MS platform, and the only specialized instrumentation required is a relatively inexpensive KrF excimer laser. The data generated are sufficient to identify any substantial changes in conformation, and if substantial changes are detected, the samples can be archived and probed more deeply at a later time by more sophisticated separations and MS/MS techniques in order to develop models of conformational changes for troubleshooting.
MATERIALS AND METHODS
Reagents
Neupogen® Filgrastim (Lot 1025877, expiration date 08/13) was purchased from Amgen® (Thousand Oaks, CA, USA) and stored under manufacturer’s recommended conditions until analysis. Acetonitrile, catalase from bovine liver, ammonium bicarbonate, and l-glutamine were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO, USA). Thirty percent hydrogen peroxide and formic acid were purchased from J.T. Baker (Phillipsburg, NJ, USA). Methionine amide was purchased from Bachem (Torrace, CA, USA). Dithiothreitol (DTT) was purchased from Fisher Biotech (Fair Lawn, NJ, USA). Sequencing-grade modified trypsin was purchased from Promega Corporation (Madison, WI, USA). Purified water (18 MΩ) was obtained from a Milli-Q Synthesis system (Millipore, Billerica, MA, USA). Recombinant samples of the therapeutic proteins IFN (20 mM potassium phosphate buffer, pH 5.7), EPO (50 mM sodium acetate, pH 5.8), and GCSF (100 mM sodium acetate, 0.005% Tween 80, 5% sorbitol, pH 4) that are beyond their expected shelf life were generous gifts from a donor who wishes to remain anonymous.
Circular Dichroism Spectroscopy
The circular dichroism (CD) spectra were acquired at 25°C using a Jasco 710/810 spectropolarimeter. Samples concentrations were determined by ultraviolet (UV) absorbance on a Thermo NanoDrop 2000c spectrophotometer (17; Thermo Scientific, Bremen, Germany). Data points were collected from 190 to 260 nm using a 0.1 cm path length cuvette. Data were analyzed and plotted using Microsoft Excel.
FPOP Labeling
FPOP labeling of the protein pharmaceuticals were performed using the proteins as received, without further purification or buffer exchange. All proteins were combined with glutamine and nanopure water to give a final protein concentration of 10 μM in 20 μL final volume. The final glutamine and hydrogen peroxide concentrations were 20 and 100 mM, respectively. Concentrated hydrogen peroxide was diluted to 1 M solution and was added to each replicate to give a final concentration of 100 mM just prior to infusion into the tubing for oxidative modification by FPOP. FPOP was performed as previously described (14,16). The protein samples flowed through the beam path of an EX100 KrF excimer laser at 248 nm, with a laser power of 50 mJ/pulse (GAM Laser Inc., Orlando, FL, USA). The flow rate of the syringe pump and the pulse frequency of the laser were set such that each segment of protein sample was irradiated with a single ~20 ns UV pulse with a 10% calculated buffer region between irradiated segments to help account for sample diffusion and laminar flow (14). Methionine amide (0.5 μg/μL) and catalase (0.5 μg/μL) were in a collection tube with ammonium bicarbonate (50 mM) to immediately quench oxidation by destroying excess hydrogen peroxide.
Proteolysis
Hot DTT (64 mM, 95°C) was added to each irradiated sample to give a final concentration of 5 mM and incubated at 37°C for 30 min to denature and reduce the protein. Sequencing-grade modified trypsin (0.2 μg) was added and incubated at 37°C for 30 min while rotating to digest the protein samples. A 30-min digestion was sufficient to digest all three protein pharmaceuticals with no detectable intact protein remaining when examined by LC-MS.
Mass Spectrometry
Samples were analyzed by electrospray time of flight mass spectrometry on a Synapt G2 HDMS in sensitivity mode controlled by Mass Lynx 4.1 software (Waters Corporation, Milford, MA, USA). Using the nanoAcquity UPLC system (Waters Corporation) the samples are cleaned by a Symmetry C18 trap column (5 μm, 180 μm × 20 mm) prior to injection on to the nanoAcquity UPLC BEH130 C18 column (1.7 μm, 100 μm × 100 mm, Waters Corporation). The gradient was pumped at 600 nL/min from 3% to 40% buffer B (99.9% acetonitrile, 0.1% formic acid) for 9 min, 40–95% buffer B over 1 min, held at 95% buffer B for 4 min, and re-equilibrated to 97% buffer A (99.9% water, 0.1% formic acid) for 15 min. MS was performed with the following settings: capillary voltage of 3.0 kV, cone voltage of 45 V, m/z range of 100–2,000, desolvation temperature of 150°C, and desolvation gas flow of 100 L/h.
Abbreviated FPOP Data Analysis
In silico digestion of GCSF, IFN, and EPO was performed using PROWL (http://prowl.rockefeller.edu). Only MS was performed so all data analysis was done manually using MassLynx 4.1 and quantitation was done on the peptide-level only. Using accurate masses, the tryptic peptides and corresponding oxidation products were identified from the LC-MS runs manually to calculate the average oxidation events per peptide in each of the protein samples. Average oxidation events per peptide is calculate by summing the ion intensities of all the oxidized peptide masses multiplied by the number of oxidation events required for the mass shift (e.g., one event for +16, two events for +32) divided by the sum of the ion intensities of all unoxidized and oxidized peptide masses. Two-tailed independent Student’s t tests were used for statistical analysis, with an α of 0.01 selected for statistical significance.
RESULTS
For each therapeutic protein, the protein sample was aliquoted into three replicates, and each sample was oxidized by FPOP. As the protein is exposed to the short burst of hydroxyl radicals, the radicals react with the amino acid side chains in predictable ways, with an apparent rate that is a function of the amino acid sequence and the exposure of the side chain to the solvent. As all of the therapeutic proteins had the same protein sequence, changes in the apparent rate of oxidation can be attributed to changes in the accessibility of the side chain, which changes as the population of protein conformations in the sample changes. After FPOP oxidation, the samples are quenched to eliminate less-reactive oxidants like hydrogen peroxide and superoxide, digested with trypsin for 30 min, and analyzed by a short LC-MS run. Tryptic peptides were identified by mass based on the in silico digest of the protein, and oxidation products were identified based on mass shifts and relative LC retention time shifts from the unoxidized peptide. The average number of oxidation events per peptide were calculated as described in the “Materials and Methods” section. Control samples of unoxidized protein were also analyzed intact by LC-MS, as well as digested and analyzed to detect alterations in primary structure that occurred outside of the FPOP oxidation process. In order to ensure that changes in the HRF detected were due to changes in the conformation of the protein rather than the primary structure, one GCSF sample with an unexpected three-amino acid truncation of the N-terminus in ~70% of the protein population was identified during the screening and excluded from analysis (data not shown). All other samples showed near-identical primary structure, with the only differences arising from minor changes in the amount of native methionine oxidation in the sample.
Granulocyte Colony-Stimulating Factor
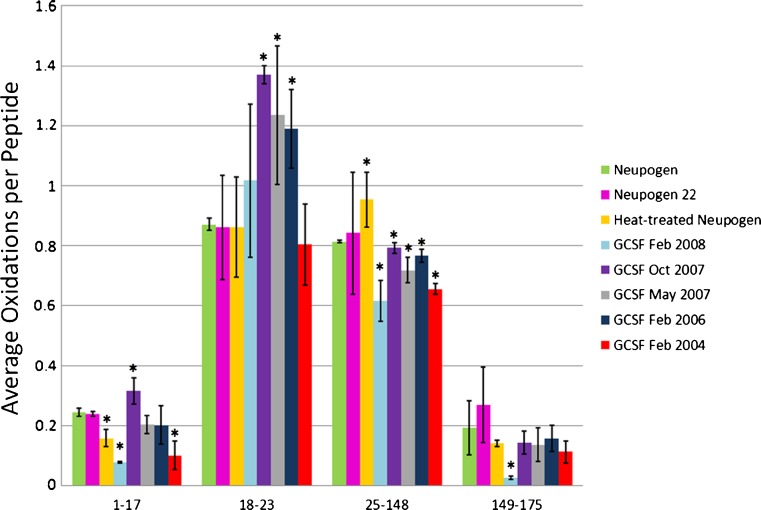
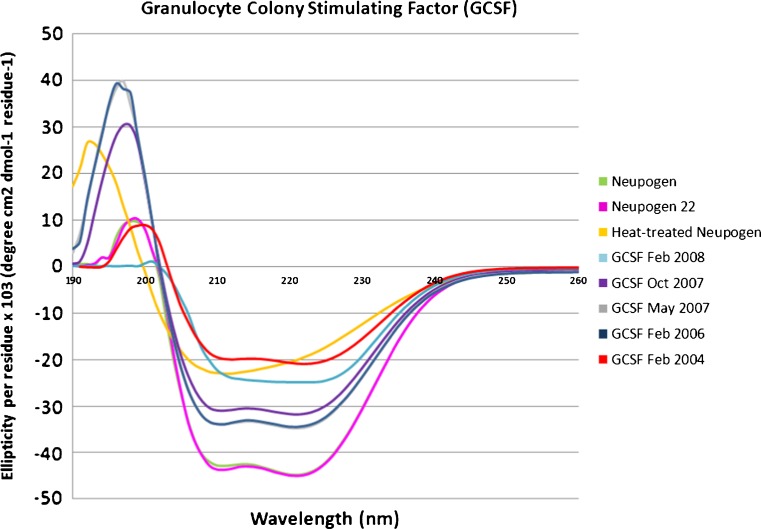
One of the most pressing needs for rapid conformational analysis in therapeutic proteins is the comparison of patented pharmaceuticals with biosimilars, which are follow-on therapeutic protein formulations that seek to establish equivalency with the patented drug to avoid the thorough clinical trials usually necessary for regulatory approval of a drug. It is essential for patient safety to ensure that any follow-on therapeutic recombinant protein has not only a primary structural equivalence (amino acid sequence, disulfide bonds, glycosylations, etc.) but also a conformational equivalence. In order to test the sensitivity of HRF to misfolding, aggregation, or other conformational differences between a potential biosimilar and a patented recombinant therapeutic protein, we compared properly handled Neupogen®, the Amgen Inc. brand name pharmaceutical for GCSF, with Neupogen® that had been heated well beyond manufacturer’s specifications and with Neupogen® that was left at room temperature for 22 h (Neupogen22), which is within the manufacturer’s storage guidelines. We also compared Neupogen® with recombinant GCSF samples that were manufactured at different dates, and were all well past their expected shelf life. These expired recombinant GCSF samples should give us a variety of different populations of non-native conformations, which should differ by varying extents from the patented therapeutic protein. After FPOP oxidation and tryptic digestion, GCSF is resolved into four peptides that cover >99% of the protein sequence, all of which yield a measurable hydroxyl radical protein footprint. To determine the ability of the HRF method to detect known changes in conformation that occur upon mishandling of the therapeutic protein, a portion of the Neupogen® sample was heated to 95°C for 1 h, cooled to room temperature, and analyzed by HRF. To determine the capability of the HRF method to identify no conformational differences in identical samples, a portion of the Neupogen® sample was stored at room temperature for 22 h (Neupogen22) and analyzed by HRF. Neupogen22 sample should be conformationally identical to cold-stored Neupogen®, as manufacturer storage specifications state that Neupogen® can be left at room temperature for up to 24 h and still be effective. We also irradiated and analyzed five expired recombinant GCSF preparations of unknown conformation, expressed at different times. The fraction oxidized was calculated for each peptide of each sample and the average fraction oxidized is graphed in Fig. 1. The gross fingerprints of the seven samples are all similar, with the two internal peptides being more heavily oxidized than the N- or C-terminus. However, comparison of each peptide in the Neupogen® sample with the corresponding peptide in each other preparation reveals measurable differences in the conformation of the samples. Student’s t tests were used to determine the statistical difference in the amount of peptide oxidation between each sample and Neupogen® (Electronic Supplementary Material (ESM) Table S1). Differences were considered significant if the calculated p value was less than or equal to 0.01. The heat-treated Neupogen® and all expired recombinant GCSF preparations saw a statistical difference in at least one of the four peptides by HRF, indicating by our technique that the heat-treated sample and each of the expired recombinant GCSF samples had different conformations than Neupogen®. Circular dichroism spectra were also taken of each unoxidized protein formulation in order to verify any differences identified by HRF with a biophysical technique that is established and in widespread use for therapeutic proteins (Fig. 2), with spectra qualitatively interpreted based on changes in features and movement of maxima and minima (18). GCSF is 61% helical, consisting of five helices (19). The heat-treated Neupogen® and all expired recombinant GCSF samples saw a notable difference by circular dichroism, verifying the differences noted by HRF, while the Neupogen® held at room temperature for 22 h showed an identical CD spectrum to the Neupogen® kept at 4°C.
Fig. 1.
Hydroxyl radical footprinting of GCSF samples. Each set of bars represents one peptide from Neupogen® (green), Neupogen22 (pink), heat-treated Neupogen® (yellow), or recombinant GCSF samples generated in Feb 2008 (light blue), Oct 2007 (purple), May 2007 (gray), Feb 2006 (navy), or Feb 2004 (red). The y-axis represents the average number of oxidation events per peptide in the sample. Error bars 2 SD from a triplicate set of FPOP oxidations and analyses. Asterisks peptides with oxidation levels that significantly different than Neupogen® (p ≤ 0.01)
Fig. 2.
Circular dichroism analysis of GCSF samples. The near-UV CD spectra are presented from Neupogen® (green), Neupogen22 (pink), heat-treated Neupogen® (yellow), or recombinant GCSF samples generated in Feb 2008 (light blue), Oct 2007 (purple), May 2007 (gray), Feb 2006 (navy), or Feb 2004 (red)
Neupogen® vs. Neupogen22
According to the manufacturer, Neupogen® is still potent when left at room temperature for up to 24 h; therefore, our sample stored at room temperature for 22 h should be conformationally identical to the Neupogen® sample stored at 4°C. Looking at the HRF results in Fig. 1, no statistical differences are detected on the peptide level between the Neupogen® and Neupogen22 samples (ESM Table S1); the CD spectra are also identical, as expected (Fig. 2). This comparison establishes the capability of the HRF method to detect conformational equivalency between samples and adds a powerful aspect to the technique.
Neupogen® vs. heat-treated Neupogen®
As shown in Fig. 1, two peptides exhibit significant changes in the hydroxyl radical footprint after heating of Neupogen® to 95°C and cooling back to room temperature. Peptide 25–148 contains 124 amino acids and represents almost 71% of the protein. The native protein structure shows 80% alpha helical content in peptide 25–148 by NMR. The heat treated Neupogen® sample is the only sample that had a relative increase in the amount of oxidation on peptide 25–148, and this increase was substantial. The increase in hydroxyl radical oxidation of this peptide is probably due to the loss of structural stability here upon heating. A small decrease in oxidation at peptide 1–17 is possibly due to non-native structuring of the normally disordered N-terminus. CD data in Fig. 2 illustrates a loss of most of the original secondary structure in the heat treated Neupogen® (yellow) when compared to Neupogen® control (green), confirming the differences detected by HRF.
Neupogen® vs. GCSF February 2008 sample
Three of the four GCSF peptides (all except for 18–23) representing 96% of the whole protein are less oxidized in the February 2008 sample. A reduced amount of oxidation indicates a reduced solvent accessibility, which could be due to aggregation or oligomerization, or collapse of flexible loops into a non-native secondary structure. The CD spectrum for GCSF Feb 2008 sample suggests a shift in alpha helical to more beta sheet and random coil secondary structure. The reduced amount of oxidation and the appearance of beta-sheet secondary structure, which is often associated with protein aggregation (20), may indicate that the GCSF Feb 2008 is aggregating.
Neupogen® vs. GCSF October 2007 sample
Two GCSF peptides representing 13% of the protein are reported to have increased oxidation in the October 2007 sample compared to the Neupogen® sample; a slight increase in oxidation for peptide 1–17, and a much larger increase in oxidation for peptide 18–23. Peptide 1–17 consists of a highly mobile N-terminus and the first part of helix 1, while peptide 18–23 is highly structured; an increase in oxidation suggests a loss of structural stability in the N-terminal and helix 1 region of the protein. GCSF Oct 2007 CD spectrum corresponds to a modest decrease in alpha helical structure that dominates peptides 1–17 and 18–23, without the appearance of substantial beta-sheet structure observed in the GCSF Feb 2008 sample.
Neupogen® vs. GCSF May 2007 and Feb 2006 sample
May 2007 and Feb 2006 samples have identical CD spectra to each other in the near-UV region (minor differences measured in the UV region around 197 nm are probably due to increased noise stemming from buffer interferences in this region), indicating that the two proteins should have identical conformations to each other (Fig. 2). Both expired recombinant GCSF samples are similar to Neupogen®, but show measurably less helical structure. If HRF is a reliable technique, these two separate expired GCSF recombinant samples should show no differences in hydroxyl radical footprint when compared to each other, but both should show significant and similar differences when compared to Neupogen®. Using a Student’s t test (p values ≤ 0.01) to determine the statistical difference in the amount of oxidation from the HRF experiments, the 2007 and 2006 samples are not statistically different from one another on the peptide level (ESM Table S1). When comparing the two expired recombinant GCSF samples to Neupogen®, two peptides show small but statistically significant differences from Neupogen® (Fig. 1, ESM Table S1). Peptide 25–148 is slightly less oxidized than Neupogen®; this reduced amount of oxidation may represent a subpopulation that is beginning to oligomerize in solution, or it may represent a non-native conformation that is more compact in this region. Peptide 18–23 is more oxidized, suggesting a loss of structural stability in this region and confirming the results found by comparison of the CD spectra. These samples demonstrate clearly that HRF is capable of successfully identifying both identical conformations and small differences in conformation with sensitivity and robustness.
Neupogen® vs. GCSF February 2004 sample
The 2004 sample differs from Neupogen® by decreased oxidation on two peptides representing 81% of the protein. Both these peptides show a substantial decreases in oxidation similar to the Feb 2008 recombinant GCSF sample, suggesting the GCSF Feb 2004 sample has undergone significant conformational changes that protect surfaces that should be solvent accessible. Of all of the expired recombinant GCSF samples, the Feb 2004 sample has the CD spectrum that has the greatest loss of helical content and appearance of beta-sheet content, along with an increase in the random coil content. The HRF data would be consistent with a beta-sheet-mediated oligomer or aggregate of the recombinant GCSF sample.
Overall, the expired recombinant GCSF samples are all different from Neupogen®, although the extent of the differences varies. Hydroxyl radical footprinting was able to identify all of the differences in the recombinant GCSF samples as confirmed by CD. HRF was also able to confirm the identical conformations shared between the Neupogen® and the Neupogen22 sample held at room temperature for 22 h prior to analysis, as well as the Feb 2006 and May 2007 GCSF samples, which were confirmed by identical CD spectra. Unlike the CD data, the HRF data identified the general regions of the protein that changed conformationally, and the samples could be reanalyzed more thoroughly to identify the regions of conformational change with higher spatial resolution. Our data support the use of HRF for comparison of therapeutic recombinant proteins and potential biosimilars to their patented counterparts to test for conformational equivalence, as well as describe differences detected. HRF is well-suited to the analysis of conformational changes that occur due to sample aging and mishandling, as it is sensitive to relatively small conformational changes, rapid, and able to analyze conformational differences using a single analytical platform that can simultaneously analyze differences in primary structure.
While the analysis of GCSF was in excellent agreement with the data generated by CD, in order to test the robustness and sensitivity of HRF across the range of therapeutic proteins, we need to test more than different preparations of one protein. In order to do this, we examined different preparations of two other therapeutic proteins, interferon α-2A and erythropoietin, that were generated at different times during the last 12 years in order to identify conformational differences between the samples generated at different times.
Interferon
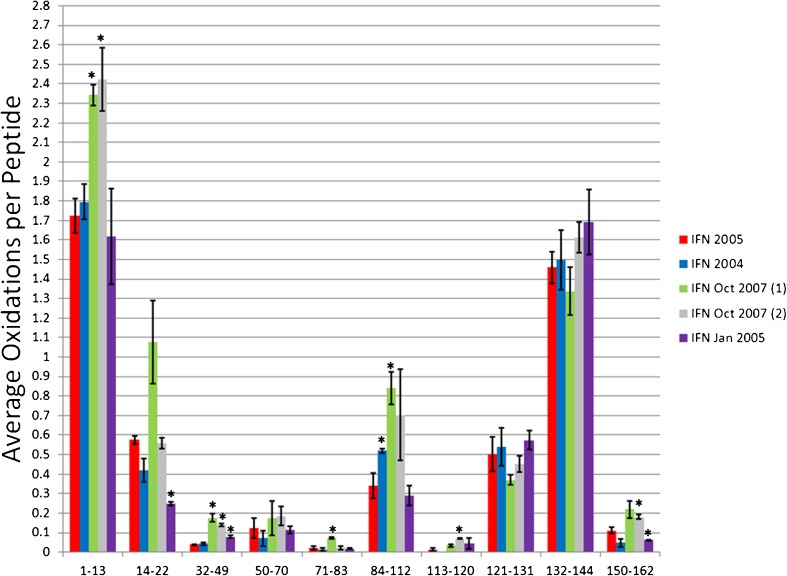
IFN has 165 residues with seven helices constituting 61% helical secondary structure (21). Ten tryptic peptides were analyzed giving 91% sequence coverage. The IFN 2005 sample was used as our “reference” conformation, and all other samples were compared to this conformation by HRF (Fig. 3), with observed differences verified by CD (Fig. 4). Student’s t test was used to calculate significance of differences in the hydroxyl radical footprint between the IFN 2005 reference and the other recombinant IFN protein sample (p value ≤ 0.01; ESM Table S2).
Fig. 3.
Hydroxyl radical footprinting of IFN samples. Each set of bars represents one peptide from recombinant IFN samples generated in 2005 (red), 2004 (blue), two samples from Oct 2007 (green and gray), and Jan 2005 (purple). The y-axis represents the average number of oxidation events per peptide in the sample. Error bars 2 SD from a triplicate set of FPOP oxidations and analyses. Asterisks peptides with oxidation levels that significantly different than IFN 2005 (p ≤ 0.01)
Fig. 4.
Circular dichroism analysis of IFN samples. The near-UV CD spectra are presented from recombinant IFN samples generated in 2005 (red), 2004 (blue), Oct 2007 (green), and Jan 2005 (purple)
IFN 2005 vs. IFN 2004
IFN 2004 shows a very similar hydroxyl radical footprint to IFN 2005, with very few quantitative differences in the amount of oxidation of any of the 10 peptides. The only peptide that shows a statistically significant change in the amount of oxidation is peptide 84–112, which consists of a helix–loop–turn structure. This region exhibits a modest increase in oxidation in the 2004 sample, suggestive of either a minor change in conformation in this region; probably in the loop region as the helices that pack against this helix do not exhibit changes in solvent accessibility. Comparison of the CD spectra of IFN 2005 and IFN 2004 reveal a small but measurable change in the secondary structure content indicated by a small shift in the local minima around 207 and 222 nm, and a change in the maxima and amplitude around 197 nm (although the extent of these farther UV changes are often obscured by buffer interferences). Even though the conformational change was small as measured by CD, we were able to clearly detect it using hydroxyl radical footprinting.
IFN 2005 vs. IFN Oct 2007 (1)
The IFN Oct 2007 (1) sample has four peptides that are more heavily oxidized (and therefore more solvent accessible) to a statistically significant extent than the IFN 2005 reference sample, as well as one additional peptide that is much more oxidized on average, but on which the replicate statistics are relatively poor. A significant increase in oxidation for the IFN Oct 2007 (1) samples occurs on peptides 1–13, 32–49, 71–83 and 84–112, along with an apparently large (but statistically insignificant) increase in oxidation of peptide 14–22. Peptide 1–22 represents the flexible N-terminus and the first helix of the IFN native structure, and the other two peptides, 71–112, stretch from the start of helix 4 through helix 5, turn 2 and 2 residues in helix 6. The substantial increase in solvent accessibility in these regions rich in secondary structure probably indicates destabilization of the structure of the protein, and should be reflected by a substantial loss of helical structure in the CD spectrum. The CD spectra for the IFN Oct 2007 (1) sample is much different than the other recombinant IFN samples. There is a large decrease in the amount of alpha helical secondary structure in the IFN Oct 2007 (1), consistent with the data from the HRF indicating a loss of structural stability in regions known to contain a large amount of helical content.
IFN 2005 vs. IFN Oct 2007 (2)
Due to lack of sample we were unable to obtain an accurate CD spectrum of the IFN Oct 2007 (2) sample. IFN 2007 (2) gave an almost identical HRF as IFN Oct 2007 (1), with the sole differences being reduced oxidation and improved statistics for peptide 14–22, yielding results similar to IFN 2005, and improved statistics for the C-terminal peptide 150–162, indicating a slight increase in oxidation compared to IFN 2005. All other differences were essentially identical to IFN Oct 2007 (1) described above.
IFN 2005 vs. IFN Jan 2005
In the HRF analysis, two peptides of the IFN Jan 2005 sample showed statistically significant changes in the amount of oxidation compared to the IFN 2005 sample, peptide 14–22 (representing the latter half of helix 1) and 150–162 (representing the latter half of the helix 7 and a post-helix turn). Unlike the previous samples, the IFN Jan 2005 sample shows a decrease in the solvent accessibility from the IFN 2005 sample, indicating either an increased stability of the structure, and alternate, more compact conformation, or an oligomerization/aggregation event. The IFN Jan 2005 sample has a similar CD spectrum to the IFN 2005 reference, but exhibits an increase in alpha helical structure. An increase in secondary structure content caused by stabilization and/or extension of helixes 1 and 7 of the protein would explain both the CD spectra and the HRF data.
Erythropoietin
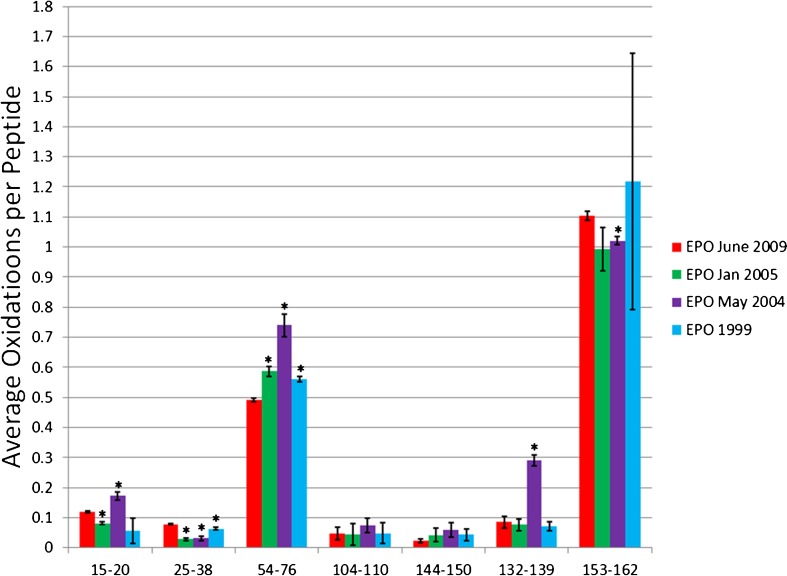
EPO is a therapeutic protein with 166 residues, with 56% of the protein forming five alpha helices (22). Trypsin digestion of EPO yields eight detectable peptides resulting in 53% coverage. The relatively low sequence coverage was due to the primary sequence of EPO, which has a large number of lysines and arginines resulting in much of the protein being cleaved by trypsin into very small peptides that are very difficult to detect by LC-MS. Of the eight peptides, one had no oxidation modifications after HRF. We compared the seven peptides that were modified for each of the five recombinant EPO samples using HRF (Fig. 5). EPO June 2009 sample is the most recently made sample and will be used as a reference to compare the difference in conformation among the other EPO samples. A difference in the hydroxyl radical footprint was considered significant if the p value by Student’s t test was ≤ 0.01 (ESM Table S3). Differences in conformation identified by HRF were confirmed by CD (Fig. 6).
Fig. 5.
Hydroxyl radical footprinting of EPO samples. Each set of bars represents one peptide from recombinant EPO samples generated in 2009 (red), Jan 2005 (green), May 2004 (purple), and 1999 (blue). The y-axis represents the average number of oxidation events per peptide in the sample. Error bars represent 2 SD from a triplicate set of FPOP oxidations and analyses. Asterisks peptides with oxidation levels that significantly different than EPO 2009 (p ≤ 0.01)
Fig. 6.
Circular dichroism analysis of EPO samples. The near-UV CD spectra are presented from recombinant EPO samples generated in 2009 (red), Jan 2005 (green), May 2004 (purple), and 1999 (blue)
EPO June 2009 vs. EPO Jan 2005
EPO Jan 2005 is more oxidized at peptide 54–76 compared to the 2009 reference sample. Peptide 54–76 is essentially helix three of native EPO. An increase in oxidation probably signifies a loss of structural stability, causing the peptide to become more solvent exposed and increasing peptide oxidation. The change in the amount of oxidation is minimal, but statistically significant. The sample also exhibits a small decrease in the solvent accessibility of peptide 15–20 and 25–38. All other peptides are not oxidized differently between the EPO 2009 and EPO 2005 samples to statistical significance. The CD spectra for EPO Jan 2005 appears to have the similar shape of EPO 2009, but with a slight shift of the minimum and maximum to shorter wavelengths, a decrease in ellipticity amplitude in the 200–240 nm range, and an increase in ellipticity amplitude in the 190–200 nm range.
EPO June 2009 vs. EPO May 2004
EPO May 2004 shows statistically significant increases in solvent accessibility in three peptides; peptide 15–20 exhibits a small increase in solvent accessibility, while peptides 54–76 and 132–139 show fairly large increases in solvent accessibility. Additionally, EPO May 2004 exhibits small but statistically significant decreases in solvent accessibility in two other peptides, 25–38 and 153–162. The increased protection of some regions and increased exposure of other regions argues against a global structural destabilization, and rather suggests a different but relatively stable conformation for EPO May 2004 compared to EPO June 2009. These results are confirmed by the CD spectra, which show the EPO May 2004 sample with a clearly structured, but different CD spectrum than the EPO June 2009 reference. The minimum is shifted to lower wavelengths, with a small decrease in the amplitude of ellipticity, while the maximum is shifted to lower wavelengths with a substantial increase in ellipticity amplitude.
EPO June 2009 vs. EPO 1999
The hydroxyl radical footprint of the EPO 1999 sample was almost identical to the 2009 sample, with the only statistically significant difference in the hydroxyl radical footprint appearing as a very small decrease in oxidation on peptide 25–38, which has a random coil structure in native EPO, and a small increase in oxidation in peptide 54–76, which contains the majority of helix 3. These data correlate very well with the CD data, where the 1999 sample, by far, has the most similar CD spectrum to the 2009 reference, with almost identical minima and maxima. However, the CD spectrum does indicate small differences in the secondary structure, which are confirmed by small differences in the hydroxyl radical footprint. The data indicate quite clearly that even small differences in conformation as measured by CD can be clearly observed in the hydroxyl radical footprint.
All of the EPO samples were different from the 2009 reference sample by HRF, although the differences varied in magnitude from the almost-identical 1999 sample to the significantly different 2004 sample. The differences were also confirmed in the CD spectrum, with the 1999 sample being nearly identical and the 2004 and 2005 samples being substantially more varied.
DISCUSSION
Hydroxyl radical footprinting was capable of identifying differences in the conformation between 12 different pairings of samples across three different therapeutic proteins. HRF proved to be able to detect differences due to loss of solvent accessibility (probably due in some cases to aggregation or oligomerization) or due to increases in solvent accessibility (probably due to structural destabilization). HRF was also able to successfully detect two pairs of samples that had no differences in conformation. All analyses were carried out in technical triplicate (i.e., the therapeutic protein was aliquoted into three samples which were independently oxidized by FPOP and independently analyzed by LC-MS). The overall time for a single analysis is approximately 2 h (~10 min for FPOP and quenching, 1 h for denaturation and digestion, 30 min for LC-MS), with all of the FPOP steps being capable of automation with currently available commercial technology. Sample requirements are light, with only ~90 pmol of sample required for the triplicate analysis. HRF is capable of accommodating almost any buffer or formulation component and is largely insensitive to pH (12), although some components like carrier proteins can complicate the resulting LC-MS spectra and the inclusion of high concentrations of radical scavengers will require the generation of higher burst concentrations of hydroxyl radicals to generate sufficient hydroxyl radical footprinting data to differentiate conformations. The method is also capable of handling heterogeneous protein conformations and oligomerization/aggregation states, as potentially demonstrated by the GCSF samples with non-native beta sheet content and substantially more protected surface areas, and also as demonstrated in other studies on confirmed polydisperse protein oligomers (23). The reproducibility of the oxidation chemistry by FPOP and the quantitation of oxidation by LC-MS were more than sufficient for the purposes of routine analysis. The use of only MS data from short LC gradients, as opposed to MS/MS and long, careful separations to isolate oxidation isomers, limits the spatial resolution of the footprinting information obtained. However, interpretation of MS/MS spectra of peptides oxidized by hydroxyl radicals is difficult and time consuming, requiring at the very least manual verification of all identified oxidized peptides. Conversely, quantification at the peptide level is fast and can be performed without special expertise, as the products of the complicated hydroxyl radical protein chemistry resolve into a limited number of mass shifts, most of which represent net additions of one or more oxygen atoms (11). Once a sample has been initially characterized, the data analysis can also be easily automated, as the hydroxyl radical footprint will boil down to a measurement of the abundance of specific peak abundances eluting at specific times from the LC. Similarly, the quantification by LC-MS does not require high-end LC or MS instrumentation, as the experiment is quite compatible with almost any electrospray instrument that is suitable for peptide LC-MS.
These useful analytical qualities make HRF an attractive technique for the characterization of therapeutic proteins, both during the development and production stage and for quality control afterwards. The ability to use a single analytical platform to characterize both primary structure and three-dimensional conformation in a single analysis is a powerful option for screening of therapeutic proteins. The small sample quantities, the potential for automation of analysis, and the relatively short analysis times makes HRF suitable for insertion into the production process as a quality control measure, as well as for spot checks of protein shelf life during the storage and transportation chain. Similarly, HRF is a very useful analytical technique for the development and validation of biosimilars to patented therapeutic proteins; as abbreviated, HRF is capable of confirming conformational identity as well as capable of not only identifying conformational differences, whether they be in regions with stable secondary structure or in more dynamic loops, but also roughly localizing these conformational differences for either more thorough HRF analysis with UPLC-MS/MS or for thorough analysis by more demanding high-resolution structural techniques. The abbreviated HRF technique demonstrated here can fill an important niche as an intermediate resolution and rapid structural technique for the conformational analysis of therapeutic proteins.
While HRF performed well in all cases examined here, there are instances that should be handled with care when using HRF for comparing therapeutic proteins. One important factor that must be carefully considered in designing an HRF experiment for the analysis of therapeutic protein formulations is composition of the formulation other than the therapeutic protein. Different formulation components (buffers, carrier proteins, etc.) will scavenge radicals to different extents, which can result in global reductions in the hydroxyl radical footprint of a protein even without any conformational change. This reduction is due to competition between the therapeutic protein and buffer components for the hydroxyl radical during the radical burst, resulting in less diffusing hydroxyl radical available to oxidize the therapeutic protein and a lower apparent rate of oxidation. However, it is quite possible with careful planning to apply HRF to different formulations with widely differing radical scavenging properties. In order to correct for different radical scavenging properties of the buffer, mixtures matching the formulations to be tested, without the therapeutic protein, should be prepared and spiked with a reporter. This reporter can either be a radical-sensitive chromophore, or a small unstructured peptide that can be monitored for oxidation by LC-MS. The radical dose (as controlled by laser pulse energy and hydrogen peroxide concentration) is then adjusted until the amount of oxidation of the reporter is equal between the two samples. These adjusted conditions are then compared to compensate for the radical scavenging properties of the different formulations.
CONCLUSION
Here, we describe an abbreviated hydroxyl radical footprinting technique for the conformational comparison of therapeutic protein samples using hydroxyl radical protein chemistry and LC-MS analysis. Using this method, we are able to identify conformational differences between Neupogen® and expired recombinant GCSF samples, as well as conformational differences between different samples of the therapeutic proteins erythropoietin and interferon α-2A. We are also able to confirm conformational equivalence between two different samples of GCSF produced 9 months apart, as well as two Neupogen® samples handled differently, but still retaining the same conformation. All conformational HRF analyses were successfully confirmed by circular dichroism analysis, demonstrating the accuracy and robustness of this technique for the comparison of conformations of therapeutic proteins.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Independent two-tailed Student’s t test of HRF between GCSF samples (DOC 34 kb)
Independent two-tailed Student’s t test of HRF between IFN samples and IFN 2005 (DOC 28 kb)
Independent two-tailed Student’s t test of HRF between EPO samples and EPO June 2009 (DOC 26 kb)
ACKNOWLEDGMENTS
This project was supported by grants from the National Center for Research Resources (5P41RR005351-23) and the National Institute of General Medical Sciences (8 P41 GM103390-23) from the National Institutes of Health. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would also like to thank Dr. Jeffrey Urbauer of The University of Georgia for use of his CD instrumentation. JSS would also like to thank Dr. Marshall Bern for numerous productive conversations on the topics explored here.
References
- 1.Locatelli F, Del Vecchio L, Pozzoni P. Pure Red-cell aplasia “epidemic”—mystery completely revealed? Perit Dial Int. 2007;27(Supplement 2):S303–S307. [PubMed] [Google Scholar]
- 2.McKoy JM, Stonecash RE, Cournoyer D, Rossert J, Nissenson AR, Raisch DW, et al. Epoetin‐associated pure red cell aplasia: past, present, and future considerations. Transfusion. 2008;48(8):1754–1762. doi: 10.1111/j.1537-2995.2008.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessler M, Goldsmith D, Schellekens H. Immunogenicity of biopharmaceuticals. Nephrol Dial Transplant. 2006;21(suppl 5):v9. doi: 10.1093/ndt/gfl476. [DOI] [PubMed] [Google Scholar]
- 4.Srebalus Barnes CA, Lim A. Applications of mass spectrometry for the structural characterization of recombinant protein pharmaceuticals. Mass Spectrom Rev. 2007;26(3):370–388. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- 5.Bobst CE, Abzalimov RR, Houde D, Kloczewiak M, Mhatre R, Berkowitz SA, et al. Detection and characterization of altered conformations of protein pharmaceuticals using complementary mass spectrometry-based approaches. Anal Chem. 2008;80(19):7473–7481. doi: 10.1021/ac801214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol Adv. 2012;30(1):210–222. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 8.Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol Adv. 2012;30(1):210–222. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendoza VL, Vachet RW. Probing protein structure by amino acid‐specific covalent labeling and mass spectrometry. Mass Spectrom Rev. 2009;28(5):785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom Rev. 2010;29(4):651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 11.Xu G, Chance MR. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal Chem. 2005;77(14):4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 12.Buxton GV, Greenstock CL, Helman WP, Ross AB. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals (.Oh/.O−) in aqueous solution. J Phys Chem Ref Data. 1988;17(2):513–886. doi: 10.1063/1.555805. [DOI] [Google Scholar]
- 13.Charvátová O, Foley B, Bern M, Sharp J, Orlando R, Woods R. Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1. J Am Soc Mass Spectrom. 2008;19(11):1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem. 2009;81(16):6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson C, Janik I, Zhuang T, Charva Tova O, Woods R, Sharp J. Pulsed electron beam water radiolysis for submicrosecond hydroxyl radical protein footprinting. Anal Chem. 2009;81(7):2496–2505. doi: 10.1021/ac802252y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16(12):2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Desjardins P, Hansen JB, Allen M (2009) Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. J Visualized Experiments: JoVE (33): 1610 [DOI] [PMC free article] [PubMed]
- 18.Woody RW. Circular dichroism. Biochem Spectrosc. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 19.Zink T, Ross A, Lueers K, Cieslar C, Rudolph R, Holak TA. Structure and dynamics of the human granulocyte colony-stimulating factor determined by NMR spectroscopy. Loop mobility in a four-helix-bundle protein. Biochemistry. 1994;33(28):8453–8463. doi: 10.1021/bi00194a009. [DOI] [PubMed] [Google Scholar]
- 20.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 21.Klaus W, Gsell B, Labhardt AM, Wipf B, Senn H. The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution. J Mol Biol. 1997;274(4):661–675. doi: 10.1006/jmbi.1997.1396. [DOI] [PubMed] [Google Scholar]
- 22.Cheetham JC, Smith DM, Aoki KH, Stevenson JL, Hoeffel TJ, Syed RS, et al. NMR structure of human erythropoietin and a comparison with its receptor bound conformation. Nat Struct Biol. 1998;5(10):861–866. doi: 10.1038/2302. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Watson C, Sharp JS, Handel TM, Prestegard JH. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure. 2011;19(8):1138–1148. doi: 10.1016/j.str.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Independent two-tailed Student’s t test of HRF between GCSF samples (DOC 34 kb)
Independent two-tailed Student’s t test of HRF between IFN samples and IFN 2005 (DOC 28 kb)
Independent two-tailed Student’s t test of HRF between EPO samples and EPO June 2009 (DOC 26 kb)