Abstract
Recent studies suggest CD133, a surface protein widely used for isolation of colon cancer stem cells, to be associated with tumor angiogenesis and recurrence. We hypothesized that gene expression levels and germline variations in CD133 will predict clinical outcome in patients with mCRC, treated in first-line setting with 5-FU, oxaliplatin and bevacizumab and we investigated whether there is a correlation with gene expression levels of CD133, vascular endothelial growth factor (VEGF) and its receptors.
We evaluated intra-tumoral gene expression levels by quantitative RT-PCR from 54 patients and 3 germline variants of the CD133 gene by PCR-RFLP from 91 patients with genomic DNA. High gene expression levels of CD133 (>7.76) conferred a significantly greater tumor response (RR=86%) than patients with low expression levels (≤7.76, RR=38%, adjusted p=0.003), independent of VEGF or its receptor gene expression levels. Gene expression levels of CD133 were significantly associated with VEGF and its receptors mRNA levels (VEGFR-1 (p<.01), -2, and -3, p<0.05). Combined analyses of two polymorphisms showed a significant association with PFS (18.5 months vs 9.8 months, p=0.004), in multivariate analysis as an independent prognostic factor for PFS (adjusted p=0.002). These results suggest CD133 is a predictive marker for standard first-line bevacizumab-based treatment in mCRC.
Keywords: CD133, Prominin-1, bevacizumab, predictive markers, molecular markers, colorectal cancer
Introduction
The cancer stem cell hypothesis proposes that only a subpopulation of cancer cells is responsible for tumor initiation, disease progression and chemoresistance. Cancer stem cells (CSC) have many shared characteristics with normal stem cells including the ability to self-renew, the capacity for homeostatic control thus sustaining tumor growth and the potential to produce more differentiated progenitors, albeit aberrant differentiation in the case of CSCs [1, 2]. Prospective identification of cancer stem cells was first reported in leukemia. They have subsequently been identified in a wide range of tumors including glioblastoma, liver-, breast, prostate- and colon cancer[3–8]. Several cell surface markers have been proposed for the identification of these CSC. Among these, CD133, a pentaspan transmembrane protein, has been used by several groups to isolate CSC in colorectal cancer [6, 7, 9, 10].
CD133 has been associated both in vitro and in vivo with tumor growth and progression [6, 7, 9, 10]. In vitro studies demonstrated the ability of human CD133+ colon cancer cells to initiate and maintain tumorspheres, whereas CD133− cells did not grow in cell culture. In xenograft-models, human CD133+ colon cancer cells have demonstrated a higher clonogenic capacity and engraftment rate for tumor-growth in immunocompromised mice than CD133− cells [6, 7]. Recent work by Zhu et al. highlighted the importance of CD133 in colorectal cancer. They demonstrated that the murine analogue of CD133, Prominin-1, marks intestinal adult stem cells, which are susceptible to malignant transformation [11]. However, there has been significant controversy concerning CD133 expression as a marker of CSC. Shmelkov et al proposed, based upon the use of a knock-in LacZ transgenic mouse model that in fact CD133 is mainly a marker of mature colonic epithelial cells. In human colons, they observed that expression of CD133 message overlapped with the expression of an epithelial cell adhesion molecule [12]. Although this seems at first contradictory, in fact the issue is not only whether CD133 is transcriptionally active in colon cancer ‘stem cells’, but also which alternatively spliced and glycosylated epitopes of the protein can be recognized by specific monoclonal antibodies, in this case Prom1/CD133 antibodies. Additionally, CD133 mRNA levels may not originate from the colon cancer stem cells or more differentiated colon cancer epithelium but from endothelial cells.
However, studies investigating characteristics of CD133+-cells suggest a subpopulation among these cells demonstrating attributes of endothelial progenitors, leading towards maturation into endothelial cells and thereby proving angiogenic potential [13, 14]. Bevacizumab, a monoclonal human antibody targeting Vascular Endothelial Growth Factor (VEGF), has been proven its efficacy in colorectal cancer and is given as first line chemotherapy in patients with mCRC.
The rationale for this study was to investigate if gene expression levels and potentially functional germline variations (rs3130, rs2286455 and rs2240688) in CD133 could predict clinical outcome in patients with mCRC, treated in first-line setting with 5-FU, Oxaliplatin and the anti-angiogenic agent bevacizumab. For further confirmation of our thesis, we investigated whether there is a correlation between gene expression levels of CD133, VEGF and its receptors (VEGFR-1, -2 and -3).
Material and methods
Patients
Ninety-one patients with primary colorectal adenocarcinoma, either metastatic or recurrent, were eligible for this study (registered at www.clinicaltrials.gov, NCT00070122). The patients received first-line treatment with FOLFOX or XELOX and bevacizumab (BV) between April 2004 and January 2009 at the University of Southern California/Norris Comprehensive Cancer Center (USC/NCCC) or the Los Angeles County/University of Southern California Medical Center (LAC/USCMC). Primary tumor samples were available from 54 patients; whereas whole blood samples for genotyping were available from all participating 91 patients. This study was conducted at the USC/NCCC and approved by the Institutional review board of the University of Southern California for Medical Sciences. All patients signed an informed consent; follow-up information and clinical data were collected through a prospectively started database and in a retrospective attempt through chart review.
Tumor response evaluation
Baseline evaluations were conducted within 1 week prior to administration of study drug. Scans and x-rays were conducted ≤4 weeks prior to start of therapy. The Response Evaluation Criteria In Solid Tumors (RECIST) was used. Tumor response was evaluated every six weeks. Response was defined as follows: Complete Response (CR), disappearance of all target lesions; Partial Response (PR), at least a 30% decrease in the sum of LD (longest diameter) of target lesions taking as reference the baseline sum LD; Stable Disease (SD), neither sufficient shrinkage to qualify for PR nor sufficient increase to qualify for PD taking as references the smallest sum LD since the treatment started; Progression of Disease (PD), at least a 20% increase in the sum of LD of target lesions taking as references the smallest sum LD recorded since the treatment started or the appearance of one or more new lesions. Disease progression was also recorded by clinicians even without radiologic assessment when patient symptoms deteriorated.
Micro-dissection
For the assessment of gene expression levels, formalin-fixed paraffin-embedded tissue samples (FFPE) from the primary tumor from 54 patients were available. After a representative review of hematoxylin- and eosin- stained slides by a pathologist, 10-μm-thick sections were obtained for laser-captured microdissection (P.A.L.M. Microlaser Technologies AG, Munich, Germany) from the regions with the highest amount of tumor cells according to a standard procedure.
Isolation of RNA and cDNA synthesis
The sections were then transferred to a reaction tube containing 400 μl of RNA lysis buffer; RNA-isolation from FFPE-samples was performed according to a patented procedure of Response Genetics Inc. (Los Angeles, CA, U.S. Patent No 6,248,535). After RNA-isolation, cDNA synthesis was performed as previously described [15].
Real-Time Polymerase Chain Reaction Quantification of mRNA expression
The quantification of gene expression levels of CD133, VEGF and VEGFR1, -2 and -3 was performed using β-actin as an internal housekeeping gene and the gene-set for above named genes in a fluorescence-based real-time detection method (ABI prism 7900 Sequence Detection System, [TaqMan] Perkin-Elmer Applied Biosystem, Foster City, CA). RT-PCR was executed as previously described by Lord et al [15], primers and probes are listed in table 1. Quantification of gene expression levels is validated with the assessment of Cycle threshold (Ct) values. These Ct values are inversely correlated with the amount of cDNA in each sample and imply number of PCR cycles, until the fluorescent signal exceeds the threshold and is therefore detected. The relative messenger RNA levels (gene expression levels) are expressed as the quotient between the gene of interest and the internal housekeeping gene, which is utilized as a normalization factor for the amount of RNA isolated from the specimen. For quality assurance purposes, samples were run in triplicates.
Table 1.
Primers and Probes for gene expression in CD133 and VEGF, VEGFR-1, -2,- 3 and genotyping in CD133
| Gene | Gen Bank accession | Forward (5′-3′) | Reverse (5′-3′) | Taqman probe (5′-3′) |
|---|---|---|---|---|
| β-actin | NM_001101 | GAGCGCGGCTACAGTT | TCCTTAATGTCACGCACGATTT | ACCACCACGGCCGAGCGG |
| CD133 | NM_006017 | CAAGGACAAGGCGTTCACAG | GTTGGGTCTCAGTCGGTCAA | TTCCGCCTCCTAGCACTGAATTGA |
| VEGF | NM_003376 | AGTGGTCCCAGGCTGCAC | TCCATGAACTTCACCACTTCGT | ATGGCAGAAGGAGGAGGGCAGAATCA |
| VEGFR1 | NM_002019 | CGCATATGGTATCCCTCAACCT | AGTCACACCTTGCTTCGGAATG | TGGTTCTGGCACCCCTGTAACCATAA |
| VEGFR2 | NM 002253 | CCTGTGGCTCTGCGTGGA | CTGAGCCTGGGCAGATCAAG | CACTAGGCAAACCCACAGAGGCGGC |
| VEGFR3 | NM_182925 | GGACACCCTGCAAGATGTTTG | TCACGGCACTGTGGCATGA | CGCCGCCGGAGACTACGCTGG |
| Polymorphism RS number | Annealing temperature | Forward (5′-3′) | Reverse (5′-3′) | Digesting enzyme |
| rs3130 | 60° | AGAACTGCAATCTGCACATGA | TGATCAGCAATGAAGAACTGG | EcoR1 |
| rs2286455 | 60° | ACGCCTCTTTGGTCTCCTTG | TCCATCCCAAGTCCCTTTAG | Mbo1 |
| rs2240688 | 60° | TCAAGATCTCTCTCTCTCTTTTGA | GTGGAACATGGCCAATCTTT | Cvikl-1 |
Candidate polymorphisms
Candidate polymorphisms were picked with the assistance of the Ensembl homepage. Two main criteria were chosen for selecting the germline variations:
That the polymorphism has some degree of likelihood to alter the function of the gene in a biological relevant manner. All three polymorphisms are located in the 3′UTR region of the CD133 gene, where they might alter gene expression levels of CD133 i.e. by influencing microRNA-binding sites.
That the frequency of the polymorphism is sufficient enough that its impact in clinical outcome would be meaningful on a population level [16].
Isolation of genomic DNA and genotyping
Peripheral blood was available from 91 patients. Genomic DNA was extracted from white blood cells using the QiaAmp kit (Qiagen, Valencia, CA, USA). Forward and reverse primers were used for PCR amplification. Samples were analyzed by PCR-RFLP assays. Forward/reverse primers, digesting enzymes and annealing temperatures are listed in table 1.
Statistical analysis
Primary endpoints of this study were progression -free survival (PFS) and response rate (RR). The PFS was calculated from the date of the first treatment with FOLFOX/BV or XELOX/BV at USC medical facilities until the first observation of disease progression or death from any cause. If no disease progression occurred and the patient was still alive at the time of the last follow-up, PFS was censored at the date of the last follow-up.
The association between CD133 gene expression value and tumor response was assessed using maximal χ2 method [17, 18]. The optimal cut-off value of CD133 was used to separate patients into 2 groups in terms of likelihood of responding to the therapy. The p-value for the association was adjusted with 2000 bootstrap like simulations that estimated the distribution of the maximal χ2 statistics. The maximal χ2 method had been used in our previous studies to examine the associations between the gene expression and clinical outcome [19-21]. The differences in CD133 gene expression value by CD133 polymorphisms were tested using Wilcoxon two-sample test.
Allelic distribution of CD133 polymorphisms by each race was tested for deviation from Hardy-Weinberg equilibrium (HWE) using χ2 test. The associations of individual CD133 polymorphisms with PFS were analyzed using Kaplan-Meier curves and log-rank test assuming codominant, dominant, or recessive genetic model. The associations between genomic polymorphisms and clinicodemographic parameters and tumor response were assessed using contingency tables and the Fisher’s exact test. The estimate of the hazard ratio (HR) with 95% CIs was based on the log-rank test for the univariate analysis [22]. The cumulative effect of CD133 polymorphisms on clinical outcome was examined by combining 2 or more CD133 alleles.
The COX proportional hazards regression model was used to assess the association between CD133 polymorphisms and PFS when adjusting for gender, the number of metastatic sites, and race. The Spearman correlation coefficient method was used to investigate the correlations between gene expression levels of CD133, VEGF and VEGF- receptor genes.
Finally, we performed leave-one-out cross-validation, in which one patient was removed from the analysis and the association between CD133 and PFS adjusted for covariates was assessed using the Cox proportional hazards conditional survival function in the remaining patients. The process was repeated with each patient left out at a time. This method provides an approximation of the unbiased estimate of the concordance probability, an index for discrimination and the predictive accuracy of models [21, 23]. Ninety five percent bootstrap confidence intervals of the concordance probability were calculated using the bias correct method with 1999 bootstrap samples [24]. The concordance probability ranges from 0.5 (no discrimination) to 1.0 (perfect discrimination).
The level of significance was set to p < 0.05, and all tests were 2-sided. Analyses were performed using the SAS statistical package version 9.2 (SAS Institute Inc. Cary, North Carolina, USA) and SAS %MACRO (%cpe developed by Gönen&Heller).
Results
A total of 91 patients (54 men, 37 women) with a median age of 56 (range 28-81) participated in this study. The racial/ethnic distributions of study participants were as follows: 37 whites, (41%), 21 Asians (23%), 28 Hispanics (31%), and 5 African American (5%). At a median follow-up of 28.7 months (range 3.3-53.8 months), the median PFS was 12.4 months (95% CI: 8.3-15.2) in these patients. One metastatic site was observed in 51 patients, whereas 40 patients had 2 or more metastatic sites. Tumors of 67 patients showed moderate differentiation, 21 patients had poor differentiation and 3 patients had missing data on differentiation. Of the 91 patients, 31 patients have died; whereas the median overall survival has not been reached. Patient characteristics are described in table 2.
Table 2.
Baseline characteristics among patients whose specimens were available for genotyping
| Patients with CD133 gene expression data (n=54) | All patients (n=91) | |||
|---|---|---|---|---|
| Frequency | % | Frequency | % | |
| Median age, yr (range) | 57 (30-81) | 56 (28-81) | ||
| Sex | ||||
| Female | 26 | 48 | 37 | 41 |
| Male | 28 | 52 | 54 | 59 |
| Race | ||||
| Asian | 12 | 22 | 21 | 23 |
| Black | 1 | 2 | 5 | 5 |
| Caucasian | 25 | 46 | 37 | 41 |
| Hispanic | 16 | 30 | 28 | 31 |
| Primary site | ||||
| Colon | 39 | 75 | 62 | 70 |
| Rectal | 12 | 23 | 24 | 27 |
| Recto-sigmoid | 1 | 2 | 3 | 3 |
| Differentiation | ||||
| Moderate | 42 | 82 | 66 | 76 |
| Poor | 9 | 18 | 21 | 24 |
| No. of metastatic sites | ||||
| 1 | 32 | 59 | 51 | 56 |
| ≥2 | 22 | 41 | 40 | 44 |
Gene expression levels of CD133 and tumor response and PFS
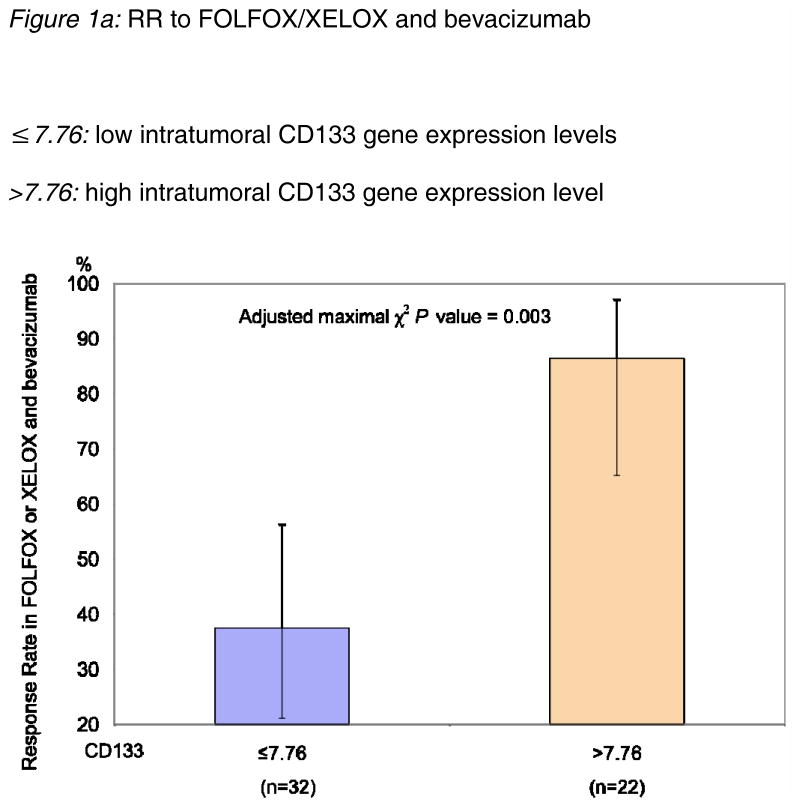
Gene expression levels of CD133 were quantifiable in 54 patients with a median mRNA level for CD133 of 6.31 (table 3a). The gene expression levels of CD133 were significantly associated with tumor response (adjusted p=0.003, maximal χ2 method). A cut-off value for CD133, 7.76, was determined as the optimum value to divide patients into poor- and good- prognosis subgroups in terms of response to treatment. Patients with high gene expression levels of CD133 (>7.76, n=22) showed a significantly better tumor response (86%) than patients with low expression levels (≤7.76, n=32, RR=38%, figure 1a). However, there was no significant difference in PFS between high and low CD133 expression levels (data not shown).
Table 3a.
Median mRNA levels of tested genes
| Variable | N | Median | (range) |
|---|---|---|---|
| CD133 | 54 | 6.31 | (0.03-35.09) |
| VEGF | 53 | 9.70 | (3.63-28.70) |
| VEGFR1 | 52 | 7.27 | (1.32-52.68) |
| VEGFR2d | 53 | 1.86 | (0.45-14.25) |
| VEGFR3 | 52 | 0.32 | (0.001-1.56) |
Figure 1.
Figure 1a: RR to FOLFOX/XELOX and bevacizumab
≤7.76: low intratumoral CD133 gene expression levels
>7.76: high intratumoral CD133 gene expression level
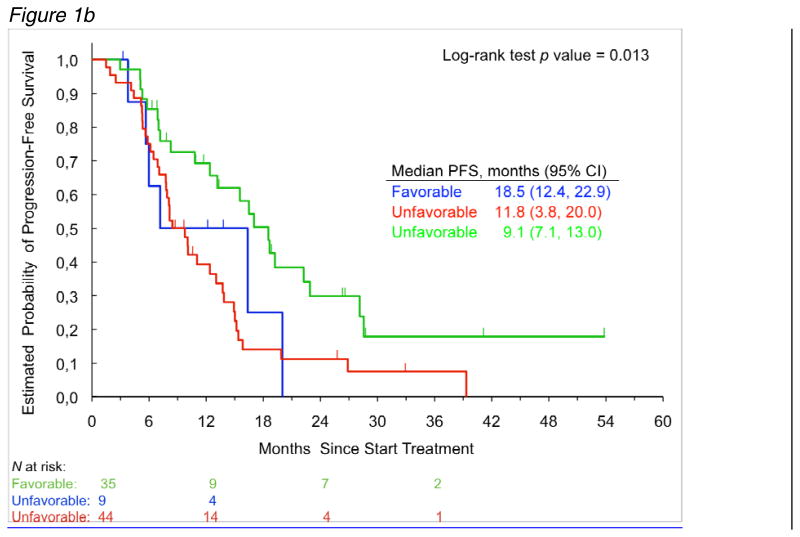
Figure 1b: PFS for the combination of single nucleotide polymorphisms rs3130 and rs2086455. Patients with the favorable combination of alleles (either homozygous for C/C in both polymorphisms or the combination of C/T in rs2086455 with either C/T or T/T in rs3130) showed a significantly increased PFS, compared to a decreased PFS for patients with C/C in one polymorphism and C/T or T/T in the other polymorphism (unfavorable alleles).
Gene expression levels of CD133 and VEGF-pathway genes
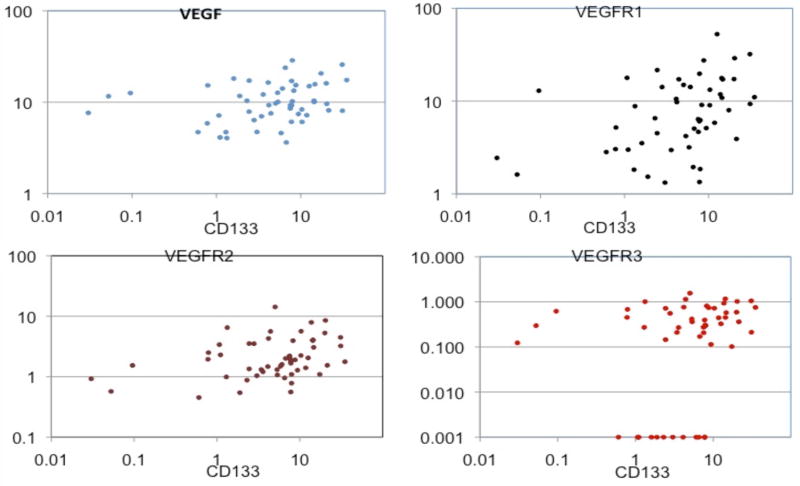
Gene expression levels for VEGF and VEGFR1 were quantifiable in 53 respectively 52 patients (table 3a). VEGF and VEGFR gene expression levels were not significantly associated with PFS or RR (data not shown). Gene expression levels of CD133 were significantly associated with VEGF, VEGFR 1, -2 and -3 (Figure 2a,-b,-c,-d). Patients with high intratumoral CD133 gene expression levels also showed high VEGF or VEGFR-receptor gene expression levels (VEGF, VEGFR-2 and -3 = p<0.05, VEGFR1= p<0.01, table 3b).
Figure 2.
a,b,c,d: Scatterplots demonstrating the relationship between CD133 and VEGF (a), VEGFR1 (b), VEGFR2 (c) and VEGFR3 (d)
Table 3b.
Relationship (Spearman Correlation Coefficients) between Gene expression levels of CD133 and genes in VEGF pathway
P <.01;
P<.05
Genetic variants in CD133 and PFS
A total of 91 patients were successfully genotyped for rs3130, rs2240688 or rs2286455. The allelic frequencies observed for rs2240688 and rs2286455 variants were within the probability limits of Hardy-Weinberg equilibrium (P > 0.05, χ2 test for HWE) in each race group. Rs3130 allelic distribution in white patients significantly departed from HWE (p=0.035 χ2 test). There were no significant differences in the distribution of 3 CD133 genetic variants by race (table 4). None of the 3 CD133 polymorphisms were significantly associated with tumor response or PFS. In a combination analysis we found rs3130 and rs2286455 significant for PFS. Patients who carried C/C in rs2286455 and rs3130 or the combination of C/T with either C/T or T/T showed a significantly increased PFS of 18.5 months, compared to 9.8 months PFS for patients with CC in one polymorphism and C/T or T/T in the other polymorphism (p=0.004, log-rank test, figure 1b). Allele frequencies were 15.3% for rs2286455 and 54.1% for rs3130 for each variant allele in the white study population. After adjustment for sex and number of metastatic sites, multivariate analysis showed the combination analysis of rs2286455 and rs3130 to be an independent prognostic factor for PFS (adjusted p=0.002). The concordance probability (0.675, 95% bootstrap CI 0.653-0.726) estimated using the leave-one-out cross validation indicated good discrimination and predictive accuracy of the multivariate model including CD133 and covariates (gender and number of metastatic sites). The association between favorable CD133 genetic variants and tumor response was not significant (data not shown).
Table 4.
Allelic distribution of CD133 polymorphisms by race
| Polymorphisms | Race | |||||
|---|---|---|---|---|---|---|
| rs2286455 | Total N | African American | Asian | Hispanic | White | P value* |
| C/C | 60 | 4 | 14 | 15 | 27 | |
| % | 80.00 | 66.67 | 53.57 | 75.00 | ||
| C/T | 27 | 1 | 6 | 13 | 7 | 0.26 |
| % | 20.00 | 28.57 | 46.43 | 19.44 | ||
| T/T | 3 | 0 | 1 | 0 | 2 | |
| % | 0.00 | 4.76 | 0.00 | 5.56 | ||
| T allele frequency | 0.100 | 0.190 | 0.232 | 0.153 | ||
| rs3130 | ||||||
| C/C | 24 | 2 | 3 | 8 | 11 | |
| % | 50.00 | 14.29 | 29.63 | 29.73 | ||
| C/T | 36 | 1 | 13 | 10 | 12 | 0.38 |
| % | 25.00 | 61.90 | 37.04 | 32.43 | ||
| T/T | 29 | 1 | 5 | 9 | 14 | |
| % | 25.00 | 23.81 | 33.33 | 37.84 | ||
| T allele frequency | 0.375 | 0.548 | 0.519 | 0.541 | ||
| rs2240688 | ||||||
| T/T | 51 | 4 | 13 | 16 | 18 | |
| % | 80.00 | 65.00 | 61.54 | 60.00 | ||
| T/G | 25 | 0 | 6 | 7 | 12 | 0.22 |
| % | 0.00 | 30.00 | 26.92 | 40.00 | ||
| G/G | 5 | 1 | 1 | 3 | 0 | |
| % | 20.00 | 5.00 | 11.54 | 0.00 | ||
| G allele frequency | 0.200 | 0.200 | 0.250 | 0.200 |
Based on Fisher’s exact test.
Combination of gene expression and genetic variants
Gene expression values of CD133 and genotypes in rs2286455 were significantly associated with each other (p=0.041, Wilcoxon two-sample test). Patients carrying C/C had lower CD133 gene expression levels (median=4.43, range 0.03-31.17) compared to patients carrying C/T (median=9.07, range: 0.05-35.09). There were no patients homozygous for the T-allele in the patients with gene expression samples. In addition, high intra-tumoral CD133 gene expression levels were also significantly associated with the favorable alleles by the combination of 2 CD133 polymorphisms (p=0.044), implying that patients who respond to 5-FU/BV show an increased PFS.
Discussion
Despite an understanding of its molecular function, the cell surface epitope defined by Prominin1/CD133 has garnered significant notoriety as a marker that is associated with stem cells, cancer stem cells and symmetric cell division. However, the expression of the CD133 gene is clearly not restricted to a stem/progenitor population and CD133 gene expression is also associated with more differentiated epithelium. Our data demonstrate for the first time, to the best of our knowledge, that patients with mCRC and high intra-tumoral CD133 gene expression levels benefit from treatment with the angiogenesis-inhibitor bevacizumab. Furthermore, we show that patients with high CD133 gene expression also express high levels of VEGF and its receptors. Additionally, the combination analysis of two polymorphisms in the CD133 gene (rs2286455 and rs3130) was associated with favorable benefit in PFS.
CD133 has been shown to play a role in postnatal physiologic and pathologic angiogenesis [25-27]. Several studies have demonstrated that tumor growth and -aggressiveness is directly correlated with the degree of neovascularization [28]. The angiogenic process is controlled by the release of pro-angiogenic factors such as VEGF and interleukin 8 (IL-8).
In a mouse model of diabetic ischemic ulcers, Barcelos et al. showed that CD133 positivity was associated with expedited wound healing. The authors associated this effect with stimulation of endothelial cell proliferation and migration. Further analysis revealed high gene expression levels of angiogenic factors such as VEGF-A and IL-8. Not surprisingly, the pro-angiogenic and pro-migratory effects of CD133+ cells or the conditioned medium could be weakened by anti-angiogenic antibodies [25]. Consistent with these findings, we report a correlation of high CD133 gene expression levels with high gene expression levels of VEGF and VEGFR 1, -2 and -3, associated with a role in endothelial cell proliferation and migration.
Willett and colleagues demonstrated that the effect of a single infusion with bevacizumab decreases tumor perfusion, vascular volume, microvascular density, interstitial fluid pressure and circulating endothelial and progenitor cells. Patients with locally advanced rectal cancer received neoadjuvant treatment with bevacizumab 5 mg/kg alone, followed two weeks later by concurrent administration of bevacizumab, 5-FU and external beam radiation to the pelvis before surgery. Three days after the initial bevacizumab treatment, the amount of viable CD133+ cells in peripheral blood was significantly decreased (p<0.05, Wilcoxon-signed ranked test) [29]. In our study, patients with high intra-tumoral CD133 gene expression levels showed an increased response rate to treatment with bevacizumab, indicating a predictive role for CD133 to anti-VEGF treatment.
To our knowledge, there are no studies that describe a relationship with germline variations in CD133 in relation to clinical outcome. The fact that the polymorphisms which are significantly associated with a prolonged PFS are also linked with gene expression levels of CD133 might mirror the effect of an increased response rate to bevacizumab therapy and further supports a functional significance for these polymorphisms. Germline variations may impact the expression not only in the tumor but also in the tumor enviroment and normal tissue such as endothelial cells.
Nonetheless, this study has several limitations. Patients were all treated with bevacizumab and FOLFOX/XELOX without a control arm receiving only FOLFOX/XELOX. Second, based on the fact that gene expression levels were significantly associated with polymorphisms in CD133, we assumed a functional significance for these polymorphisms. However, a function for the CD133 gene product, no less the significance of these polymorphisms, is presently unknown and the preliminary nature of this pilot study has to be considered. Thirdly, in an attempt to reduce the likelihood of overanalyzing this dataset, we conducted an internal validation analysis. Forthly, several studies suggested a difference in CD133-expression in primary tumor- and metastasis- tissue samples. Our gene-expression data was generated only from primary tumor samples and may thereby lack heterogeneity. Nonetheless, we are aware of the fact that our findings are preliminary and require confirmation in an independent, larger dataset.
Importantly however, these are the first data to show that patients with high intra-tumoral CD133 gene expression levels benefit from treatment with the angiogenesis-inhibitor bevacizumab; indicating on a preliminary level a predictive role for CD133 in anti-VEGF treatment, data that warrant validation in large prospective studies.
Acknowledgments
This work was funded by the NIH grant 5 P30CA14089-27I and in honor of Jerome Comet Klein MD by the Irving & Estelle Levy Foundation and performed in the Sharon A. Carpenter Laboratory at USC/Norris CCC
References
- 1.Ferrandina G, et al. Targeting CD133 antigen in cancer. Expert Opin Ther Targets. 2009;13:823–37. doi: 10.1517/14728220903005616. [DOI] [PubMed] [Google Scholar]
- 2.Pohl A, et al. Stem cells in colon cancer. Clin Colorectal Cancer. 2008;7:92–8. doi: 10.3816/CCC.2008.n.012. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins AT, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 5.Ma S, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132(7):2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien CA, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Yu SC, et al. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265(1):124–34. doi: 10.1016/j.canlet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Haraguchi N, et al. CD133+CD44+ population efficiently enriches colon cancer initiating cells. Ann Surg Oncol. 2008;15(10):2927–33. doi: 10.1245/s10434-008-0074-0. [DOI] [PubMed] [Google Scholar]
- 10.Puglisi MA, et al. Isolation and characterization of CD133+ cell population within human primary and metastatic colon cancer. Eur Rev Med Pharmacol Sci. 2009;13(Suppl 1):55–62. [PubMed] [Google Scholar]
- 11.Zhu L, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457(7229):603–7. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmelkov SV, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118(6):2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 14.Elkhafif N, et al. CD133(+) human umbilical cord blood stem cells enhance angiogenesis in experimental chronic hepatic fibrosis. APMIS. 119(1):66–75. doi: 10.1111/j.1600-0463.2010.02693.x. [DOI] [PubMed] [Google Scholar]
- 15.Lord RV, et al. Telomerase reverse transcriptase expression is increased early in the Barrett’s metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000;4(2):135–42. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- 16.Lurje G, et al. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res. 2008;14(23):7884–95. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
- 17.Halpern J. Maximally selected X2 statistics for small samples. Biometrics. 1982;38:1017–23. [Google Scholar]
- 18.Siegmund R. Maximally selected X2 statistics. Biometrics. 1982;38:1011–6. [Google Scholar]
- 19.Leichman CG, et al. Quantitation of intratumoral thymidylate synthase expression predicts for disseminated colorectal cancer response and resistance to protracted-infusion fluorouracil and weekly leucovorin. J Clin Oncol. 1997;15(10):3223–9. doi: 10.1200/JCO.1997.15.10.3223. [DOI] [PubMed] [Google Scholar]
- 20.Schneider S, et al. Gene expression in tumor-adjacent normal tissue is associated with recurrence in patients with rectal cancer treated with adjuvant chemoradiation. Pharmacogenet Genomics. 2006;16(8):555–63. doi: 10.1097/01.fpc.0000220563.44724.6d. [DOI] [PubMed] [Google Scholar]
- 21.Gonen M. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965–70. [Google Scholar]
- 22.Berry G, Kitchin RM, Mock PA. A comparison of two simple hazard ratio estimators based on the logrank test. Stat Med. 1991;10(5):749–55. doi: 10.1002/sim.4780100510. [DOI] [PubMed] [Google Scholar]
- 23.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21(15):3301–7. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 24.Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19(9):1141–64. doi: 10.1002/(sici)1097-0258(20000515)19:9<1141::aid-sim479>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 25.Barcelos LS, et al. Human CD133+ progenitor cells promote the healing of diabetic ischemic ulcers by paracrine stimulation of angiogenesis and activation of Wnt signaling. Circ Res. 2009;104(9):1095–102. doi: 10.1161/CIRCRESAHA.108.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehling UM, et al. In vitro differentiation of endothelial cells from AC133-positive progenitor cells. Blood. 2000;95(10):3106–12. [PubMed] [Google Scholar]
- 27.Senegaglia AC, et al. In vitro formation of capillary tubules from human umbilical cord blood cells with perspectives for therapeutic application. Rev Bras Cir Cardiovasc. 2008;23(4):467–73. doi: 10.1590/s0102-76382008000400003. [DOI] [PubMed] [Google Scholar]
- 28.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1(1):27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 29.Willett CG, et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat Med. 2004;10(2):145–7. doi: 10.1038/nm988. [DOI] [PMC free article] [PubMed] [Google Scholar]