Abstract
Genetic factors clearly contribute to exceptional longevity and healthy aging in humans, yet the identification of the underlying genes remains a challenge. Longevity is a complex phenotype with modest heritability. Age-related phenotypes with higher heritability may have greater success in gene discovery. Candidate gene and genome-wide association studies (GWAS) for longevity have had only limited success to date. The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium conducted a meta-analysis of GWAS data for longevity, defined as survival to age 90 years or older, that identified several interesting associations but none achieved genome-wide significance. A recent GWAS of longevity conducted in the Leiden Longevity Study identified the ApoE E4 isoform as deleterious to longevity that was confirmed in an independent GWAS of long-lived individuals of German descent. Notably, no other genetic loci for longevity have been identified in these GWAS. To examine the conserved genetic mechanisms between the mouse and humans for life span, we mapped the top Cohorts for Heart and Aging Research in Genomic Epidemiology GWAS associations for longevity to the mouse chromosomal map and noted that eight of the ten top human associations were located within a previously reported mouse life-span quantitative trait loci. This work suggests that the mouse and human may share mechanisms leading to aging and that the mouse model may help speed the understanding of how genes identified in humans affect the biology of aging. We expect these ongoing collaborations and the translational work with basic scientists to accelerate the identification of genes that delay aging and promote a healthy life span.
Keywords: Longevity, Genetics, Epidemiological studies
IT is well established that both genetic factors and health-related behaviors influence survival to old age and survival to old age in good health. Longitudinal cohort studies demonstrate that lower levels of cardiovascular risk factors measured in midlife or early older years predict survival and healthy survival to 85 years of age (1,2) and beyond (3,4). Longevity has been observed to cluster within families so that parents and siblings of centenarians have a greater likelihood of attaining advanced age (5–7), and offspring of centenarians appear to have a delay in age-related disease (8,9). Studies of families clustered for longevity both in the United States and Europe (Long Life Family Study and Leiden Longevity Study) have demonstrated that offspring of long-lived participants has more favorable midlife risk factor profiles and less age-related disease (10,11). Similarly in the community-based Framingham Heart Study, adults with at least one parent surviving to old age have lower risk factor levels compared with individuals whose parents died younger and the risk factor advantage persists over time (12). The genetic contribution to longevity and human aging is likely to result from many genes each with modest effects. Some genes will likely affect longevity by increasing susceptibility to age-related disease and early death, whereas other genes are likely to slow the aging process itself leading to a long life. How genetic factors and their interaction with modifiable behavioral and environmental factors contribute to longevity remains unknown.
LONGEVITY AND HEALTHY AGING PHENOTYPES: DEFINITIONS AND HERITABILITY
Longevity is often defined as age at death or survival to an exceptional age such as 90 years or older or 100 years or older. Because life expectancy has improved dramatically across birth cohorts since 1900, care must be taken when study designs compare long-lived with younger cohorts. Women live longer than men and make up a larger proportion of the older population especially at exceptional old ages. For example, among the original 5,209 Framingham Heart Study participants with follow-up through 2011, there are 43 centenarian women and only six centenarian men, whereas at study entry (1948–1953), 55% of participants were women. Men are more likely to attain extreme old age escaping common age-related disease, whereas women are more likely to attain 100 after surviving common morbidities (13). While these observations raise the hypothesis that genetic and environmental factors influence the path to longevity differently in men and women, whether genetic factors play a greater or lesser role in men than in women is an area of debate (3,14). In a study of centenarians (100–104 years), semisupercentenarians (105–109 years), and supercentenaians (110–119 years), there was a progressive delay in the onset of age-related disease and onset of physical and cognitive function impairment with increasing age (15). Whether genes that influence survival to these extreme ages also play a role in survival to age older than 90 years is unknown.
The genetic contribution to longevity (age at death) has been estimated using both large twin registries and population-based samples (Table 1). Most heritability estimates from twin registries range between 20% and 30% (16,17), whereas estimates from population-based samples are slightly lower, ranging from 15% to 25% (18,19), suggesting a significant but modest genetic contribution to the human life span. One study conducted among an ethnically diverse group suggests that genetic influences on life span may vary by ethnicity with heritabilities ranging from a low of 4% for African Americans to 29% and 26% for Caribbean Hispanics and Caucasians, respectively (21). Using data from the GenomeEUtwin project that included more than 20,000 Nordic twins, Hjelmborg and coworkers (42) noted that genetic effects on life span were minimal prior to 60 years of age, but genetic effects on life spans greater than 60 years of age were significant and constant to increasing with advancing age. Starting at about age 60 years, the relative recurrence risk of an individual living past a specified age given that his/her cotwin also lived past that age increased with increasing age cut point in both men and women so that at age 92, the recurrence risk was 4.8 and 1.8 in monozygotic and dizygotic male twins and 2.5 and 1.6 in monozygotic and dizygotic female twins. Notably, recurrence risks similar to men occurred in women at a 5- to 10-year older age perhaps reflecting the longer average longevity in women. Using the Framingham Heart Study cohorts, we explored whether genetic influences on life span increase with achievement of older ages by examining age at death as a dichotomous trait using a liability threshold model adjusting for sex and birth year (20). In contrast to the modest heritability estimate for continuous age at death (16%), heritability of surviving past 65 years and surviving past 85 years was substantial at 36% (p = 4.2 × 10−10) and 40% (p = 9.0 × 10−10), respectively. Thus, genetic effects appear to be greater for survival to more advanced ages. In the Framingham Heart Study cohorts, heritability appears to increase with each 10-year increment in survived age (65 years, 75 years, and 85 years) for men, but not women, again suggesting that genetic effects on aging may be more substantial for men than women (20).
Table 1.
Familiality of Aging Phenotypes
| Exceptional survival: centenarians | Sibling survival probability |
| New England Centenarian Study, likelihood of achieving age 100 (6) | Women eightfold; men 17-fold |
| Okinawa Centenarian Study (7), likelihood of achieving age 90 | Women 2.6-fold; men 5.4-fold |
| Age at death | Heritability |
| Twin registries (16,17) | 20%–30% |
| Old Order Amish (18) | 25% |
| Utah Population Database (19) | 15% |
| Framingham Heart Study (20) | 16% |
| Medicare recipients, New York City (21) | |
| European ancestry | 26% |
| African American | 4% |
| Caribbean Hispanic | 29% |
| Age at death ≥65 y | |
| Framingham Heart Study ≥65, ≥75, and ≥85 (20) | 36%–40% |
| Healthy aging and morbidity-free survival | |
| Male twins at age 70 y (22) | 50% |
| Framingham Heart Study ≥65, ≥75, and ≥85 (20) | 20%–25% |
| Physical function, disability, and self-report | |
| Danish twins, aged 80 and older, women (23) | 34%–47% |
| Danish twins, aged 75–79, women (23) | 15%–34% |
| Rate of change in functional ability* (24) | ns |
| Framingham Heart Study, disability free, aged 75‡ | 44% |
| Frailty | |
| Framingham Heart Study, frailty/prefrailty vs no frailty, aged >60 y‡ | 19% |
| Danish twins, cluster analysis approach† (25) | 43% |
| Handgrip | |
| Twin studies (26–28) | 40%–65% |
| Long Life Family Study (29) | ∼40% |
| Framingham Heart Study‡ | 38% |
| Walking speed | |
| Female twins (30) | 16% |
| Male twins (31) | 42% |
| Framingham Heart Study, usual pace walk‡ | 38% |
| Framingham Heart Study, quick walk‡ | 36% |
| Long Life Family Study (29) | 10% |
| Bone mineral density (32,33) | 50%–70% |
| Alzheimer’s disease (34,35) | 58%–79% |
| Reproductive aging | |
| Age at menarche (36,37) | 50%–80% |
| Age at natural menopause (38–41) | 44%–87% |
Note: ns = nonsignificant.
60% of sample did not participate in 4 year follow-up, half lost to mortality.
Cluster analysis approach based on mini-mental state examination, activities of daily living, self-reported health status, and handgrip strength.
Not previously published.
The longevity phenotype measures overall life span without consideration of health and physical or cognitive function and hence is a very heterogeneous phenotype that may be affected by many environmental and other nongenetic factors. The relative contribution of additive genetic effects may be greater for more homogeneous phenotypes that describe specific aspects of aging and in turn may result in greater success in gene discovery. The heritability of reproductive aging phenotypes is at least 50%, and heritability is even higher for age-related diseases such as osteoporosis (low bone mineral density) and Alzheimer’s disease (Table 1). Genetic association studies have been successful in identifying genetic variants for these aging phenotypes and have the potential to uncover new biologic insights into the associated underlying aging processes (43–49).
Epidemiological studies that have followed participants over adulthood and collected a wealth of information in a standardized fashion have been important sources for development of aging-related phenotypes. Alternative aging phenotypes include disease-free survival, preservation of high levels of function including maintenance of cognitive function (50) and avoidance of bone loss (51), and successful aging (reaching advanced age with intact cognitive ability, physical function, and social engagement) (52). An index of physiologic age developed in the Cardiovascular Health Study by combining data across multiple systems was found to be a better predictor of death and disability than age itself (53). A frailty phenotype defined by five criteria, including unintentional weight loss, exhaustion, weakness, low physical activity, and slow walking speed (54), is distinct from physical disability and itself is predictive of mortality and other adverse outcomes. Although the frailty phenotype was developed in the Cardiovascular Health Study sample, it was found to be applicable across diverse studies (55). Many components of the multidimensional aging phenotypes developed in longitudinal studies are heritable, such as weakness (defined using handgrip strength) and lower extremity function, suggesting the potential for a genetic contribution to the overall phenotype (26,31).
In the family-based Framingham Heart Study, we estimated the heritability of several of the age-related phenotypes including longevity, morbidity-free survival, physical function, and frailty as well as walking speed and handgrip strength (Table 1). For quantitative traits, we used the variance components model, and for dichotomous traits, we used the liability model implemented in the software Sequential Oligogenic Linkage Analysis Routines (56). For both, we defined heritability as the proportion of phenotypic variance due to additive genetic effects only. The heritability of the physical function and frailty phenotypes in the Framingham sample has not previously been reported. For physical disability, three items from the Rosow–Breslau Functional Health Scale (are you able to walk a half mile without help? are you able to walk up and down one flight of stairs without help? are you able to do heavy work around the house without help? [57]) and five items from the Katz Activities of Daily Living Scale (can you do the following five activities independently: dressing, bathing, eating, toileting, and transferring) (58) were used. Physical disability was defined as present if the participant was unable to do any of the items. We examined the presence of physical disability at age 75 years using the exam at which the participant was closest to and within 5 years from age 75 using both the original cohort and offspring samples. Among the 2,614 individuals included in the analysis, 42% reported physical disability at age 75, and the heritability was 44% (p = .0002). We estimated heritability of frailty and two of its components handgrip strength and walking speed in the Framingham Offspring cohort participants who attended the last completed research examination (2005–2008) during which the short physical performance battery was administered including a timed 4-m usual paced and quick walk (59). Frailty was defined if three of the five criteria proposed by the Cardiovascular Health Study investigators were present and prefrailty if one to two criteria were present (54). The analysis was adjusted for age and sex and included only participants aged 60 and older. The prevalence of frailty and prefrailty among the 2,207 individuals in this sample was 5% and 41%, respectively, and the combined trait of prefrailty and frailty was modestly heritable (h2 = 19%, p = .05). The usual and fast paced walking times in participants aged 65 years and older were rank normalized to reduce skewness and adjusted for age, sex, body mass index, and height. In contrast to frailty, both the usual and quick walk had a substantial genetic contribution with heritabilities of nearly 40% (usual walk: h2 = 0.38, p = .0002; quick walk: h2 = 0.36, p = .0003). Next, we estimated heritability of handgrip strength in all offspring participants (mean age 67, range 43–93 years). Handgrip strength was measured three times in each hand with a JAMAR dynamometer. The maximum of the six trials was used in the analysis. Consistent with reports from twin studies, handgrip strength adjusted for age and sex had a heritability of 38% (p = 5 × 10−15). Aging phenotypes are associated with varied heritabilities (Table 1), and thus, the genetic contribution to the phenotype may be quite modest. Populations that differ in environmental factors may produce different heritability estimates even if the genetic factors influencing the trait are the same. Hence, it is remarkable that the heritability estimates for many of the age-related phenotypes are similar. Longevity and age-related phenotypes with higher heritability are of higher priority for genetic association studies, as these phenotypes are more likely to result in multiple genetic associations.
GENETIC ASSOCIATION STUDIES
Genome-wide association studies (GWAS) test genetic variants across the entire genome for association with a phenotype and have proven highly successful for discovery of novel genes and biologic pathways involved in many common complex conditions (Table 2). However, few GWAS of longevity have been conducted to date. The Framingham Heart Study 100K project was the first investigation of the GWAS approach for longevity and aging traits (76). The project was relatively small in size including just 1,345 Framingham participants from the largest 310 families and limited in coverage of the genome as the genotyping was conducted with the 100K Affymetrix GeneChip. Modest associations between longevity (defined as age at death) and single nucleotide polymorphisms (SNPs) in or near FOXO1a, a gene important for life span in animal models, as well as other candidate genes were observed but failed to reach genome-wide statistical significance. Results of this investigation are considered hypothesis-generating and remain to be replicated. Lending some support to the Framingham 100K longevity investigation, a genome-wide linkage study looking for chromosomal regions linked to successful aging in the Amish Study identified a linkage region near one of the SNPs associated with age at death (52). In 2007, more than 9,300 Framingham Heart Study participants were genotyped with the Affymetrix 500K mapping array plus 50K gene centric supplemental array as part of the National Heart, Lung, and Blood Institute’s SNP Health Association Resource project (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000342.v2.p6, accessed March 9, 2012). At the same time, multiple large population-based longitudinal cohort studies in the United States and Europe with richly phenotyped participants planned to conduct genome-wide genotyping. Thus, in 2008, the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium was formed to facilitate GWAS meta-analyses and replication opportunities to enhance gene discovery for many phenotypes (77). The CHARGE aging and longevity working group conducted a meta-analysis of GWAS results for longevity defined as survival to age 90 and older from four cohort studies (Age, Gene/Environment, Susceptibility-Reykjavik Study, the Cardiovascular Health Study, the Framingham Heart Study, and the Rotterdam Study) (64). The CHARGE collaboration permitted the assembly of one of the largest samples of long-lived individuals with genome-wide genotyping available at that time (1,836 individuals achieved aged ≥90 years). The comparison group was drawn from the same cohorts and included only deceased participants to ensure that no individual achieved longevity. The investigation detected 273 SNP associations for longevity that achieved p < .0001, but none of the associations achieved genome-wide significance (p < 5 × 10−8). In the next stage of the discovery analysis, among the 24 strongest independent SNP associations in the CHARGE meta-analysis, 16 SNPs were successfully genotyped in the Leiden Longevity Study and the Danish 1905 cohort, and one SNP near the MINPP1 gene was associated with longevity with p = 6.8 × 10−7 in the combined stage 1 and stage 2 discovery samples. The minor (less frequent) allele was associated with a lower odds of achieving longevity (odds ratio 0.8). MINPP1 is a highly conserved gene involved in cellular proliferation. The CHARGE aging and longevity working group now includes investigators from over 15 cohort studies permitting future investigations of even larger samples of long-lived individuals with genome-wide genotyping and additional aging phenotypes that may improve our power to detect age-related genetic variation and provide support to our initial findings. We have subsequently conducted a meta-analysis of GWAS data from nine studies in more than 25,000 individuals aged 55 years and older for two age-related phenotypes, all-cause mortality and survival free of major disease and death (63). Although none of the SNP associations for either phenotype achieved genome-wide significance, 14 independent SNPs were associated with mortality, and 8 independent SNPs were associated with event-free survival. The SNPs were in or near genes highly expressed in the brain, genes involved with neural function, and genes associated with a variety of age-related diseases. Thus, our findings suggest that neural processes may be important in regulating aging.
Table 2.
Genetic Association Studies for Human Longevity
| Genome-Wide Association Studies With Discovery and Replication Samples in Humans | |||||||
| Year | Discovery Sample | Replication Sample | Region | Gene | SNP | p Value | Odds Ratio |
| 2011 (60) | 763 long-lived German individuals (mean age 99.7) | 754 long-lived German individuals (mean age 96.9) | 19q13.32 | APOC1* | rs4420638 | 1.8 × 10−10 | 0.53 |
| 1,085 young German Individuals (mean age 60.2 y) | 860 young German individuals (mean age 67.3 y) | ||||||
| 2011 (61) | Leiden Longevity Study: 403 Long-lived (mean age 94); 1,760 younger controls (mean age 58) | Rotterdam Study: 960 long-lived (mean age 94); 1,825 younger controls (mean age 62) | 19q13.32 | TOMM40† | rs2075650 | 3.4 × 10−17 | 0.71 |
| Leiden 85+ Study: 1,208 long-lived (mean age 92); 2,090 younger controls (mean age 35) | |||||||
| Danish 1905 cohort: 1,598 long-lived (mean age 93); 1,997 younger controls (mean age 57) | |||||||
| 2011 (62) | 410 long-lived individuals from Southern Italy (90–109 y); 553 younger controls (18–48 y) | 116 long-lived individuals (90–109 y); 160 younger controls (18–44 y) | 5q22.1 | CAMKIV | rs10491334 | 1.7 × 10−6 | 0.55 |
| 2011‖ (63) | CHARGE cohorts (AGES, ARIC, BLSA, CHS, FHS, HABC, InCHIANTI, RS, and SHIP), 25,007 participants age ≥55 y at baseline (55% women), European origin, 8,444 deaths (mean age 81.1); average follow-up 10.6 y | Four independent samples of European origin; N = 10,411, deaths = 1,295 | 3q26.1 | OTOL1 | rs1425609 | 1.6 × 10−6 | — |
| 2010‖ (64) | CHARGE cohorts (AGES, CHS, FHS, and RS) 1,836 individuals age >90 y; 1,955 individuals who died between ages 55–80 y | Leiden Longevity Study: 940 long-lived (mean age 94); 744 partners of offspring (mean age 60); Danish 1905 cohort: 1,644 long-lived (mean age 93); 2,007 younger Danish twins (mean age 57) | 10q23.2 | MINPPI | rs9664222 | 6.8 × 10−7 | 0.82 |
| Highly Replicated Candidate Gene Association Studies in Humans‡ | |||||||
| 2008 (65) | HHP/HAAS: 203 men of Japanese descent who survived to age 95; 402 “average-lived” men who died prior to 81 y | No replication sample FOXO3A association subsequently replicated in: German Centenarian study (66); Southern Italian Centenarian study (67); Han Chinese Study in both men and women Southern Chinese Centenarians (68); Danish 1905 cohort of long-lived individuals (69); CHS and Ashkenazi Jewish Centenarians (70) | 6q21 | FOXO3a | rs2802292 | .00009 | 2.75§ |
| 1994 (71) | 338 French Centenarians; n = 160 French adults aged 20–70 y | No replication sample ApoE E 4 association with longevity subsequently reported in: Danish Centenarians (72); Danish 1905 cohort (73,74) | 19q13.32 | Apo E | E4 allele | <.001 | 0.43 |
Notes: AGES = Age, Gene/Environment Susceptibility-ReyKjavik Study; ARIC = Atherosclerosis Risk in Communities Study; BLSA = Baltimore Longitudinal Study of Ageing; CHARGE = Cohorts for Heart and Aging Research in Genomic Epidemiology; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study; HAAS = Honolulu Asia Aging Study; HABC = Health, Aging and Body Composition Study; HHP = Honolulu Heart Program; InCHIANTI = Invescchiare nel Chianti; RS = Rotterdam Study; SHIP = Study of Health in Pomerania; SNP = single nucleotide polymorphism.
Explained by linkage equilibrium with the ApoE E4 allele (r2 = .72).
Explained by moderate linkage disequilibrium with ApoE E4 (r2 = .55, rs429358).
For additional candidate genes that may be associated with longevity, see Christensen and coworkers (73) and Barzilai and Gabriely (75).
Homozygous minor (GG) versus homozygous major (TT) alleles between cases and controls.
None of the associations achieved genome-wide significance; only the most significant association in the discovery plus replication stage is provided in the table.
A GWAS conducted in 403 nonagenarians from the Leiden Longevity Study, and 1,670 younger population controls identified 62 SNPs associated with longevity at p < 1 × 10−4 (61). Successful genotyping of 58 of these SNPs was conducted in three independent studies: the Rotterdam Study, Leiden 85-plus Study, and Danish 1905 cohort. A meta-analysis of the 58 SNPs in all four studies that included more than 4,000 nonagenarians and 7,500 younger controls identified only one genome-wide significant SNP rs2075650 in TOMM40 at chromosome 19q13.32 close to the ApoE gene. The minor allele was associated with lower odds of longevity (odds ratio 0.71, p = 3.4 × 10−17). No other SNPs were associated with longevity. SNP rs2075650 was noted to be in moderate linkage disequilibrium with rs429358, the SNP that defines the ApoE E4 isoform and in very low linkage disequilibrium with rs7412, the SNP that defines the ApoE E2 isoform. In conditional analysis, with all three SNPs in the model, rs2075650 was no longer associated with longevity, whereas the minor allele of rs429358 had a deleterious effect on longevity, and rs7412 a protective effect leading the authors to conclude that rs2075650 effect on longevity was most likely mediated through the isoforms of the ApoE gene. A case-control GWAS conducted in 763 German nonagenarians and centenarians, and 1,085 controls (mean age 60 years) identified rs4420638 near the APOC1 gene and replicated the finding in an independent sample (60). This finding was also fully explained by linkage disquilibrium with ApoE E4 isoform confirming the prior report. These results are intriguing as the ApoE gene is one of only two candidate genes with consistent evidence for association with longevity in humans (73). The ApoE E4 isoform has been linked to elevated cholesterol, cardiovascular disease, age-related cognitive decline, and dementia. The ApoE E4 isoform is more strongly associated with Alzheimer disease than longevity and other conditions. Homozygosity for the Apo E E4 allele confers up to a 15-fold risk for Alzheimer’s disease in whites and an 8-fold risk in African Americans compared with the most common ApoE genotype (E3/E3 [78]). Thus, ApoE may influence longevity through premature atherosclerosis and age-related diseases. Notably, the CHARGE study did not observe genome-wide significant associations between the ApoE gene region and longevity. However, neither of the two SNPs (rs429358 and rs7412) that define the ApoE E4 polymorphism nor any strong proxies appear on any of the chips used by the CHARGE consortium studies. In the CHARGE meta-analysis, the odds of living past age 90 years associated with the minor allele of rs2075650 was 0.85 ( p = .046); hence, the effect is consistent with the prior reports.
FOXO3a was first noted to be associated with longevity in a candidate gene study conducted in male centenarians of Japanese descent (65) and subsequently replicated in diverse samples of centenarians and long-lived individuals (66–69). Remarkably neither the Leiden or German studies nor the CHARGE GWAS identified an association between longevity and FOXO3a. Finally, a recent GWAS of 410 long-lived individuals and 553 young controls from Southern Italy identified an SNP in an intron of the CAMKIV gene among the top associations. This association was replicated in a sample of 116 long-lived and 160 young controls (p < 10−4 discovery analysis, joint replication analysis p = 1.7 × 10−6) (62). Interestingly, in vitro work suggests that this gene activates proteins in candidate genes for longevity (AKT, SIRT1, and FOXO3a). About 300 genetic variants in 30 genes in the insulin/insulin-like growth factor 1 (IGF-1) signaling pathway were genotyped in older women participating in the Study of Osteoporotic Fracture. Replication studies were conducted in the Cardiovascular Health Study and an Ashkenazi Jewish Centenarian Study (70). SNPs in two genes in this pathway (AKT1 and FOXO3a) were significantly associated with human life span. A better understanding of the biological mechanisms by which the FOXO3a and AKT1 variants influence human longevity will be important to development of interventions to delay aging (79).
Individuals with a family history of longevity have lower mortality for most age-related diseases (80). Therefore, researchers tested the hypothesis that this may be due to the absence of susceptibility alleles for common diseases in the Leiden Longevity Study and the Leiden 85 Plus Study. They examined whether the long-lived individuals had fewer copies of 30 alleles discovered through GWAS to be associated with coronary artery disease, cancer, and type 2 diabetes compared with a younger comparison group (81). Notably, no difference in the number of risk alleles was detected, suggesting that at least in these populations, survival to old age is not determined by the absence of risk alleles identified to date for age-related disease.
The effort to identify genes that affect longevity through candidate gene and GWAS studies has had only modest success to date. This is likely due to a combination of factors including the heterogeneity of the phenotype, the influence of environmental and dietary factors, which vary widely across populations and the relatively small sample sizes of published longevity GWAS. Many of the successful GWAS with many replicating genome-wide significant signals have sample sizes of more than 10,000, and some published studies of quantitative traits such as age at menarche have had sample sizes of more than 80,000 (43). It is clear that defining more homogeneous phenotypes through age-related traits such as age at menopause and bone mineral density leads to greater success in identifying aging-related genes. Another potential explanation for the lack of identification of risk variants for longevity is that the bulk of the genetic effects are due to rare variants or structural variation in the genome. The GWAS chips used to date have focused on common SNP variants, which typically do not tag rare variants well. Recent work within the CHARGE consortium studies suggests that copy number variants are associated with mortality (82). With the advent of low-cost exome and whole genome sequencing as well as higher-density SNP chips with 5 million or more variants, we will soon have the opportunity to determine whether rare or structural variants explain a substantive proportion of the heritability of longevity and other aging traits.
Translation
Comparison across genomes of different organisms may greatly facilitate the process of gene identification and testing of homologous genes in humans (83). Genome-wide mapping studies of longevity have been conducted in animal models, including Caenorhabditis elegans and Drosophila (84). Through the National Institute of Aging’s Longevity Consortium (http://longevityconsortium.org/, accessed August 15, 2011) that brings together scientists from multiple disciplines, investigators from the CHARGE consortium, and the Aging Center at the Jackson Laboratory in Maine began to examine the conserved genetic mechanisms between humans and the mouse for life span (85). The laboratory mouse is an excellent model organism for understanding mammalian physiology and genetics for several reasons, including the availability of extensive genetic resources, such as hundreds of inbred, congenic, consomic, and recombinant inbred strains, as well as the ability to add and delete genes via transgenesis and targeted mutagenesis (86,87). Sequence analyses have demonstrated that mice and humans share more than 99% of their genes and that these genes are arranged in a homologous fashion on chromosomes, a phenomenon termed synteny. The appearance of quantitative trait loci (QTL) for a given phenotype in syntenic regions of two different species is evidence that the same gene regulates the phenotype in both species (88). In the past 30 years, seven mouse life-span QTL studies have been carried out in two groups of recombinant inbred strains and 3 four-strain crosses (85).
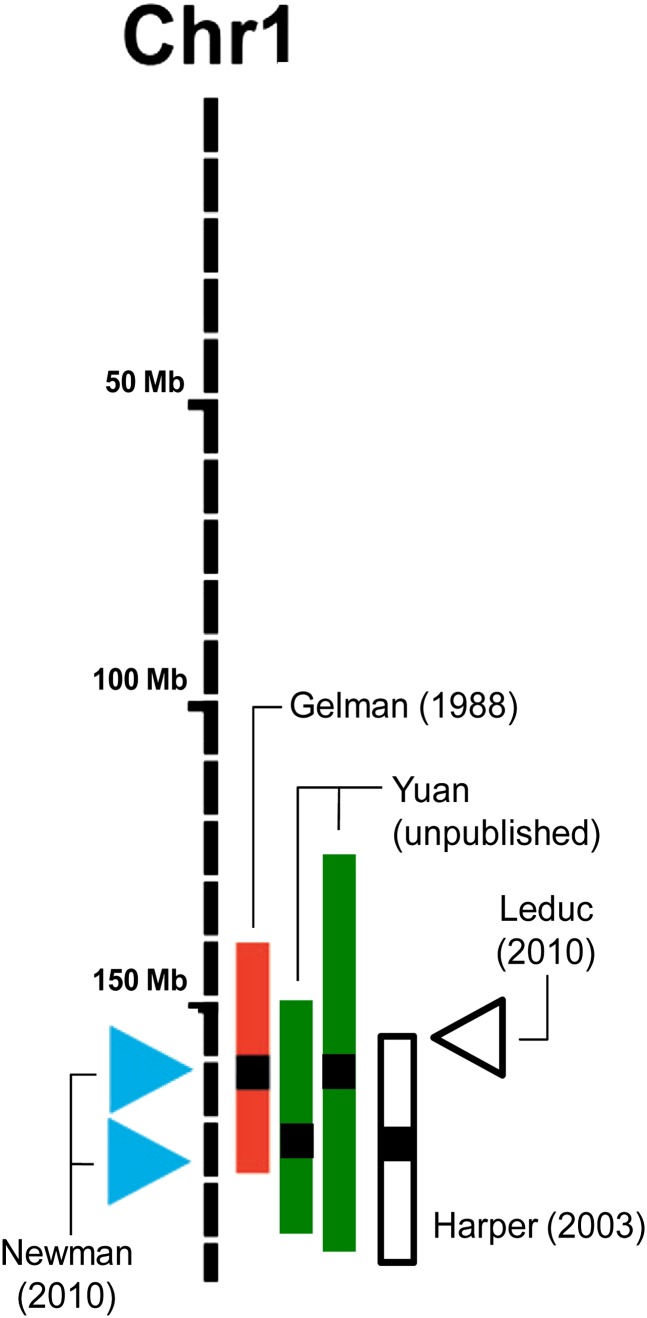
Although none of the SNP associations for longevity in the CHARGE GWAS achieved genome-wide significance (64), 8 of the top 10 hits were located within a mouse QTL, and five of the human signals were located within 10 Mb of a mouse QTL peak; the probability that this is due to chance is very low (p = .0025 using Fisher’s exact test, based on life-span QTL covering 860 Mb of the 2,700 Mb genome and each human peak being 1 Mb in size) (85). The colocalizations of the human peaks and mouse longevity QTL are especially notable in the distal region of mouse chromosome 1 (summarized in Figure 1). In this region, Gelman (90) first reported a longevity QTL that was identified in recombinant inbred lines of C57BL/6J X DBA/2. Recently, Yuan and coworkers identified two longevity QTL in this region in a backcross mouse population (POHN/DehJ × C57BL/6J) × POHN/DehJ (unpublished data, 2011). The peak of Gelman’s QTL overlaps with the peak of one of the Yuan and coworkers QTL, around 160 Mb–163 Mb. The peak of the other Yuan and coworkers QTL was 14 Mb apart, around 175 Mb. Syntenic regions of 2 of the 10 highest peaks (rs16850255 and rs4443878) identified in the CHARGE study are found around 161 Mb and 176 Mb, colocalizing with the mouse longevity QTL. This suggests that human and mouse may share some mechanisms that regulate life span.
Figure 1.
Quantitative trait loci (QTL) for mouse longevity and insulin-like growth factor 1 (IGF-1) as well as genome-wide association peaks for human longevity and mouse IGF-1, both depicted on the mouse genome, mouse chromosome 1 (mapped in Mb). Colored bars are longevity QTL; the open bar is an IGF-1 QTL. The height of the bars represents the 95% confidence interval if reported or an estimated 40 Mb if not reported; the black squares in the bars represent the QTL peaks. We determined the Mb position using a recently revised mouse map (89) and the Mouse Map Converter from the Center for Genome Dynamics (http://cgd.jax.org/mousemapconverter/). Arrows to the left of the chromosome represent human genome-wide association peaks at the homologous mouse genome locations. The arrow on the right of the chromosome is the mouse genome-wide association peak of IGF-1. (Figure is modified from Figure 1 of Yuan et al [85]). Chr = chromosome. Gelman et al (90), Harper et al (91), Leduc et al (92), Newman et al (64).
Interestingly, around 160 Mb and 175 Mb, one peak from a mouse genome–wide association study and one QTL of IGF-1 have been reported (Figure 1). Yuan and coworkers reported that across mouse inbred strains, lower IGF-1 levels are associated with longer life span. The overlap between longevity QTL and IGF-1 QTL suggests that this region may contain genes that could regulate longevity through the regulation of IGF-1 level. Although this hypothesis must be further verified, the combination of such human and mouse genetic studies establishes a foundation for a powerful translational strategy. We plan to integrate data for additional age-related traits and use bioinformatics and genetic resources in the mouse to test promising candidate genes for longevity. Although genetic association studies in humans can help identify potential genes linked to longevity, the mouse model may be very useful in uncovering the underlying biologic mechanisms that lead to aging.
Other investigators have examined human–chimpanzee orthologous gene pairs to explore evolutionary forces on genes related to aging (93). Genes that appeared to have a pattern of selection tended to have important biological functions that were conserved among mammals (93). Of interest, the study findings suggest that one gene that may have undergone rapid evolution is WRN. Defects in WRN cause Werner’s syndrome characterized by premature aging.
Ultimately, we hope that the identification of genes influencing human longevity will provide insights into the biology of aging and in turn new therapeutics to slow aging and improve health. Evidence from both human and animal studies has identified genes in the IGF1/insulin signaling pathway influencing life span. Therefore, interest in resveratrol, a compound shown to extend life span in animals and improve insulin secretion and insulin sensitivity among many other benefits, is not surprising. A recent pilot study conducted in 10 older adults suggests that resveratrol improved insulin sensitivity in individuals with impaired glucose tolerance (94). These data combined with animal studies provide support for larger studies of the benefits of resveratrol in humans (95).
FUTURE DIRECTIONS
Technological advances now permit the complete sequencing of all protein-coding portions of the genome (the “exome”), and the ability to sequence the whole genome of individuals quickly and efficiently is just beginning. Sequencing represents an opportunity to detect rare potentially functional genetic variants unlikely to be discovered with the GWAS that focus on common genetic variation (minor allele frequencies of >5%). The National Human Genome Research Institute and National Heart, Lung, and Blood Institute (NHLBI) funded the Exome project with the goal of identifying genes contributing to heart, lung, and blood disorders (http://www.nhlbi.nih.gov/resources/exome.htm, accessed August 15, 2011). This innovative technology and the analytic tools under development will be able to be extended to longevity and age-related traits. The sequencing of centenarian genomes may uncover rare genetic variants underlying human extreme longevity and provide insights into the basic biology of aging.
Changes in gene expression that occur as a result of molecular mechanisms that do not change the primary DNA sequence are referred to as epigenetics (http://www.ncbi.nlm.nih.gov/books/NBK45788/#epi_sci_bkgrd.About_Epigenetics, accessed March 9, 2012). Epigenetic mechanisms influence phenotypic expression and are affected by development, the environment, diet, drugs, and aging. One of the best studied epigenetic mechanisms called DNA methylation usually results in suppression of nearby genes. A study of global DNA methylation in an Icelandic cohort and a family-based Utah cohort demonstrated familial clustering of DNA methylation changes and changes in DNA methylation over time (96). The changes in DNA methylation that occur with aging may alter normal gene expression and in turn contribute to development of age-related disease and functional decline (97). Epigenetic changes can be identified using genome-wide analysis with microarrays (ChIP-chip) or next generation sequencing (ChIP-Seq). These new technologies may be used in longitudinal cohort studies in the future to uncover the role of epigenetics in human longevity.
CONCLUSIONS
Genetic factors undoubtedly contribute to human aging and longevity, yet candidate gene and GWAS have yielded few replicated longevity-gene associations to date with the exceptions of the ApoE and FOXO3A genes. Genome-wide genotyping of participants in longitudinal cohort studies, family-based studies, and special populations of long-lived individuals such as centenarians along with unprecedented collaboration among investigators in the United States, Europe, and worldwide provide the opportunity for the assembly of the large discovery and replication samples needed for genetic discoveries. Existing consortia that include both population-based and laboratory-based scientists may speed the translation of newly discovered genetic associations by uncovering the function of the identified genetic variants and ultimately the biologic mechanisms leading to human aging. Many epidemiological studies are poised to use new technologies to move beyond common genetic variants to explore the contribution of low frequency and rare genetic variants, structural changes, and epigenetic changes to human longevity and aging.
FUNDING
This work was funded through grants from the National Institute on Aging (R01AG29451, Drs J.M.M. and K.L.L.; P30 AG038070, Drs J.M.M. and R.Y.; AG034349, Dr R.Y.).
References
- 1.Terry DF, Pencina MJ, Vasan RS, et al. Cardiovascular risk factors predictive for survival and morbidity-free survival in the oldest-old Framingham Heart Study participants. J Am Geriatr Soc. 2005;53(11):1944–1950. doi: 10.1111/j.1532-5415.2005.00465.x. [DOI] [PubMed] [Google Scholar]
- 2.Willcox BJ, He Q, Chen R, et al. Midlife risk factors and healthy survival in men. JAMA. 2006;296(19):2343–2350. doi: 10.1001/jama.296.19.2343. [DOI] [PubMed] [Google Scholar]
- 3.Dutta A, Henley W, Lang I, et al. Predictors of extraordinary survival in the Iowa established populations for epidemiologic study of the elderly: cohort follow-up to “extinction.”. J Am Geriatr Soc. 2011;59(6):963–971. doi: 10.1111/j.1532-5415.2011.03451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yates LB, Djousse L, Kurth T, Buring JE, Gaziano JM. Exceptional longevity in men: modifiable factors associated with survival and function to age 90 years. Arch Intern Med. 2008;168(3):284–290. doi: 10.1001/archinternmed.2007.77. [DOI] [PubMed] [Google Scholar]
- 5.Perls T, Shea-Drinkwater M, Bowen-Flynn J, et al. Exceptional familial clustering for extreme longevity in humans. J Am Geriatr Soc. 2000;48(11):1483–1485. [PubMed] [Google Scholar]
- 6.Perls TT, Wilmoth J, Levenson R, et al. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci U S A. 2002;99(12):8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willcox BJ, Willcox DC, He Q, Curb JD, Suzuki M. Siblings of Okinawan centenarians share lifelong mortality advantages. J Gerontol A Biol Sci Med Sci. 2006;61(4):345–354. doi: 10.1093/gerona/61.4.345. [DOI] [PubMed] [Google Scholar]
- 8.Atzmon G, Schechter C, Greiner W, Davidson D, Rennert G, Barzilai N. Clinical phenotype of families with longevity. J Am Geriatr Soc. 2004;52(2):274–277. doi: 10.1111/j.1532-5415.2004.52068.x. [DOI] [PubMed] [Google Scholar]
- 9.Terry DF, Wilcox MA, McCormick MA, et al. Lower all-cause, cardiovascular, and cancer mortality in centenarians’ offspring. J Am Geriatr Soc. 2004;52(12):2074–2076. doi: 10.1111/j.1532-5415.2004.52561.x. [DOI] [PubMed] [Google Scholar]
- 10.Newman AB, Glynn NW, Taylor CA, et al. Health and function of participants in the Long Life Family Study: A comparison with other cohorts. Aging (Albany NY) 2011;3(1):63–76. doi: 10.18632/aging.100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozing MP, Westendorp RG, de Craen AJ, et al. Favorable glucose tolerance and lower prevalence of metabolic syndrome in offspring without diabetes mellitus of nonagenarian siblings: the Leiden longevity study. J Am Geriatr Soc. 2010;58(3):564–569. doi: 10.1111/j.1532-5415.2010.02725.x. [DOI] [PubMed] [Google Scholar]
- 12.Terry DF, Evans JC, Pencina MJ, et al. Characteristics of Framingham offspring participants with long-lived parents. Arch Intern Med. 2007;167(5):438–444. doi: 10.1001/archinte.167.5.438. [DOI] [PubMed] [Google Scholar]
- 13.Evert J, Lawler E, Bogan H, Perls T. Morbidity profiles of centenarians: survivors, delayers, and escapers. J Gerontol A Biol Sci Med Sci. 2003;58(3):232–237. doi: 10.1093/gerona/58.3.m232. [DOI] [PubMed] [Google Scholar]
- 14.Franceschi C, Motta L, Valensin S, et al. Do men and women follow different trajectories to reach extreme longevity? Italian Multicenter Study on Centenarians (IMUSCE) Aging (Milano) 2000;12(2):77–84. doi: 10.1007/BF03339894. [DOI] [PubMed] [Google Scholar]
- 15.Andersen SL, Sebastiani P, Dworkis DA, Feldman L, Perls TT. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J Gerontol A Biol Sci Med Sci. 2012 doi: 10.1093/gerona/glr223. doi:10.1093/gerona/glr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGue M, Vaupel JW, Holm N, Harvald B. Longevity is moderately heritable in a sample of Danish twins born 1870–1880. J Gerontol. 1993;48(6):B237–B244. doi: 10.1093/geronj/48.6.b237. [DOI] [PubMed] [Google Scholar]
- 17.Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97(3):319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell BD, Hsueh WC, King TM, et al. Heritability of life span in the Old Order Amish. Am J Med Genet. 2001;102(4):346–352. doi: 10.1002/ajmg.1483. [DOI] [PubMed] [Google Scholar]
- 19.Kerber RA, O’Brien E, Smith KR, Cawthon RM. Familial excess longevity in Utah genealogies. J Gerontol A Biol Sci. 2001;56(3):B130–B139. doi: 10.1093/gerona/56.3.b130. [DOI] [PubMed] [Google Scholar]
- 20.Murabito JM, Lunetta KL. Genetics of human longevity and healthy aging. In: Newman AB, Cauley JA, editors. The Epidemiology of Aging. New York, NY: Springer; 2011. In press. [Google Scholar]
- 21.Lee JH, Flaquer A, Costa R, et al. Genetic influences on life span and survival among elderly African-Americans, Caribbean Hispanics, and Caucasians. Am J Med Genet A. 2004;128(2):159–164. doi: 10.1002/ajmg.a.30062. [DOI] [PubMed] [Google Scholar]
- 22.Reed T, Dick DM. Heritability and validity of healthy physical aging (wellness) in elderly male twins. Twin Res. 2003;6(3):227–234. doi: 10.1375/136905203765693889. [DOI] [PubMed] [Google Scholar]
- 23.Christensen K, McGue M, Yashin A, Iachine I, Holm NV, Vaupel JW. Genetic and environmental influences on functional abilities in Danish twins aged 75 years and older. J Gerontol A Med Sci. 2000;55(8):M446–M452. doi: 10.1093/gerona/55.8.m446. [DOI] [PubMed] [Google Scholar]
- 24.Christensen K, Gaist D, Vaupel JW, McGue M. Genetic contribution to rate of change in functional abilities among Danish twins aged 75 years or more. Am J Epidemiol. 2002;155(2):132–139. doi: 10.1093/aje/155.2.132. [DOI] [PubMed] [Google Scholar]
- 25.Dato S, Montesanto A, Lagani V, Jeune B, Christensen K, Passarino G. Frailty phenotypes in the elderly based on cluster analysis: a longitudinal study of two Danish cohorts. Evidence for a genetic influence on frailty. Age (Dordr) 2011 doi: 10.1007/s11357-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carmelli D, Reed T. Stability and change in genetic and environmental influences on hand-grip strength in older male twins. J Appl Physiol. 2000;89(5):1879–1883. doi: 10.1152/jappl.2000.89.5.1879. [DOI] [PubMed] [Google Scholar]
- 27.Reed T, Fabsitz RR, Selby JV, Carmelli D. Genetic influences and grip strength norms in the NHLBI twin study males aged 59–69. Ann Hum Biol. 1991;18(5):425–432. doi: 10.1080/03014469100001722. [DOI] [PubMed] [Google Scholar]
- 28.Frederiksen H, Gaist D, Petersen HC, et al. Hand grip strength: a phenotype suitable for identifying genetic variants affecting mid-and late-life physical functioning. Genet Epidemiol. 2002;23(2):110–122. doi: 10.1002/gepi.1127. [DOI] [PubMed] [Google Scholar]
- 29.Matteini AM, Fallin MD, Kammerer CM, et al. Heritability estimates of endophenotypes of long and health life: the Long Life Family Study. J Gerontol A Biol Sci Med Sci. 2010;65(12):1375–1379. doi: 10.1093/gerona/glq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega-Alonso A, Pedersen NL, Kujala UM, et al. A twin study on the heritability of walking ability among older women. J Gerontol A Biol Sci Med Sci. 2006;61(10):1082–1085. doi: 10.1093/gerona/61.10.1082. [DOI] [PubMed] [Google Scholar]
- 31.Carmelli D, Kelly-Hayes M, Wolf PA, et al. The contribution of genetic influences to measures of lower-extremity function in older male twins. J Gerontol A Biol Sci. 2000;55(1):B49–B53. doi: 10.1093/gerona/55.1.b49. [DOI] [PubMed] [Google Scholar]
- 32.Karasik D, Cupples LA, Hannan MT, Kiel DP. Age, gender, and body mass effects on quantitative trait loci for bone mineral density: the Framingham Study. Bone. 2003;33(3):308–316. doi: 10.1016/s8756-3282(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 33.Karasik D, Myers RH, Cupples LA, et al. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham Study. J Bone Miner Res. 2002;17(9):1718–1727. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- 34.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 35.Ashford JW, Mortimer JA. Non-familial Alzheimer’s disease is mainly due to genetic factors. J Alzheimers Dis. 2002;4(3):169–177. doi: 10.3233/jad-2002-4307. [DOI] [PubMed] [Google Scholar]
- 36.Anderson CA, Duffy DL, Martin NG, Visscher PM. Estimation of variance components for age at menarche in twin families. Behav Genet. 2007;37(5):668–677. doi: 10.1007/s10519-007-9163-2. [DOI] [PubMed] [Google Scholar]
- 37.Towne B, Czerwinski SA, Demerath EW, Blangero J, Roche AF, Siervogel RM. Heritability of age at menarche in girls from the Fels Longitudinal Study. Am J Phys Anthropol. 2005;128(1):210–219. doi: 10.1002/ajpa.20106. [DOI] [PubMed] [Google Scholar]
- 38.Murabito JM, Yang Q, Fox C, Wilson PW, Cupples LA. Heritability of age at natural menopause in the Framingham Heart Study. J Clin Endocrinol Metab. 2005;90(6):3427–3430. doi: 10.1210/jc.2005-0181. [DOI] [PubMed] [Google Scholar]
- 39.van Asselt KM, Kok HS, Pearson PL, et al. Heritability of menopausal age in mothers and daughters. Fertil Steril. 2004;82(5):1348–1351. doi: 10.1016/j.fertnstert.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 40.de Bruin JP, Bovenhuis H, van Noord PA, et al. The role of genetic factors in age at natural menopause. Hum Reprod. 2001;16(9):2014–2018. doi: 10.1093/humrep/16.9.2014. [DOI] [PubMed] [Google Scholar]
- 41.Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman’s reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab. 1998;83(6):1875–1880. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- 42.Hjelmborg JV, Iachine I, Skytthe A, et al. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119(3):312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- 43.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42(12):1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He C, Kraft P, Chen C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41(6):724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stolk L, Zhai G, van Meurs JB, et al. Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat Genet. 2009;41(6):645–647. doi: 10.1038/ng.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coon KD, Myers AJ, Craig DW, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007;68(4):613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 48.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivadeneira F, Styrkarsdottir U, Estrada K, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaffe K, Lindquist K, Vittinghoff E, et al. The effect of maintaining cognition on risk of disability and death. J Am Geriatr Soc. 2010;58(5):889–894. doi: 10.1111/j.1532-5415.2010.02818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cauley JA, Lui LY, Barnes D, et al. Successful skeletal aging: a marker of low fracture risk and longevity. The Study of Osteoporotic Fractures (SOF) J Bone Miner Res. 2009;24(1):134–143. doi: 10.1359/JBMR.080813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edwards DR, Gilbert JR, Jiang L, et al. Successful aging shows linkage to chromosomes 6, 7, and 14 in the Amish. Ann Hum Genet. 2011;75(4):516–528. doi: 10.1111/j.1469-1809.2011.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman AB, Boudreau RM, Naydeck BL, Fried LF, Harris TB. A physiologic index of comorbidity: relationship to mortality and disability. J Gerontol A Biol Sci Med Sci. 2008;63(6):603–609. doi: 10.1093/gerona/63.6.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Med Sci. 2001;56(3):M146–M156. doi: 10.1093/gerona/56.3.m146. [DOI] [PubMed] [Google Scholar]
- 55.Bandeen-Roche K, Xue QL, Ferrucci L, et al. Phenotype of frailty: characterization in the women’s health and aging studies. J Gerontol A Biol Sci Med Sci. 2006;61(3):262–266. doi: 10.1093/gerona/61.3.262. [DOI] [PubMed] [Google Scholar]
- 56.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosow I, Breslau NA. Guttman health scale for the aged. J Gerontol. 1966;21(4):556–559. doi: 10.1093/geronj/21.4.556. [DOI] [PubMed] [Google Scholar]
- 58.Katz S. Assessing self-maintenance: activities of daily living, mobility, and instrumental activities of daily living. J Am Geriatr Soc. 1983;31(12):721–727. doi: 10.1111/j.1532-5415.1983.tb03391.x. [DOI] [PubMed] [Google Scholar]
- 59.Guralnik JM, Simonsick EM, Ferrucci L, et al. A short physical performance battery assessing lower extremity function: association with self-reported disability and prediction of mortality and nursing home admission. J Gerontol. 1994;49(2):M85–M94. doi: 10.1093/geronj/49.2.m85. [DOI] [PubMed] [Google Scholar]
- 60.Nebel A, Kleindorp R, Caliebe A, et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132(6–7):324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Deelen J, Beekman M, Uh HW, et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10(4):686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malovini A, Illario M, Iaccarino G, et al. Association study on long-living individuals from Southern Italy identifies rs10491334 in the CAMKIV gene that regulates survival proteins. Rejuvenation Res. 2011;14(3):283–291. doi: 10.1089/rej.2010.1114. [DOI] [PubMed] [Google Scholar]
- 63.Walter S, Atzmon G, Demerath EW, et al. A genome-wide association study of ageing. Neurobiol Aging. 2011;32(11):2109e15–28. doi: 10.1016/j.neurobiolaging.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newman AB, Walter S, Lunetta KL, et al. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J Gerontol A Biol Sci Med Sci. 2010;65(5):478–487. doi: 10.1093/gerona/glq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105(37):13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flachsbart F, Caliebe A, Kleindorp R, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106(8):2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anselmi CV, Malovini A, Roncarati R, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12(2):95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Wang WJ, Cao H, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18(24):4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soerensen M, Dato S, Christensen K, et al. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9(6):1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pawlikowska L, Hu D, Huntsman S, et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8(4):460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schachter F, Faure-Delanef L, Guenot F, et al. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6(1):29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 72.Gerdes LU, Jeune B, Ranberg KA, Nybo H, Vaupel JW. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: apolipoprotein E gene is a “frailty gene,” not a “longevity gene.”. Genet Epidemiol. 2000;19(3):202–210. doi: 10.1002/1098-2272(200010)19:3<202::AID-GEPI2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 73.Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7(6):436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bathum L, Christiansen L, Jeune B, Vaupel J, McGue M, Christensen K. Apolipoprotein e genotypes: relationship to cognitive functioning, cognitive decline, and survival in nonagenarians. J Am Geriatr Soc. 2006;54(4):654–658. doi: 10.1111/j.1532-5415.2005.53554.x. [DOI] [PubMed] [Google Scholar]
- 75.Barzilai N, Gabriely I. Genetic studies reveal the role of the endocrine and metabolic systems in aging. J Clin Endocrinol Metab. 2010;95(10):4493–4500. doi: 10.1210/jc.2010-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lunetta KL, D’Agostino RB, Sr, Karasik D, et al. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet. 2007;8(suppl 1):S13. doi: 10.1186/1471-2350-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Logue MW, Schu M, Vardarajan BN, et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. 2011;68(12):1569–1579. doi: 10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tazearslan C, Cho M, Suh Y. Discovery of functional gene variants associated with human longevity: opportunities and challenges. J Gerontol A Biol Sci Med Sci. 2011 doi: 10.1093/gerona/glr200. doi:10.1093/gerona/glr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Brien E, Kerber R, Smith K, Mineau G, Boucher K, Reed DL. Familial mortality in the Utah population database: characterizing a human aging phenotype. J Gerontol A Biol Sci Med Sci. 2007;62(8):803–812. doi: 10.1093/gerona/62.8.803. [DOI] [PubMed] [Google Scholar]
- 81.Beekman M, Nederstigt C, Suchiman HE, et al. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc Natl Acad Sci U S A. 2010;107(42):18046–18049. doi: 10.1073/pnas.1003540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuningas M, Estrada K, Hsu YH, et al. Large common deletions associate with mortality at old age. Hum Mol Genet. doi: 10.1093/hmg/ddr340. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 2008;180(4):2227–2235. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shmookler Reis RJ, Kang P, Ayyadevara S. Quantitative trait loci define genes and pathways underlying genetic variation in longevity. Exp Gerontol. 2006;41(10):1046–1054. doi: 10.1016/j.exger.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 85.Yuan R, Peters LL, Paigen B. Mice as a mammalian model for research on the genetics of aging. ILAR J. 2011;52(1):4–15. doi: 10.1093/ilar.52.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paigen K, Eppig JT. A mouse phenome project. Mamm Genome. 2000;11(9):715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- 87.O’Brien T, Woychik R. Our small relative. Nat Genet. 2003;1:3–4. doi: 10.1038/ng1069. [DOI] [PubMed] [Google Scholar]
- 88.Sugiyama F, Churchill GA, Higgins DC, et al. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics. 2001;1:70–77. doi: 10.1006/geno.2000.6401. [DOI] [PubMed] [Google Scholar]
- 89.Cox A, Ackert-Bicknell CL, Dumont BL, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182(4):1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gelman R, Watson A, Bronson R, Yunis E. Murine chromosomal regions correlated with longevity. Genetics. 1988;118(4):693–704. doi: 10.1093/genetics/118.4.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harper JM, Galecki AT, Burke DT, Pinkosky SL, Miller RA. Quantitative trait loci for insulin-like growth factor I, leptin, thyroxine, and corticosterone in genetically heterogeneous mice. Physiol Genomics. 2003;15(1):44–51. doi: 10.1152/physiolgenomics.00063.2003. [DOI] [PubMed] [Google Scholar]
- 92.Leduc MS, Hageman RS, Meng Q, et al. Identification of genetic determinants of IGF-1 levels and longevity among mouse inbred strains. Aging Cell. 2010;9(5):823–836. doi: 10.1111/j.1474-9726.2010.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Magalhaes JP, Church GM. Analyses of human-chimpanzee orthologous gene pairs to explore evolutionary hypotheses of aging. Mech Ageing Dev. 2007;128(5–6):355–364. doi: 10.1016/j.mad.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crandall JP, Oram V, Trandafirescu G, et al. Pilot study of resveratrol in older adults with impaired glucose tolerance. J Gerontol A Biol Sci Med Sci. 2012 doi: 10.1093/gerona/glr235. doi:10.1093/gerona/glr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smoliga JM, Vang O, Baur JA. Challenges of translating basic research into therapeutics: resveratrol as an example. J Gerontol A Biol Sci Med Sci. 2011;67(2):158–167. doi: 10.1093/gerona/glr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gravina S, Vijg J. Epigenetic factors in aging and longevity. Pflugers Arch. 2010;459(2):247–258. doi: 10.1007/s00424-009-0730-7. [DOI] [PubMed] [Google Scholar]