Summary
Colonization by methicillin-resistant Staphylococcus aureus (MRSA) may be persistent in people, and is horizontally transmissible. The scientific literature suggests that domestic pets may also participate in cross-transmission of MRSA within households. The objectives of this study were to evaluate the prevalence of and risk factors for MRSA carriage by pets residing in households with an MRSA-infected person. From 66 households in which an MRSA infected patient resided, we screened 47 dogs and 52 cats using a swab protocol. Isolates from pets and humans were genotyped using two techniques, and compared for concordance. Human participants completed a 22-question survey of demographic and epidemiologic data relevant to staphylococcal transmission. Eleven of 99 pets (11.5%) representing 9 (13.6%) of households were MRSA-positive, but in only 6 of these households were the human and animal-source strains genetically concordant. Human infection by strain USA 100 was significantly associated with pet carriage [OR = 11.4 (95% C.I. 1.7, 76.9); p=0.013]. Yet, for each day of delay in sampling the pet after the person’s MRSA diagnosis, the odds of isolating any type of MRSA from the pet decreased by 13.9% [(95% C.I. 2.6%, 23.8%); p=0.017)]. It may be concluded that pets can harbor pandemic strains of MRSA while residing in a household with an infected person. However, the source of MRSA to the pet cannot always be attributed to the human patient. Moreover, the rapid attrition of the odds of obtaining a positive culture from pets over time suggests that MRSA carriage may be fleeting.
Keywords: Pets, Staphylococcus, MRSA, zoonosis, Bacterial Typing Techniques
Bacteria in the genus Staphylococcus are the most common pathogens associated with nosocomial and community-onset skin and soft tissue infections (SSTI), of both humans and animals, world-wide (Werkenthin et al., 2001; Diederen et al., 2006). An increase in the prevalence of methicillin-resistance (MR) in Staphylococcus aureus isolates (MRSA) over the past four decades has led to higher morbidity, mortality, and hospital expenditures associated with S. aureus infections of people (Cosgrove et al., 2005). It is known that persons who live in close contact with MRSA patients can also become colonized, and that colonization may then persist for months to years (Marschall et al., 2006; Vriens et al., 2005). Colonized people may also serve as a source of MRSA spread to susceptible individuals (Calfee et al, 2003; Johansson et al., 2007; Huijsdens et al., 2006). Although S. aureus is not the primary staphylococcal species that causes SSTIs of dogs and cats, the current scientific literature demonstrates that domestic pets can carry or be infected by the same epidemic strains of MRSA that cause SSTI of people (Weese et al, 2006; Loeffler et al., 2010). Recommended protocols for prevention of recurrent MRSA infections of people do address pet colonization, but the supporting risk assessments are based on limited reports such as small case series and anecdotal descriptions (CLSI 2010). Additional objective data to help quantify and subsequently mitigate the risk of human-animal cross-transmission would be valuable to health care providers and public health professionals.
The objectives of this study were to test the primary hypothesis that infected people can cross-transmit MRSA to domestic pets (dogs and cats) with which they live in close contact, and to identify risk factors associated with this cross-transmission. It was also hypothesized that the proximity of a pet to a person with MRSA infection (defined as “close” versus “casual” contact) would be the primary risk factor associated with pet carriage of MRSA.
Materials and methods
Regulatory approvals
Approval for human specimen collection and processing and administration of the survey questionnaire, was granted by the University of Pennsylvania’s Institutional Review Board. Approval for pet sampling was granted by the Institutional Animal Care and Use Committee. Adult participants signed informed consent documents for themselves, their pets, and minor children under the age of 18. Verbal assent was obtained from children 5 years of age and older.
Human-source specimens
Participants were recruited from the University of Pennsylvania Health System (UPHS) and the Children’s Hospital of Philadelphia (CHOP) Health Network. All patients 3 years of age or greater, evaluated at each of the participating institutions in various outpatient settings (e.g., emergency department, offices, clinics) for SSTI over an 18-month time period, and who had MRSA isolated from aerobic cultures, were identified as potentially eligible to participate in the study. Laboratory personnel stored MRSA isolates on cotton-tipped swabs (Bacti swab, Remel, Lenexa, KS, USA), with no patient identifiers other than a numeric code to link the specimen to the laboratory information system. Assent to contact patients was obtained from submitting physicians. Patients (or guardians in the case of minors) were then contacted by phone to determine if a dog or cat resided in the household. Those who shared a household with a pet were eligible for inclusion in the study. Participants agreed to visit either the Veterinary Hospital of the University of Pennsylvania (VHUP) or their primary veterinary care provider for pets to be sampled. Informed consent was obtained at that time for access to the participant’s MRSA isolate, and these were transferred to the Clinical Microbiology Laboratory at the VHUP, where they were cryopreserved (Microbank, Pro-Lab Diagnostics, Austin, TX, USA) for subsequent molecular testing.
Survey instrument and data collected
At the time of pet screening, each participant completed a survey questionnaire which captured epidemiologic data for the 12 month period prior to MRSA diagnosis, and covered all members of the household. Data were collected with regard to several risk factors for community-based acquisition of MRSA (participation in team sports, utilization of a gym/health club, residence in group housing) versus nosocomial acquisition (recent surgery, hospitalization, emergency room visits, repeated visits to outpatient treatment facilities, employment in human health care practice), in an attempt to correlate these risk factors with the strain types isolated from individual households. The survey also captured data regarding pet-related factors, such as living conditions (indoor/outdoor/mixed), presence of other pets in the household, and the nature of person-pet contact. Animal health questions documented the pet’s history of pyoderma, otitis externa, or any other types of infection by staphylococci, and comorbid conditions such as diabetes mellitus, renal insufficiency, hepatic insufficiency, neoplasia, and retroviral infection (cats). Glucocorticoid use and other immunosuppressive therapies administered to pets within the preceding 90 days, as well as all antimicrobial therapy of human and animal participants within the 90 days that preceded study enrollment, was documented. The antibiotic class and the specific antimicrobial agent were noted. Data were provided by pet owners and confirmed by review of the primary care provider’s veterinary medical record (when available).
The nature of human and animal contact was defined as either “close” or “casual”. This definition was extrapolated from the medical literature on interpersonal MRSA transmission within households (Johansson et al., 2007). Participants were asked several questions about person-pet interaction, and points were assigned as follows: (1). Human participant is the primary care-provider for the pet (feeding, grooming, bathing, medicating, exercising): yes = 1 point, no = 0; (2). The pet sleeps in/on the human participant’s bed: yes = 2 points, no = 0; (3). Human participant allows the pet to lick the face or hands: daily = 4 points, weekly = 2 points, less than weekly = 1 point, no = 0; (4). The pet is housed: exclusively indoors = 2 points, indoor/outdoor = 1 point, exclusively outdoors = 0. The maximum point total was 9. The contact score was dichotomized for statistical analysis, and “close” contact was defined as a score of 6 or greater, while “casual” contact was defined as a score less than 6. Other data collected included the date of the person’s culture, site of infection, admitting outpatient service, and the date of pet sampling. The lag time between the human participants’ MRSA isolation dates and the collection of samples from the pets was calculated.
Animal-source specimens
Pets were screened for MRSA carriage using cotton-tipped swabs. Pets were sampled with a single swab at four body sites that have been established as primary colonization sites in dogs and cats: anal mucosa, groin, distal nares, and oral mucosa (Abraham et al, 2007; Griffeth et al., 2008). The oral cavity was swabbed first, followed by each naris, then the groin, and finally the anus. If evidence of active staphylococcal infection was present on the pet, as determined by the veterinarian, a representative site was sampled with an additional swab.
Each swab was placed into a tube of mannitol salt broth (Northeast Laboratory Services, Winslow, ME) and incubated at 35°C overnight as a selective enrichment for the isolation of S. aureus. One microliter of MSB was subcultured to a BBL CHROMagar™ MRSA plate (Remel) and incubated for a minimum of 18 h at 35°C. Plates were read against a white background and mauve colonies were selected for further processing. If no mauve colonies were recovered, plates were incubated for an additional 24 ± 4 h as described by the manufacturer, and then discarded if no colony growth was apparent. Potential MRSA isolates were sub-cultured to blood agar plates for additional confirmatory testing. If colonies of more than one morphologic type were present, one representative of each was sub-cultured. Isolates were tested for clumping factor and protein A (associated with S. aureus) using the Staphaurex test (Remel). S. aureus isolates were tested for the PBP2’ protein product of the mec-A gene to confirm identity as MRSA (Oxoid - Remel). The rapid PBP2’ test has been shown to be equal in specificity and sensitivity to commercial real-time PCR (polymerase chain reaction) for the mec-A gene (Lee et al., 2004). Biochemical confirmation of bacterial species and verification of resistance to oxacillin, using breakpoints defined by the Clinical and Laboratory Standards Institute guidelines, (CLSI 2008) was performed in a MicroScan Walkaway 40 (Dade Behring, Irvine, CA, USA). Confirmed isolates were cryopreserved for subsequent molecular testing.
Molecular testing of human and animal-source specimens
Pulsed-field-gel-electrophoresis (PFGE) was performed on all human and animal MRSA isolates as described previously (McDougal et al, 2003). BioNumerics software version 5.1 (Applied Maths, Kortrijk, Belgium) was used to identify percent similarities on a dendrogram derived from the unweighted pair group method using arithmetic averages and based on Dice coefficients. A similarity coefficient of 80% was selected to define pulsed-field profile (PFP) clusters as described for S. aureus (McDougal et al., 2003). S. aureus protein A (spA) typing was performed as described previously (Shopsin et al, 1999). Briefly, amplification and sequencing was perfomed to identify sequence variation of the polymorphic region X of the spa gene. Bionumerics software, version 5.1, was used for spa type analyses. SpA type assignments are provided automatically through the spA typing plugin.
Sample size and power
Because the prevalence of MRSA colonization among animals living with a MRSA-infected person was unknown, we based our estimation on a report which suggested that 15% of humans living in a household with a patient with an MRSA infection would also be colonized with MRSA (Calfee et al, 2003). We therefore conservatively estimated that the prevalence of MRSA among animals in our study would approximate 12.5%. We predicted that approximately 50% of cases (positive pet in household) and 25% of controls would exhibit a “close” relationship with their pet, and believed it would be clinically important to be able to show an odds ratio (OR) of at least 2.0 for the association between “close” human-animal contact and MRSA colonization of the pet. Given these assumptions, we calculated that with 38 case households and 272 control households we would have 99% power to detect an association (2-sided alpha of 0.05).
Statistical analysis
Categorical variables were summarized by frequencies while continuous variables were summarized by the mean or median, standard deviation, and range. Bivariable analysis was subsequently conducted to determine the unadjusted association between potential risk factors and MRSA strain type. Categorical variables were compared using the chi square test, while continuous variables were compared using the Student’s t or Wilcoxon Rank Sum test. Unadjusted OR’s and confidence intervals (CI) were calculated to evaluate both the strength of any associations as well as the precision of the estimate of the effect. Stratified analyses were subsequently performed using the Mantel-Haenszel statistic to evaluate the effects of each variable of interest as a possible confounder. Multivariable OR estimates were calculated using multiple logistic regression analysis. Variables were initially included in the model if they were significant at the p<0.1 level in bivariable analysis. Other variables of interest were added in a forward and then backward selection approach and retained in the final model when p<0.05. All analyses were conducted with SAS version 9.2 (Cary, NC, USA).
Results
During the 18 months of open enrollment, MRSA isolates were collected from 1,135 adult and 1,051 child outpatients presenting to the UPHS and CHOP health networks, respectively. The physician response rate to requests for assent to contact patients was 43% overall. From the potential subjects contacted to ascertain pet ownership, the response rate was less than 30%. Ultimately, 80 subjects assented to participation and 66 of them presented their pets for evaluation and provided informed consent.
Age of participants ranged from 3 years to 89 years (median 18.0; sd = 20.3), and 32/66 (48.5%) were children aged 18 years or younger. Forty (60.6%) participants were female, 33 (50.0%) were white non-Hispanic, 30 (45.5%) were black non-Hispanic, 1 was white/Hispanic, and 2 participants did not self-identify race or ethnicity. There were no significant differences in MRSA strain types isolated from participants according to sex, race, or age category (data not shown). The MRSA strain type isolated from the majority of human participants was USA 300 (49/66; 74.2%), while 14/66 isolates (21.2%) were strain USA 100, and 2 isolates (3.0%) were strain USA 800 (complete data shown only for those households in which a positive pet resided; figure 1). One isolate was not susceptible to Sma1 enzyme digestion, and PFGE could not be performed. Because there was not a positive pet in the household, this human-source isolate was not evaluated further.
Figure 1.
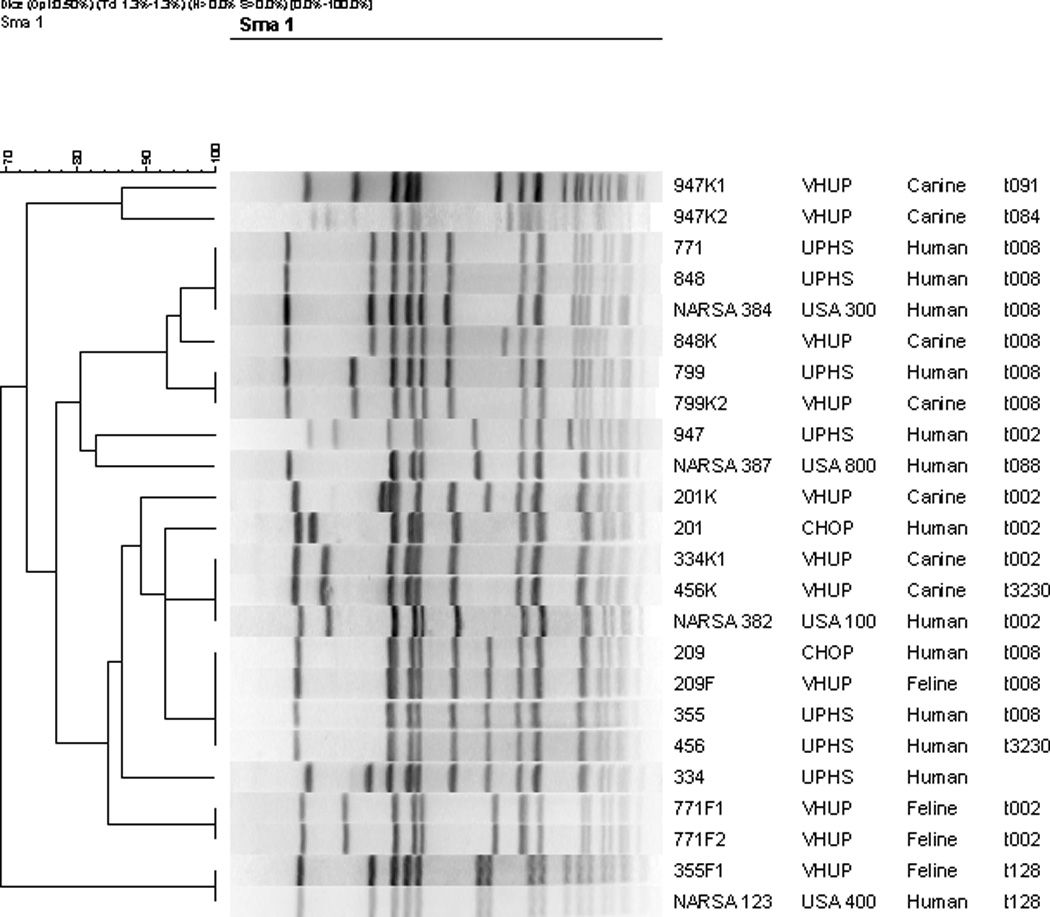
Results of PFGE after SmaI restriction of MRSA isolates derived from 20 clinical samples, which were obtained from people and their respective pets. Four USA type strains [provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA)] are included for reference. The groupings of vertical lines represent DNA fragments separated according to length and molecular weight, and the scale to the left of the figure indicates the degree of relatedness among isolates. Human-animal paired isolates share a 3-number code (left-hand column), while pet isolate codes also include a letter designation for pet type (K=canine, F=feline) and in some cases, an additional number if more than one pet of the same species is represented from the same household. spa type designations are presented in the far right-hand column. The participants’ hospitals of origin are indicated by: CHOP = Children’s Hospital of Philadelphia; UPHS = University of Pennsylvania Health System; VHUP = Veterinary Hospital of the University of Pennsylvania.
From the 66 households in which an MRSA infected patient resided, 47 dogs and 52 cats were screened. The number of pets sampled per household ranged from one to three. MRSA was isolated from 11/99 (11%) of pets (7 dogs and 4 cats) and represented 9 (13.6%) of the 66 households. One pet had clinical lesions suggestive of bacterial infection at the time of sampling, but methicillin-resistant S. pseudintermedius was isolated rather than MRSA (confirmed by biochemical methods and PFGE). The most common strain isolated from pets was USA 100 (6/11; 54.5%). Other strain types isolated from pets included USA 300 (2 isolates), USA 400 (1 isolate) and two isolates that did not cluster with a USA clonal group.
The Figure illustrates PFGE and spA types of the strains isolated from human and animal participants in the 9 case households. Within all 9 households, the participant was the first and only person with known MRSA infection. In six of the case households, the human and animal-source isolates were concordant as defined by Dice coefficients >80% (Table). In two of these concordant households (nos. 6 and 9), the participants were children under the age of 8.
Table.
Strain type data for human-animal paired MRSA isolates
| Household no. |
Human & animal case designations |
Strain type interpretation |
% Similarity (Dice coeff.) |
|---|---|---|---|
| 1 | 771 & 771F1 | USA300/100 | 77.4% |
| 1 | 771 & 771F2 | USA300/100 | 77.4% |
| 2* | 848 & 848K | USA 300/300 | 90.9% |
| 3* | 799 & 799K2 | USA300/300 | 100% |
| 4 | 947 & 947K1 | USA800/xxx | 72.7% |
| 4 | 947 & 947K2 | USA800/xxx | 74.3% |
| 5 | 355 & 355F1 | USA100/400 | 73.3% |
| 6* | C209 & C209F | USA100/100 | 100% |
| 7* | 456 & 456K | USA100/100 | 92.9% |
| 8* | 334 & 334K1 | USA100/100 | 82.8% |
| 9* | C201 & C201K | USA100/100 | 85.7% |
Concordant human-animal paired isolates
xxx = isolates that do not cluster with a designated USA strain type
In a multivariable model, significant risk factors for pet carriage of MRSA included human exposure to amoxicillin-clavulanic acid within the month prior to diagnosis [OR = 16.6 (95% C.I. 2.6, 107.9), p=0.003], and human infection by strain USA 100 [OR = 11.4 (95% C.I. 1.7, 76.9), p=0.013]. None of the positive pets had received antimicrobial or immunomodulatory drugs within the 90 days that preceded sampling, and none had any known co-morbidities that might have predisposed them to MRSA carriage. The lag in sampling pets after the person’s MRSA diagnosis ranged from 2 to 55 days (mean 22.5, sd = 10). For each day of delay in sampling the pet after the person’s positive culture, the odds of isolating MRSA from the pet decreased by 13.9% [(95% C.I. 2.6%, 23.8%); p=0.017)]. There was no correlation between the odds of MRSA isolation from a pet and the human-animal contact score (p=0.753). Neither the median contact score nor the odds of isolating MRSA from the pet varied between adult and child participants.
Discussion
The results of this study demonstrate that MRSA can be isolated from a sizable proportion of pets (11.6%) when they reside with a person that has active or recent infection. However, the observational methods used cannot discern between true biological colonization of these pets versus transient mechanical carriage. Similarly, the temporal relationship of human and pet colonization is not discernable from our study since longitudinal sampling was not conducted. Although pet owners were encouraged to return for longitudinal sampling every 2 weeks, only 1 participant complied with this request. This household had 2 positive cats identified during initial screening; 1 of the cats remained positive 2 weeks later (see figure).
The decreased yield over time (13.9% reduction in the odds of positive culture for each day delay in pet sampling) may suggest that carriage is very transitory in pets. Dogs are primarily colonized by Staphylococcus pseudintermedius, a coagulase-positive species which has also been shown to be the primary canine pathogen (Griffeth et al., 2008). It is plausible that S. pseudintermedius could out-compete a non-native species such as S. aureus, therefore preventing persistent colonization. The case is less clear for cats, as a recent cross-sectional survey has demonstrated that cats harbor S. pseud-intermedius and S. aureus with equal frequency, although co-colonization is rare (Abraham et al., 2007). Due to the low numbers of positive pets in this study, it was not possible to statistically compare the rates of MRSA carriage between the dogs and cats examined.
Strain USA 100 has long been the predominant clonal group of MRSA that causes nosocomial infections in the United States (Graham et al, 2006), but has recently migrated from hospitals into the community (Diederen et al., 2006; Mulvey et al., 2005; Stevenson et al, 2005). USA 300 and USA 400 have been associated with community-onset infections since the mid-1990’s (King et al., 2006)) but are a less commonly associated with nasal colonization of healthy people (Gorwitz et al, 2008). USA 800 has been described as a source of nosocomial pediatric infections, but it is also the second most prevalent strain type causing nasal colonization of Americans (Gorwitz et al, 2008). USA 500 strains are most commonly isolated from horses and the people who work with them (Weese et al, 2005). Although two participants reported regular contact with horses, no USA 500 strains were isolated.
The majority of pet isolates in this study were strain USA 100, while the majority of human isolates were USA 300. This dichotomy resulted in a very high odds ratio for pet carriage of USA 100. This should not be considered unusual, since USA 100 appears to be the predominant strain that causes infection of small animal patients (i.e., dogs and cats) and colonization of small animal veterinary practitioners. (Loeffler et al., 2005; Weese et al, 2006; Hanselman et al., 2006; Morris et al, 2010). The reasons for this are currently unknown, although increased affinity for dog and cat tissues may be speculated. An alternative explanation could be that the pets are simply more likely to be exposed to USA 100, as it is currently the top-ranking cause of human nasal colonization within the general population of the USA (Gorwitz et al, 2008).
We also identified a high odds ratio for pet carriage of MRSA when the person (rather than the pet) had been exposed to amoxicillin-clavulanic acid within the 90 days prior to study enrollment. We cannot provide a plausible explanation for this as by definition, all human participants enrolled in the study had harbored active or recent MRSA infections at the time of pet screening. Therefore, the patient’s exposure to any particular antibiotic would not be expected to increase the odds of cross-transmission to a pet. However, it is possible that human exposure to amoxi-clav is a surrogate for some cross-transmission risk factor that we have not considered or did not evaluate.
This study used PFGE and spA typing to infer clonal relationships between isolates. These methods were used because a recent analysis of multiple methods for strain typing of MRSA showed that the conjugation of results provided by these two techniques offers the best possible discriminatory power (Faria et al., 2008; Vainio et al, 2008). Clonal groups, as defined by PFGE, must exhibit 80% or greater homology, and may differ by as many as 3 bands; major PFP sub-types of each USA clonal group have been described (McDougal et al., 2003). This clonal inference system is useful to investigate dissemination trends through large populations and to track regional outbreaks. Although the authors are unaware of any guidelines that apply to interpretation of transmission studies within space-limited systems like households, it is known that 2 to 3 band differences in the PFP could represent a single genetic event, such as a point mutation, insertion, deletion, or chromosomal inversion (Tenover et al., 1995). Such events could conceivably occur in a single strain shared by residents within a household. Therefore, to define strain concordance for the purposes of statistical analysis, we elected to interpret ≤3-band differences between strains (correlating to a Dice coefficient of 82.8% or greater in this study) as sufficient evidence of person-pet cross-transmission.
Figure 1 shows that both of the person-pet pairs from households 3 and 6 that shared identical PFGE profiles also shared identical spA types, as expected. Within three households (nos. 7, 8, 9) isolates that belonged to the USA 100 clonal group were recovered from each person and their respective pets, but the Dice coefficients of similarity differed by 7.1–11.2%. However, each isolate within these pairs shared the same spA type. The same situation was observed in household 2 in which isolates that belonged to USA 300 were recovered from the person and the pet. In the remaining 3 households (1, 4 and 5) the isolates recovered from persons and their pets belonged to different clonal groups and different spA types (Table). We cannot exclude the possibility that when the isolates from the person and pet(s) were non-identical by PFGE, that the pet isolate(s) were acquired from a source other than the human participant in these households. Although employment of a household member in clinical healthcare practice was not associated with increased odds of isolating MRSA from pets in the multivariable model, at least one person in two of these four case households (each involving USA 100 strains) was employed in clinical healthcare practice. Therefore, it is conceivable that the source of MRSA to the pet could have been a healthcare worker residing in the household rather than the human patient.
Several technical and epidemiologic limitations of this study should be considered when evaluating the results. The study was likely underpowered to detect specific associations between the isolation of MR staphylococci and many of the risk factors proposed, as recruitment fell short of expectations. Although the prevalence of pet carriage was similar to that predicted, recruitment was hindered by a low rate of physician response to requests to contact patients, and a low rate of response from potential participants. Prior studies have suggested lower socioeconomic status and crowded living conditions to be risk factors for MRSA transmission between people (Gorwitz et al, 2008, Johannson et al, 2007). In the present study, demographic data was not collected on patients that either could not be contacted or declined to participate. We therefore cannot comment on any enrollment biases that may have existed within the source population.
In many cases, the lag between isolation of MRSA from a human SSTI and sampling of the pet was greater than 2 weeks. For each day of delay in sampling the pet after the person’s MRSA diagnosis, the odds of isolating MRSA from a pet in the household decreased by 13.9%, although this result should be interpreted with caution since not all human-animal paired isolates were concordant strains. Additionally, the methodology utilized does not allow deduction of the original source of MRSA within the household, as cross-sectional surveys cannot comment on cause-effect relationships.
Regardless, it is apparent that pets can harbor pandemic strains of MRSA that cause SSTI in humans, and it is conceivable that they could then be a source for cross-transmission back to human family members. As the factors that promote person-pet cross-transmission are not yet clearly elucidated, pet owners should be educated regarding common sense practices to help mitigate risk. Such recommendations may include both “social distancing” and hygienic practices. Examples could include: preventing pets from licking the infected person or sleeping on the person’s bed, covering wounds and abrasions, avoidance of contact with exudates or excretions, daily washing of the pet’s food and water dishes, frequent laundering of pet bedding, and proper disposal of pet waste. For the future, longitudinal, population-based cohort studies are recommended to more completely characterize the frequency and directionality of transmission, and the risk associated with contact between MRSA patients and their personal pets in the home.
Bullet points.
-
-
Domestic pets may harbor strains of methicillin-resistant Staphylococcus aureus similar to those that infect persons within their homes.
-
-
An obvious source for transmission cannot be identified for all MRSA-positive pets.
-
-
Pet Carriage of MRSA appears to be fleeting, although longitudinal studies are required to confirm the observations of this cross-sectional study.
Acknowledgments
We thank Dana Durso and Michelle Traverse for project management, Kathleen O’Shea for molecular testing of specimens, Dr. Karin McGowan for microbiology laboratory services, and Mary Wheeler and Pam Tolomeo for medical chart review.
This research was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, grant R21-AI-073328
References
- Abraham JL, Morris DO, Griffeth GC, Shofer FS, Rankin SC. Surveillance of healthy cats and cats with inflammatory skin disease for colonization of the skin by methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi ssp. schleiferi. Vet. Dermatol. 2007;18:252–259. doi: 10.1111/j.1365-3164.2007.00604.x. [DOI] [PubMed] [Google Scholar]
- Calfee DP, Durbin LJ, Germanson TP, Toney DM, Smith EB, Farr BM. Spread of methicillin-resistant Staphylococcus aureus (MRSA) amongst household contacts of individuals with nosocomially acquired MRSA. Infect. Control. Hosp. Epidemiol. 2003;24:422–426. doi: 10.1086/502225. [DOI] [PubMed] [Google Scholar]
- CLSI. Document M31-A3. No. 8. Vol. 28. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: informational supplement. [Google Scholar]
- CLSI. Document X07-R. No. 5. Vol. 30. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. Surveillance for methicillin-resistant Staphylococcus aureus: Principles, practices, and challenges; a report. [Google Scholar]
- Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control. Hosp. Epidemiol. 2005;26:166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- Diederen BMW, Kluytmans JAJAW. The emergence of infections with community-associated methicillin resistant Staphylococcus aureus. J. Infect. 2006;52:157–158. doi: 10.1016/j.jinf.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Faria NA, Carrico JA, Oliveira DC, Ramirez M, Lencastre H. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 2008;46:136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States 2001–2004. J. Infect. Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- Graham PL, Lin SX, Larson EL. A U.S. population-based survey of Staphylococcus aureus colonization. Ann. Intern. Med. 2006;144:318–325. doi: 10.7326/0003-4819-144-5-200603070-00006. [DOI] [PubMed] [Google Scholar]
- Griffeth GC, Morris DO, Abraham JL, Shofer FS, Rankin SC. Screening for skin carriage of methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi in dogs with healthy and inflamed skin. Vet. Dermatol. 2008;19:142–149. doi: 10.1111/j.1365-3164.2008.00663.x. [DOI] [PubMed] [Google Scholar]
- Hanselman BA, Kruth SA, Rousseau J, Low DE, Willey BM, McGeer A, Weese JS. Methicillin-resistant Staphylococcus aureus colonization in veterinary personnel. Emerg. Infect. Dis. 2006;12:1933–1938. doi: 10.3201/eid1212.060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijsdens XW, van Santen-Verheuvel MG, Spalburg MG, Heck E, Pluister MEOC, Eijkelkamp GN, de Neeling AJ, Wannet WJB. Multiple cases of familial transmission of community-acquired methicillin-resistant Staphylcoccus aureus. J. Clin. Microbiol. 2006;44:2994–2996. doi: 10.1128/JCM.00846-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson PJH, Gustafsson EB, Ringberg H. High prevalence of MRSA in household contacts. Scand. J. Infect. Dis. 2007;39:764–768. doi: 10.1080/00365540701302501. [DOI] [PubMed] [Google Scholar]
- King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. Emergence of community acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 2006;144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- Lee JH, Jeong JM, Park YH, Choi SS, Kim YH, Chae JS, Moon JS, Park H, Kim S, Eo SK. Evaluation of the methicillin-resistant Staphylococcus aureus (MRSA)-Screen latex agglutination test for detection of MRSA of animal origin. J. Clin. Microbiol. 2004;42:2780–2782. doi: 10.1128/JCM.42.6.2780-2782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler A, Boag AK, Sung J, Lindsay JA, Guardabassi L, Dalsgaard A, Smith H, Stevens KB, Lloyd DH. Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. J. Antimicrob. Chemother. 2005;56:692–697. doi: 10.1093/jac/dki312. [DOI] [PubMed] [Google Scholar]
- Loeffler A, Pfeiffer DU, Lloyd DH, Smith H, Soares-Magalhaes R, Lindsay JA. Meticillin-resistant Staphylcococus aureus carriage in UK veterinary staff and owners of infected pets: new risk groups. J. Hosp. Infect. 2010;74:282–288. doi: 10.1016/j.jhin.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Marschall J, Muhlemann K. Duration of methicillin-resistant Staphylococcus aureus carriage according to risk factors for acquisition. Infect. Control. Hosp. Epidemiol. 2006;27:15206–15212. doi: 10.1086/507917. [DOI] [PubMed] [Google Scholar]
- McDougal LA, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DO, Boston RC, O’Shea K, Rankin SC. Prevalence of methicillin-resistant staphylococcal carriage by veterinary dermatology practice staff and their respective pets. Vet. Dermatol. 2010;21:400–407. doi: 10.1111/j.1365-3164.2010.00866.x. [DOI] [PubMed] [Google Scholar]
- Mulvey MR, MacDougall L, Cholin B, Horsman G, Fidyk M, Woods S. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg. Infect. Dis. 2005;11:844–850. doi: 10.3201/eid1106.041146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Richman M, Naidich S, Kreiswirth BN. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson KB, Searle K, Stoddard GJ, Samore MH. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci in rural communities, western United States. Emerg. Infect. Dis. 2005;11:895–903. doi: 10.3201/eid1106.050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbett RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio A, Kardén-Lilja M, Ibrahem S, Kerttula AM, Salmenlinna S, Virolainen A, Vuopio-Varkila J. Clonality of epidemic methicillin-resistant Staphylococcus aureus strains in Finland as defined by several molecular methods. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:545–555. doi: 10.1007/s10096-008-0470-1. [DOI] [PubMed] [Google Scholar]
- Vriens MR, Blok HE, Gigengack-Baars AC, Mascini EM, van der Werken C, Verhoef J, Troelstra A. Methicillin-resistant Staphylococcus aureus carriage among patients after hospital discharge. Infect. Control. Hosp. Epidemiol. 2005;26:629–633. doi: 10.1086/502592. [DOI] [PubMed] [Google Scholar]
- Weese JS, Rousseau J, Traub-Dargatz JL, Willey BM, McGeer AJ, Low DE. Community-associated methicillin-resistant Staphylococcus aureus in horses and humans who work with horses. J. Am. Vet. Med. Assoc. 2005;226:580–583. doi: 10.2460/javma.2005.226.580. [DOI] [PubMed] [Google Scholar]
- Weese JS, Dick H, Willey BM, McGeer A, Kreiswirth BN, Innis B, Low DE. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet. Microbiol. 2006;115:148–155. doi: 10.1016/j.vetmic.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Werckenthin C, Cardoso M, Martel JL, Schwarz S. Antimicrobial resistance in staphylococci from animals with particular reference to bovine S. aureus, porcine S. hyicus, and canine S. intermedius. Vet. Res. 2001;32:341–362. doi: 10.1051/vetres:2001129. [DOI] [PubMed] [Google Scholar]


