Abstract
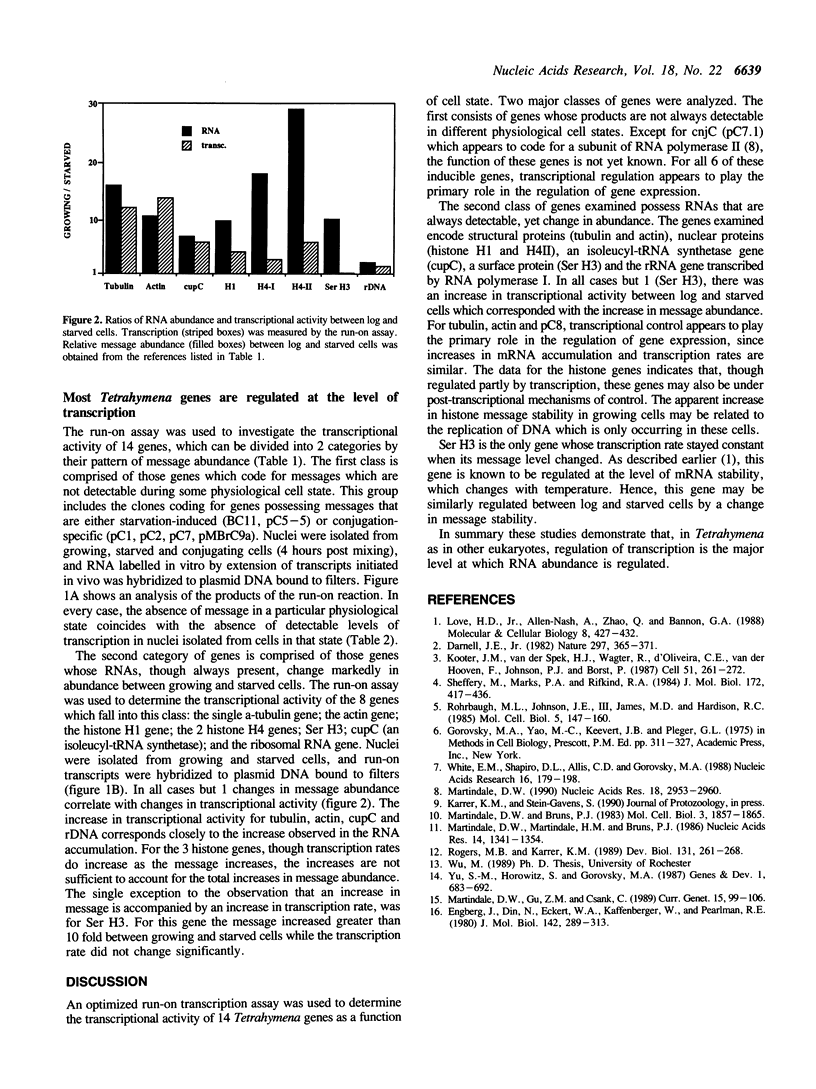
The only well-characterized study of gene expression in Tetrahymena thermophila (1) demonstrates that the temperature dependent expression of the Ser H3 gene is regulated at the level of mRNA stability. A run-on transcription assay was developed to determine if regulation of RNA stability was a major mechanism regulating gene expression in Tetrahymena or if transcriptional regulation dominates. The relative transcriptional activities of 14 Tetrahymena genes were determined in different physiological/developmental states (growing, starved and conjugating) in which many of the genes showed striking differences in RNA abundance. In every case except Ser H3, changes in transcription accompanied changes in RNA abundance. Thus differential transcription, not differential RNA degradation, is the major mechanism regulating RNA abundance in Tetrahymena.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nasir-ud-Din, Eckert W. A., Kaffenberger W., Pearlman R. E. Detailed transcription map of the extrachromosomal ribosomal RNA genes in Tetrahymena thermophila. J Mol Biol. 1980 Sep 25;142(3):289–313. doi: 10.1016/0022-2836(80)90274-0. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Kooter J. M., van der Spek H. J., Wagter R., d'Oliveira C. E., van der Hoeven F., Johnson P. J., Borst P. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell. 1987 Oct 23;51(2):261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- Love H. D., Jr, Allen-Nash A., Zhao Q. A., Bannon G. A. mRNA stability plays a major role in regulating the temperature-specific expression of a Tetrahymena thermophila surface protein. Mol Cell Biol. 1988 Jan;8(1):427–432. doi: 10.1128/mcb.8.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Bruns P. J. Cloning of abundant mRNA species present during conjugation of Tetrahymena thermophila: identification of mRNA species present exclusively during meiosis. Mol Cell Biol. 1983 Oct;3(10):1857–1865. doi: 10.1128/mcb.3.10.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Gu Z. M., Csank C. Isolation and complete sequence of the yeast isoleucyl-tRNA synthetase gene (ILS1). Curr Genet. 1989 Feb;15(2):99–106. doi: 10.1007/BF00435455. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Martindale H. M., Bruns P. J. Tetrahymena conjugation-induced genes: structure and organization in macro- and micronuclei. Nucleic Acids Res. 1986 Feb 11;14(3):1341–1354. doi: 10.1093/nar/14.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. B., Karrer K. M. Cloning of Tetrahymena genomic sequences whose message abundance is increased during conjugation. Dev Biol. 1989 Jan;131(1):261–268. doi: 10.1016/s0012-1606(89)80057-0. [DOI] [PubMed] [Google Scholar]
- Rohrbaugh M. L., Johnson J. E., 3rd, James M. D., Hardison R. C. Transcription unit of the rabbit beta 1 globin gene. Mol Cell Biol. 1985 Jan;5(1):147–160. doi: 10.1128/mcb.5.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Marks P. A., Rifkind R. A. Gene expression in murine erythroleukemia cells. Transcriptional control and chromatin structure of the alpha 1-globin gene. J Mol Biol. 1984 Feb 5;172(4):417–436. doi: 10.1016/s0022-2836(84)80015-7. [DOI] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]