Abstract
Protein misfolding and aggregation are associated with many neurodegenerative diseases, including Huntington’s disease. The cellular machinery for maintaining proteostasis includes molecular chaperones that facilitate protein folding and reduce proteotoxicity. Increasing the protein folding capacity of cells through manipulation of DNAJ chaperones has been shown to suppress aggregation and ameliorate polyglutamine toxicity in cells and flies. However, to date these promising findings have not been translated to mammalian models of disease. To address this issue, we developed transgenic mice that over-express the neuronal chaperone HSJ1a (DNAJB2a) and crossed them with the R6/2 mouse model of Huntington’s disease. Over-expression of HSJ1a significantly reduced mutant huntingtin aggregation and enhanced solubility. Surprisingly, this was mediated through specific association with K63 ubiquitylated, detergent insoluble, higher order mutant huntingtin assemblies that decreased their ability to nucleate further aggregation. This was dependent on HSJ1a client binding ability, ubiquitin interaction and functional co-operation with HSP70. Importantly, these changes in mutant huntingtin solubility and aggregation led to improved neurological performance in R6/2 mice. These data reveal that prevention of further aggregation of detergent insoluble mutant huntingtin is an additional level of quality control for late stage chaperone-mediated neuroprotection. Furthermore, our findings represent an important proof of principle that DNAJ manipulation is a valid therapeutic approach for intervention in Huntington’s disease.
Keywords: chaperones, Huntington’s disease, polyglutamine, aggregation, protein folding
Introduction
The synthesis and maintenance of correctly folded proteins are essential processes within all living cells. The accumulation of misfolded or aggregated proteins can lead to cell death or dysfunction and as such, protein homeostasis (proteostasis) must be tightly regulated (Balch et al., 2008). Members of the HSPA (HSP70) and DNAJ (HSP40) chaperone families are major mediators of protein quality control (Kampinga and Craig, 2010). HSP70 recognizes misfolded proteins and promotes their refolding or degradation in an ATP-dependent manner, while HSP40 proteins enhance this process by delivering clients and stimulating HSP70 ATPase activity through their conserved J-domain (Kampinga and Craig, 2010). As such, HSP70 and HSP40 chaperones are powerful modulators of proteome integrity and cell viability.
Despite the elegant pathways in place to prevent protein misfolding and aggregation, protein folding diseases present a major health burden in an ageing population (Balch et al., 2008). Huntington’s disease is one of nine inherited polyglutamine diseases caused by protein misfolding and characterized by the formation of detergent insoluble protein aggregates that coalesce into nuclear and cytoplasmic inclusions (Williams and Paulson, 2008). At the molecular level, Huntington’s disease is caused by a CAG expansion in exon 1 of the huntingtin gene (The Huntington’s disease collaborative research group, 1993). This leads to misfolding of the encoded huntingtin protein, which results in a toxic gain-of-function (Landles and Bates, 2004). Symptoms typically manifest around mid-adulthood and include psychiatric disturbances, weight loss and a progressive decline in motor and cognitive function. Disease progression typically occurs over 20 years before death with no disease-modifying treatment available (Novak and Tabrizi, 2010).
Given that chaperones are powerful modifiers of proteostasis, upregulation of HSP70 and HSP40 chaperones to enhance the refolding capacity, or quality control, of cells is an attractive therapeutic target for Huntington’s disease and other polyglutamine diseases. Indeed, over-expression of HSP70 or HSP40 chaperones in cells and flies can potently reduce polyglutamine aggregation and toxicity (Cummings et al., 1998; Warrick et al., 1999; Chan et al., 2000). Interestingly, over-expression of HSP40 chaperones has been reported to suppress polyglutamine aggregation more potently than HSP70 chaperones, with members of a DNAJB subfamily of HSP40 molecules proving to be the most powerful modifiers of polyglutamine aggregation in cell culture (Hageman et al., 2010).
HSJ1 is a DNAJB chaperone that is predominantly expressed in neurons (Cheetham et al., 1992) and can suppress polyglutamine aggregation and toxicity (Westhoff et al., 2005; Borrell-Pages et al., 2006; Howarth et al., 2007; Hageman et al., 2010). Alternative splicing produces two isoforms, HSJ1a and HSJ1b, which contain two ubiquitin-interacting motifs (UIMs) in addition to an N-terminal J-domain (Chapple et al., 2004). Both HSJ1 isoforms promote the sorting of ubiquitylated clients to the proteasome, thereby enhancing the degradation of misfolded or damaged proteins (Westhoff et al., 2005). HSJ1a and HSJ1b differ at the C-terminus and only HSJ1a is able to modify polyglutamine aggregation in cells, predominantly due to the prenylation-mediated association of HSJ1b with the cytosolic face of the endoplasmic reticulum (Westhoff et al., 2005; Borrell-Pages et al., 2006; Hageman et al., 2010).
Disappointingly, the over-expression of chaperones has shown limited success in ameliorating polyglutamine disease in mice (Cummings et al., 2001; Hansson et al., 2003, Hay et al., 2004; Helmlinger et al., 2004). However, the ability of HSP40 chaperones to modify polyglutamine aggregation and toxicity in mouse brain is currently unknown. In this study, we assessed the effects of HSJ1a over-expression on disease progression in the R6/2 mouse model of Huntington’s disease. R6/2 mice express exon 1 of human huntingtin with ∼ 200 CAG repeats and recapitulate many molecular and phenotypic features of Huntington’s disease over an accelerated time course (Mangiarini et al., 1996; Davies et al., 1997). Importantly, we show that HSJ1a is able to act at the level of high molecular weight mutant huntingtin complexes to reduce aggregate load and improve neurological performance through a novel chaperone mechanism. This was reliant on ubiquitin chain recognition and client protein binding ability but, surprisingly, was independent of proteasomal targeting. We conclude that our findings are an encouraging proof of principle that the manipulation of DNAJ proteins can improve disease phenotype in the context of the mammalian brain. Furthermore, this suggests that altering levels of DNAJ chaperones pharmacologically could be a promising therapeutic avenue for treatment of Huntington’s disease and other polyglutamine diseases.
Materials and methods
Production of human HSJ1a transgenic mice
Human HSJ1a transgenic mice were created by pronuclear injection of B6/CBA F1 blastocysts with the human HSJ1a complementary DNA open reading frame (Chapple and Cheetham, 2003) driven by the bovine prion protein promoter (Lemaire-Vieille et al., 2000). Positive founders were identified by polymerase chain reaction and crossed to C57Bl/6 animals. The human HSJ1a line 52a was derived and human HSJ1a expression confirmed, then backcrossed onto a C57Bl/6 background for use in this study.
Mouse maintenance, breeding, genotyping and CAG repeat sizing
All mouse experiments were performed under project and personal licenses approved and issued by the Home Office. Hemizygous R6/2 mice (Mangiarini et al., 1996) were bred by backcrossing R6/2 males to (CBA x C57Bl/6) F1 females (B6CBAF1/OlaHsd, Harlan Olac). Housing conditions and environmental enrichment were as previously described (Hockly et al., 2003).
Behavioural assessment
Mouse behavioural assessment was performed as previously described (Benn et al., 2009) with the specific exceptions that RotaRod and activity assessments were conducted over 300 s and 30 min, respectively. The data were collected and analysed by repeated measures general linear model ANOVA with Greenhouse Geisser post hoc analysis as described previously (Hockly et al., 2006).
Antibodies
The primary and secondary antibodies used in this study are summarized in Supplementary Table 3. S830, HSJ1 (16321 and S653) and huntingtin antibodies were generated as described (Ko et al., 2001; Sathasivam et al., 2001; Chapple and Cheetham, 2003).
Cell culture, plasmids and transfection
Human neuroblastoma SK-N-SH cells were transfected with 1 µg/well of pCMV-Tag3a-HSJ1a (wild-type, H31Q or UIM mutants) described previously (Chapple and Cheetham, 2003; Westhoff et al., 2005) or empty vector (Stratagene) using LipofectamineTM and Plus reagents (Invitrogen) according to the manufacturer’s instructions. Cells were lysed in HEPES (for immunoprecipitation) or AcTEVTM (for in vitro aggregation) buffer 24 h post transfection, sonicated, cleared, aliquoted and frozen at −70°C until further use.
SDS-PAGE, western blotting and immunodetection
Western blotting for chaperones, HSJ1a and mutant huntingtin was performed as previously described using 20 μg of total protein in 1× Laemmli loading buffer (Woodman et al., 2007).
Densitometry
Densitometry of western blots was performed using a BioRad GS-800 densitometer. Developed films were scanned and the average pixel density for each band was measured. The average pixel density of an area devoid of bands was subtracted from the values obtained for bands of interest in order to normalize average pixel density against background. Relative expression was determined by dividing the normalized average pixel density of bands of interest by the average pixel density of α-tubulin or mouse HSJ1a as specified for each experiment.
Seprion ligand enzyme-linked immunosorbent assay
Sample preparation and Seprion ligand enzyme-linked immunosorbent assay were performed as previously described (Sathasivam et al., 2010).
Time-resolved Förster resonance energy transfer
Sample preparation and mutant huntingtin time-resolved Förster resonance energy transfer were essentially performed as previously described (Weiss et al., 2009).
Agarose gel electrophoresis for resolving aggregates
Sample preparation and agarose gel electrophoresis for resolving aggregates (AGERA) were conducted based on previously described methods (Weiss et al., 2008).
TaqMan® real-time quantitative polymerase chain reaction
RNA extraction, complementary DNA synthesis, Taqman real-time quantitative polymerase chain reaction and ΔCt analysis were performed according to previous recommendations (Benn et al., 2008).
Immunoprecipitation, co-immunoprecipitation and sequential immunoprecipitation
One hundred microlitres of protein lysate was incubated with 25 µl of protein-G Dynabeads (Invitrogen) and antibody (Supplementary Table 3) for 1 h at room temperature on a rotating wheel. Precipitated material was eluted from beads by heating for 3 min at 98°C in 30 µl of 4× sodium dodecyl sulphate sample buffer and subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis and western blotting. In the case of sequential immunoprecipitation, material isolated after HSJ1 (S653) immunoprecipitation was eluted from beads with 1% sodium dodecyl sulphate. Samples were then diluted 10-fold and used as inputs for immunoprecipitation with S830.
Exogenous immunoprecipitation and HSJ1 competition assays
HSJ1 immunoprecipitation was performed using anti-HSJ1 (S653) (Chapple and Cheetham, 2003) or anti-myc antibodies (Sigma) in R6/2 brain lysate mixed with 1–5 µg of either affinity-purified HSJ1a (wild-type or ΔUIM) or SK-N-SH cell lysate HSJ1a (myc-tagged wild-type, H31Q or ΔUIM). For competition assays, denatured ovalbumin or ubiquitin chains were preincubated for 30 min with purified or myc-tagged HSJ1a prior to the addition of R6/2 brain lysate and antibody.
Immunohistochemistry and immunofluorescence
Mouse brain sectioning, immunohistochemistry and immunofluorescence were performed as previously described (Moffitt et al., 2009; Luthi-Carter et al., 2010). Quantification of inclusion area was performed with Zeiss Axiovision software. Further details are provided in the Supplementary material.
Seprion pull down, immuno-gold labelling and transmission electron microscopy
Sample preparation, Seprion ligand pull down and immuno-gold labelling were performed according to (Sathasivam et al., 2010).
In vitro aggregation and filter trap
Reactions were essentially performed as previously described (Tam et al., 2009). Typically, 3.0 µM of freshly purified gluathione-S transferase-tagged huntingtin Q51 fragment (GST-HTT Q51) was incubated with AcTEV™ protease (Invitrogen) in the presence of an ATP regeneration system and different ratios of HSJ1a or albumin (Sigma-Aldrich). Reactions were stopped by flash-freezing in liquid nitrogen and material was then subjected to filter trap analysis using 0.22 µm cellulose acetate (Whatman). HTT Q51 was detected using S-Protein horseradish peroxidase (Novagen) and signal was developed using ECL Plus™ reagents (GE Healthcare). Results were quantified using ImageJ software.
Purification of huntingtin seeds from mouse brain and nucleation of aggregation
Isolation of detergent insoluble material was performed similar to previously described method (Nekooki-Machida et al., 2009). For the seeding experiment, 0.3 µM of GST-HTT Q51 was incubated with AcTEV™ protease (Invitrogen) in the presence of 120 ng or 160 ng of insoluble material isolated from wild-type, R6/2 or double transgenic mouse brains in the absence of the ATP regeneration system. Reactions were stopped at 0, 2 or 5 h, then used in filter trap assay and analysed as described above.
Brain-derived neurotrophic factor enzyme-linked immunosorbent assay
BDNF Emax® Immunoassay System (Promega) was used to determine total brain-derived neurotrophic factor (BDNF) protein levels in the brain tissues of 4- and 15-week-old animals according to the manufacturer’s instructions with modifications described earlier (Szapacs et al., 2004). Brain samples were region matched and run in duplicates.
Statistical analysis
P-values were calculated by Student’s t-test in Microsoft Excel. Statistical analysis of the mouse behavioural work was determined by general linear model analysis of variance in SPSS. Full and additional methodological information is available in the Supplementary material.
Results
Spatial and temporal expression of human HSJ1a in transgenic mice
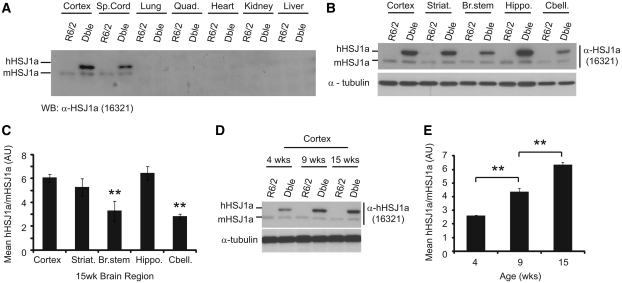
HSJ1a transgenic mice were produced that express human HSJ1a under the control of the bovine prion protein promoter. Female HSJ1a and male R6/2 mice were then crossed to generate double transgenic mice. Given that CAG repeat number is intimately coupled to aggregation and toxicity, we ensured that R6/2 and double transgenic groups were well matched for repeat size throughout (Supplementary Table 1). To determine the spatial pattern of human HSJ1a expression, protein lysates from various tissues were subjected to immunoblotting with an HSJ1a specific antibody (16321). Human HSJ1a expression was mainly in the CNS with highest levels of expression observed in brain tissue (Fig. 1A). Of the brain tissues examined, cortex, striatum and hippocampus all displayed significantly higher levels of human HSJ1a than the cerebellum or brain stem (Fig. 1B and C). Unexpectedly, human HSJ1a also exhibited differential temporal expression with levels increasing ∼3-fold between 4- and 15-weeks of age (Fig. 1D and E). Expression of human HSJ1a had no effect on levels of endogenous mouse HSJ1a (Fig. 1A, B and D).
Figure 1.
HSJ1a transgene expression is CNS specific, increases with age and is heterogeneous throughout the brain. (A and B) Representative western blots of human (hHSJ1a) and mouse (mHSJ1a) HSJ1a expression in the periphery and brain tissues of R6/2 and double transgenic (Dble) mice. (B) Cortex, striatum (Striat.), brainstem (Br Stem), hippocampus (Hippo) and cerebellum (Cbell) from 15-week-old R6/2 and double transgenic mice were western blotted for HSJ1a with antibody 16321. (C) HSJ1a levels in these brain regions were analysed by densitometry and normalized to mouse HSJ1a. Mean relative expression was plotted ±SEM for each brain region (n = 4). (D) Representative western blot of the temporal expression of human HSJ1a and mouse HSJ1a in R6/2 and double transgenic cortices. (E) Levels of human HSJ1a over-expression in 4-, 9- and 15-week double transgenic cortex were calculated relative to mouse HSJ1a by densitometry. The mean relative expression was plotted ±SEM for each age (n = 3). Statistical analysis was performed using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.005).
To ensure that mutant huntingtin exon 1 transgene messenger RNA was not altered by human HSJ1a expression, TaqMan® real-time quantitative polymerase chain reaction was performed on 15 week cortex, striatum, hippocampus and cerebellum. No significant difference in mutant huntingtin messenger RNA levels were observed between R6/2 and double transgenic mice in any of the brain regions tested (Supplementary Fig. 1A–D). In addition, no significant difference was observed in the expression of heat shock response (HSR), unfolded protein response (UPR), mitochondrial unfolded protein response or autophagy markers (Ron and Walter, 2007; Akerfelt et al., 2010; Haynes and Ron, 2010; Martinez-Vicente et al., 2010) in 15-week double transgenic mice compared with R6/2 (Supplementary Fig. 2A–C). Taken together, these data show that human HSJ1a expression in double transgenic mice is present in the CNS and increases with age without altering the expression of mutant huntingtin, mouse HSJ1a or major inducible components of the proteostasis machinery.
HSJ1a reduces aggregate load and increases levels of soluble huntingtin in a mouse model of Huntington's disease
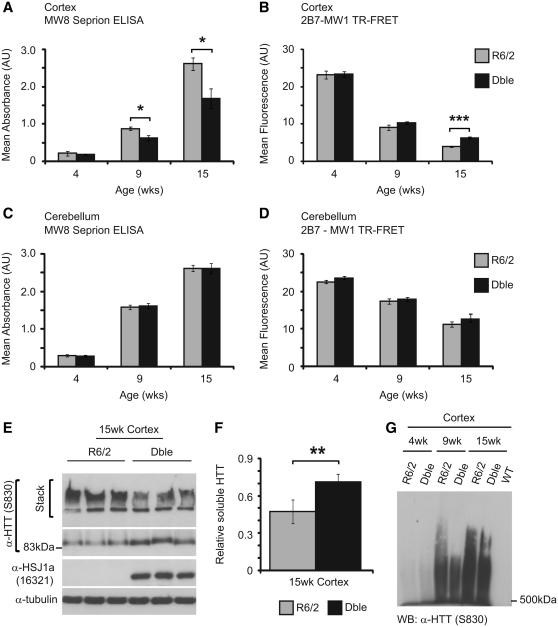
The formation of protein aggregates is a hallmark of disease progression in Huntington’s disease and other polyglutamine disorders (Williams and Paulson, 2008). To determine the molecular consequences of human HSJ1a over-expression in R6/2 brain tissue, we quantified levels of aggregated and soluble mutant huntingtin in R6/2 and double transgenic mice using Seprion enzyme-linked immunosorbent assay (Sathasivam et al., 2010) and time-resolved Förster resonance energy transfer (Weiss et al., 2009) assays, respectively. In keeping with previous studies, levels of aggregated mutant huntingtin were found to increase with age with a reciprocal reduction in levels of soluble mutant huntingtin (Weiss et al., 2009; Sathasivam et al., 2010). Seprion enzyme-linked immunosorbent assay revealed that human HSJ1a expression significantly reduced aggregate load in both the cortex (Fig. 2A) and striatum (Supplementary Fig. 3) at 15 weeks of age. Furthermore, time-resolved Förster resonance energy transfer demonstrated that human HSJ1a expression also led to increased levels of soluble mutant huntingtin in the cortices of double transgenic mice (Fig. 2B). These findings were confirmed by immunoblotting and densitometry (Fig. 2E and F). In contrast, no significant difference was observed in levels of aggregated or soluble mutant huntingtin in cerebellum at any age tested (Fig. 2C and D). This is most likely due to the much lower levels of human HSJ1a in this brain region. Taken together, these data show that human HSJ1a expression reduces aggregate load and increases levels of soluble mutant huntingtin in R6/2 brain tissue.
Figure 2.
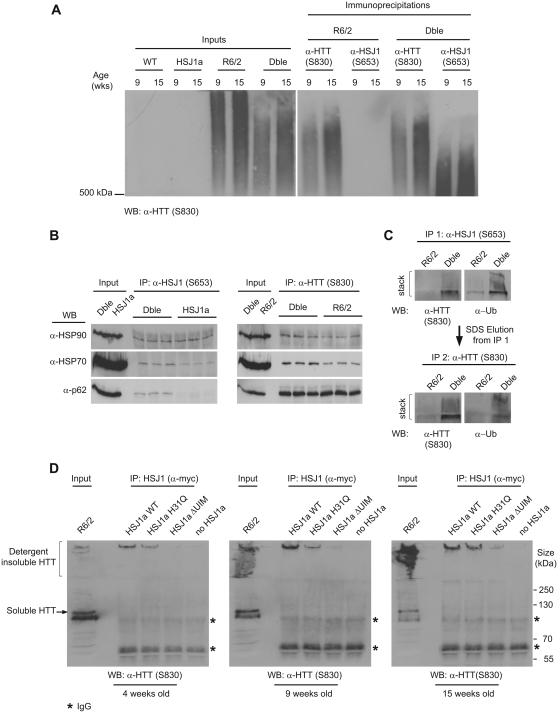
HSJ1a reduces levels of high order detergent insoluble mutant huntingtin in R6/2 mice. Levels of aggregated (A and C) and soluble mutant huntingtin (B and D) were determined in R6/2 (light grey bars) and double transgenic (black bars) cortices (A and B) and cerebellum (C and D) by MW8 seprion enzyme-linked immunosorbent assay and 2B7-MW1 time-resolved Förster resonance energy transfer (TR-FRET), respectively. Wild-type (WT) and HSJ1a signals were considered to be background and were subtracted from R6/2 and double transgenic readings, respectively. Mean values ± SEM were plotted for each group (n ≥ 6). (E) Western blotting with S830 confirmed that the double transgenic mice had reduced detergent insoluble mutant huntingtin trapped in the stacking gel and increased detergent soluble mutant huntingtin in the resolving gel. (F) Levels of detergent soluble mutant huntingtin were measured by densitometry and calculated relative to α-tubulin. Mean values were plotted ± SEM for R6/2 and double transgenic mice (n = 6). Statistical analysis was performed by Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.005). (G) Western blot of detergent insoluble high molecular weight species isolated from R6/2 and double transgenic mouse cortices at 4-, 9- and 15 weeks of age, resolved by agarose gel electrophoresis resolution of aggregates and detected using the anti-huntingtin (α-HTT) antibody S830 (representative of three experiments).
HSJ1a reduces nuclear inclusion area and aggregate size distribution in R6/2 mice
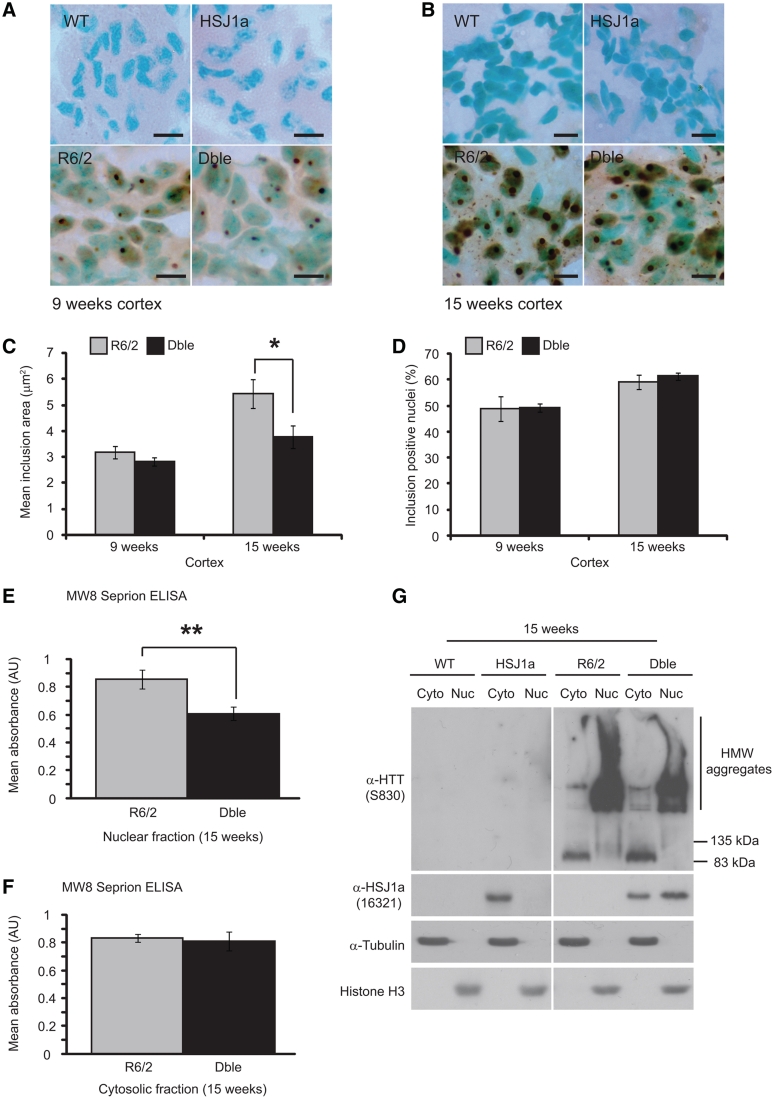
A spectrum of mutant huntingtin oligomers are formed with disease progression in Huntington’s disease (Sathasivam et al., 2010). In order to determine whether human HSJ1a reduced general aggregate load or levels of a specific aggregate species, we coupled agarose gel electrophoresis resolution of aggregates (Weiss et al., 2008) with immunoblotting. AGERA western blots probed with an antibody raised against mutant huntingtin (S830) revealed smears of detergent insoluble high molecular weight species that increased in both length and intensity with age in R6/2 mice. As expected, wild-type mice had no detectable signal. Interestingly, the high molecular weight smear observed in double transgenic mice was found to be markedly shorter than that observed in R6/2 mice at both 9- and 15 weeks of age, suggesting that HSJ1a reduces the level of high order detergent insoluble aggregates (Fig. 2G). Cytoplasmic and nuclear mutant huntingtin inclusions are formed with disease progression in the brains of patients with Huntington’s disease and mouse models of Huntington’s disease (Davies et al., 1997; DiFiglia et al., 1997). As human HSJ1a was able to reduce levels of high molecular weight aggregates, we reasoned that this might correspond to an alteration in the size of nuclear inclusions. To investigate this, mutant huntingtin immunohistochemistry was performed on 9- and 15-week brain sections and both nuclear inclusion area and incidence were determined using Zeiss Axiovision software. While no inclusions of any kind were observed in wild-type and human HSJ1a transgenic mice, R6/2 mice contained large numbers of both cytoplasmic and nuclear inclusions throughout the cortex, which increased in incidence and size between 9- and 15 weeks of age (Fig. 3A and B). Quantification of nuclear inclusion area revealed that nuclear inclusions were ∼40% smaller in double transgenic mice compared with R6/2 at 15-, but not 9-weeks of age (Fig. 3C). No significant difference in the incidence of nuclear inclusions was observed between R6/2 and double transgenic mice at either age (Fig. 3D). Collectively, these data reveal that human HSJ1a reduces levels of higher order detergent insoluble aggregates in R6/2 mice, which correlates with a reduction in nuclear inclusion size but not nuclear inclusion incidence.
Figure 3.
HSJ1a reduces nuclear aggregate load in R6/2 cortex. Representative images of S830 immunohistochemistry (brown reaction product) in motor cortex from 15 µm brain sections of wild-type (WT), HSJ1a, R6/2 and double transgenic (Dble) mice at (A) 9- and (B) 15 weeks of age. Methyl green was used as a nuclear counterstain. Scale bars: 10 µm. (C) Nuclear inclusion area and (D) percentage of inclusion positive nuclei at 9- and 15 weeks of age were determined by manually counting more than 320 nuclei and measuring more than 200 inclusions per mouse using Zeiss Axiovision software. Mean inclusion area was determined and plotted SEM for R6/2 (light grey bar) and double transgenic (black bar) mice (n = 5). Aggregate load was quantified in (E) nuclear and (F) cytosolic fractions isolated from WT, HSJ1a, R6/2 and double transgenic brain tissue at 15 weeks of age by MW8 Seprion enzyme-linked immunosorbent assay. WT and HSJ1a signals were considered to be background and were subtracted from R6/2 and double transgenic readings, respectively. Mean values ± SEM were plotted for each group (n ≥ 5). Statistical analysis was performed by Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.005). (G) Representative western blots of mutant huntingtin (S830), human HSJ1a (16321), α-tubulin and histone H3 in nuclear (nuc) and cytosolic (Cyto) fractions derived from 15-week-old WT, HSJ1a, R6/2 and double transgenic brain tissue.
HSJ1a reduces aggregate load in the nucleus but not cytoplasm of R6/2 mice
To determine whether HSJ1a was able to alter aggregate load in both the nucleus and cytoplasm, we performed cellular fractionation on 15-week brain tissue from wild-type, HSJ1a, R6/2 and double transgenic mice. We then subjected these fractions to MW8 Seprion enzyme-linked immunosorbent assay to quantify the levels of detergent insoluble aggregates present in these compartments.
We found that aggregate load was reduced in the nucleus but not cytosol of double transgenic mice at 15 weeks of age (Fig. 3E and F). These findings and the purity of the fractions was confirmed by western blotting (Fig. 3G). Interestingly, we also observed an enrichment of human HSJ1a in the nuclear fraction of double transgenic mice compared with the HSJ1a only transgenics (Fig. 3G). Soluble mutant huntingtin was detected in cytosolic but not nuclear fractions obtained from R6/2 and double transgenic brain tissues (Fig. 3G). This is consistent with our previous observations that the steady-state level of soluble mutant huntingtin is predominantly cytosolic and that any soluble nuclear mutant huntingtin is rapidly recruited into aggregates (Benn et al., 2005; Landles et al., 2010). Interestingly, there was an increase in soluble mutant huntingtin in the double transgenic cytosol, highlighting that the HSJ1a-mediated reduction in aggregation and increase in soluble mutant huntingtin occur in different compartments.
HSJ1a associates with aggregated but not soluble mutant huntingtin in 9- and 15-week-old R6/2 mice
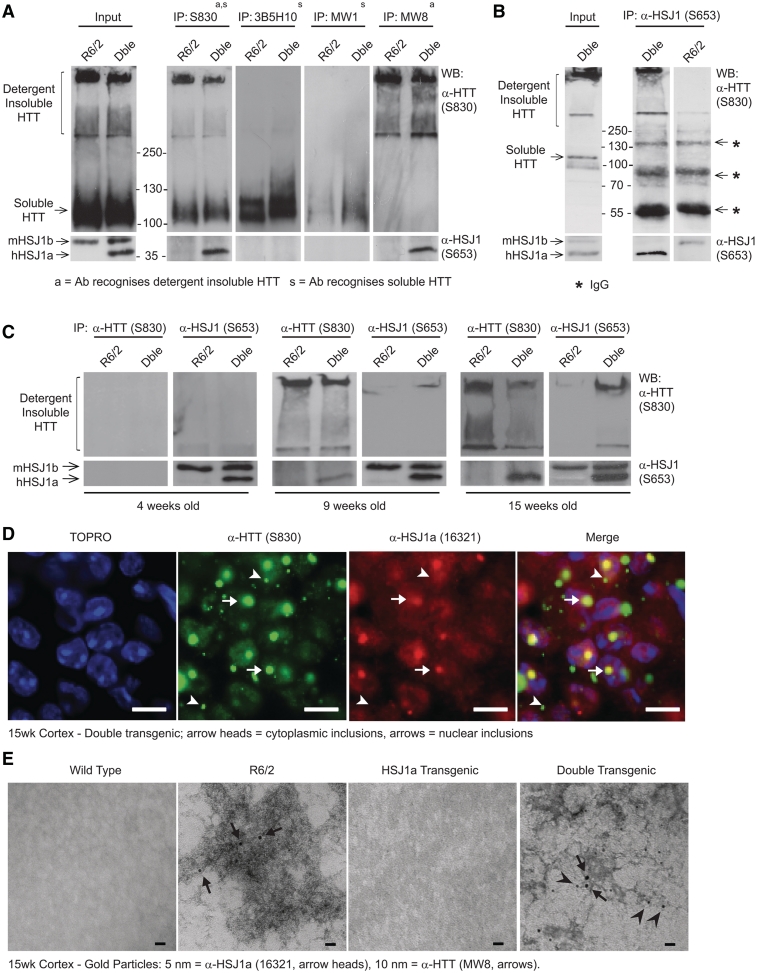
To elucidate the mechanism by which human HSJ1a was able to influence aggregate load, immunoprecipitations were performed on 15 week mouse brain tissue using antibodies that recognize detergent soluble (3B5H10 and MW1), or both soluble and aggregated (MW8 and S830) mutant huntingtin. Immunoblotting with these antibodies or anti-HSJ1 antibody (S653), revealed that human HSJ1a co-immunoprecipitated specifically with huntingtin antibodies, which precipitated detergent insoluble mutant huntingtin that was trapped in the stacking gel and well (Fig. 4A). This was further confirmed by reciprocal immunoprecipitation with anti-HSJ1, which precipitated only detergent insoluble mutant huntingtin (Fig. 4B).
Figure 4.
HSJ1a associates with detergent insoluble mutant huntingtin complexes from 9 weeks of age onwards when over-expressed in R6/2 mice. (A) Western blots probed for mutant huntingtin (HTT, S830, upper panels) or HSJ1 (S653, lower panels) after immunoprecipitation (IP) with anti-huntingtin (S830, MW1, MW8 and 3B5H10) antibodies. Huntingtin antibodies were chosen based on their ability to recognize detergent soluble (MW1, 3B5H10 and S830) or insoluble (MW8 and S830) mutant huntingtin. MW8 can recognize soluble mutant huntingtin by immunoprecipitation and western blot; however, in these experiments MW8 preferentially recognized detergent insoluble material. Immunoprecipitations were performed on 15 week brain tissue from R6/2 and double transgenic mice. (B) Western blots probed for mutant huntingtin (S830) and HSJ1 (S653) after immunoprecipitation with anti-HSJ1 (S653) from 15 week R6/2 and double transgenic brain tissue (asterisks denote IgG bands). (C) Western blots probed for mutant huntingtin (S830) and HSJ1 (S653) after immunoprecipitation with S830 or S653 from 4-, 9- and 15-week-old R6/2 or double transgenic (Dble) brain tissue. (D) Immunofluorescence images from the cortex of 15-week-old double transgenic mice stained with anti-huntingtin (S830) and anti-HSJ1a (16321) antibodies. Scale bars: 10 µm, arrowheads show the position of cytoplasmic inclusions, arrows show the position of nuclear inclusions. (E) Representative transmission electron microscopy images of aggregated material isolated from 15-week-double transgenic cortex by Seprion pull down. Aggregated material was double labelled with 5 nm [HSJ1a (16321): arrowheads] or 10 nm [mutant huntingtin (MW8): arrows] gold particles. Scale bars: 20 nm.
To ascertain the age at which human HSJ1a associates with detergent insoluble mutant huntingtin complexes, S830 immunoprecipitations were performed in 4-, 9-, and 15-week old brain tissue. Interestingly, the amount of human HSJ1a-associated mutant huntingtin high-molecular weight species increased with age (Fig. 4C). Immunohistochemistry performed on 15-week brain sections from double transgenic mice with S830 or an HSJ1a-specific antibody (16321) revealed that human HSJ1a co-localized exclusively with nuclear inclusions despite human HSJ1a’s cytoplasmic and nuclear localization (Fig. 4D and Supplementary Fig. 4). This observation is similar to previous reports in Huntington’s disease mice for other chaperones (Hay et al., 2004) and suggests that either the composition or conformation of cytoplasmic inclusions is not favourable for prolonged association with human HSJ1a and other chaperones.
To further explore the conformation/morphology of human HSJ1a–mutant huntingtin complexes, aggregated material was isolated from 15-week old R6/2 and double transgenic cortices using Seprion ligand-coated magnetic beads (Sathasivam et al., 2010). Aggregates were then subjected to transmission electron microscopy after immuno-gold labelling with anti-huntingtin (MW8) and anti-HSJ1a (16321) antibodies. Double gold labelling was only observed in aggregate material isolated from double transgenic mice (Fig. 4E), thereby confirming that HSJ1a associates directly with aggregated mutant huntingtin. Both R6/2 and double transgenic mice contained a multitude of dense MW8 positive aggregates; however, no obvious difference in aggregate morphology or conformation was observed between mice (Fig. 4E and Supplementary Fig. 5). As expected, no MW8 or HSJ1a labelled material was isolated from wild-type or HSJ1a mice (Supplementary Fig. 5). These data show that human HSJ1a associates with detergent insoluble mutant huntingtin, reduces aggregate load, and increases mutant huntingtin solubility in R6/2 mice.
HSJ1a associates with a subpopulation of aggregate species
To further understand the species of mutant huntingtin that human HSJ1a is able to associate with, aggregated material was immunoprecipitated with S830 or S653 from 9- and 15-week brain tissue. Immunoprecipitation material was then subjected to agarose gel electrophoresis resolution of aggregates and immunoblotting with anti-huntingtin (S830). The aggregate smears observed after S830 immunoprecipitation were slightly shorter than those in the input lanes, suggesting that extremely large aggregates are not efficiently magnetically separated by S830. Interestingly, the aggregate smears after HSJ1 immunoprecipitation were found to be much shorter than the smears generated by S830 immunoprecipitation (Fig. 5A). This suggests that human HSJ1a associates with a subpopulation of smaller mutant huntingtin complexes, potentially blocking their growth into higher order aggregate species.
Figure 5.
HSJ1a associates with high molecular weight mutant huntingtin complexes via a UIM-dependent mechanism. (A) Western blot of aggregated material isolated by anti-huntingtin (S830) or anti-HSJ1 (S653) immunoprecipitation and resolved by agarose gel electrophoresis resolution of aggregates. Immunoprecipitations were performed on 9- and 15-week brain tissue from HSJ1a, R6/2 or double transgenic (Dble) mice and mutant huntingtin was detected with S830. (B) Western blots showing co-immunoprecipitation of major regulators of protein homoeostasis from 15-week R6/2 and double transgenic brain tissue after immunoprecipitation with anti-huntingtin (S830) or anti-HSJ1 (S653) antibodies. (C) HSJ1 (S653) immunoprecipitation was performed on 15-week R6/2 or double transgenic brain lysates. Immunoprecipitation complexes were eluted from magnetic beads in denaturing buffer and subjected to a second immunoprecipitation with S830. Levels of ubiquitylated detergent insoluble material after each immunoprecipitation were determined by western blotting and immuno-detection with anti-huntingtin (S830) and anti-Ubiquitin (α-Ub) antibodies. (D) Western blots probed with S830 after HSJ1 immunoprecipitation (anti-myc) from 4-, 9- and 15-week R6/2 brain lysates incubated with lysates from SK-N-SH cells expressing myc-tagged wild-type, J-domain inactive (H31Q) or UIM inactive (ΔUIM) HSJ1a (asterisks denote IgG bands).
HSJ1a association with aggregates is dependent on functional ubiquitin-interacting motifs
As HSJ1a possesses an N-terminal J-domain and two C-terminal UIM motifs (UIM domain), which mediate the association with HSP70 and ubiquitin chains, respectively (Westhoff et al., 2005), we used immunoprecipitation and immunoblotting to further examine the composition of human HSJ1a:mutant huntingtin complexes. Interestingly, an increase in levels of mutant huntingtin associated HSP70 and HSP90 was observed in double transgenic mice compared with R6/2 (Fig. 5B). Furthermore, S830 immunoprecipitations performed on denatured complexes eluted after HSJ1 immunoprecipitation confirmed that HSJ1a associated with ubiquitylated mutant huntingtin complexes (Fig. 5C). No change was observed in the levels of DNAJB1 (HDJ-1), p62, GRP94 or HSP60 associated with detergent insoluble mutant huntingtin complexes (Supplementary Fig. 6). However, p62 was associated with HSJ1a–mutant huntingtin complexes but not with HSJ1a in the absence of mutant huntingtin (Fig. 5B). This most likely reflects sequestration of p62 by mutant huntingtin inclusions (Bjorkoy et al., 2005).
To confirm that the absence of an association between human HSJ1a and soluble mutant huntingtin was not a consequence of rapid mutant huntingtin degradation and to determine whether the J-domain or UIM motifs of HSJ1a are integral to HSJ1a complex association, we expressed myc-tagged HSJ1a or point mutant variants in SK-N-SH cells. These cell lysates were incubated with R6/2 brain lysates from 4-, 9- and 15-week old mice, and immunoprecipitation was performed with an anti-myc antibody. As was observed previously in double transgenic mice (Fig. 4), human HSJ1a was found to specifically co-immunoprecipitate high molecular weight mutant huntingtin, and not detergent soluble mutant huntingtin, from R6/2 brains in an age-dependent manner (Fig. 5D). To further interrogate the possibility that HSJ1a may promote the rapid degradation of soluble mutant huntingtin in mouse brain lysates, we performed ex vivo degradation assays using 4-week old R6/2 brain lysates, which contain high levels of soluble mutant huntingtin. We did not observe any degradation of mutant huntingtin in lysates over a 16-h period in the presence of bovine serum albumin or HSJ1a (Supplementary Fig. 7A and B). To confirm that the proteasome was active in the lysates we added luciferase, a previously described HSJ1a client (Westhoff et al., 2005; Howarth et al., 2007), to the lysate and measured luciferase activity over the same time course. We observed a marked decrease in luciferase activity that was enhanced by HSJ1a (Supplementary Fig. 7C), thereby confirming that HSJ1a can direct the degradation of client proteins in R6/2 lysates. These data suggest that a rapid turnover of soluble mutant huntingtin by HSJ1a is not responsible for the failure to detect an association between soluble mutant huntingtin and HSJ1a.
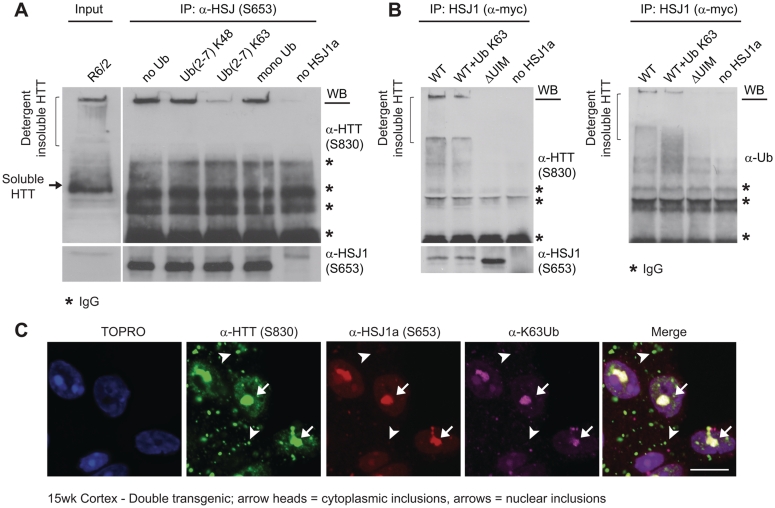
Interestingly, HSJ1a lacking a functional J-domain (H31Q) showed a reduced ability to bind aggregated mutant huntingtin; however, point mutations that disrupt the function of the UIM domains of HSJ1a [S219A/E222A/S262A/E265A (ΔUIM)] (Westhoff et al., 2005) almost completely abolished human HSJ1a mutant huntingtin binding ability (Fig. 5D). The importance of the UIM domains was further confirmed by incubating HSJ1a with R6/2 brain lysates in the presence of different ubiquitin conjugates. We observed that while K48 linked chains and mono ubiquitin were unable to compete with mutant huntingtin for HSJ1a binding (Fig. 6A), the presence of K63-linked ubiquitin chains greatly inhibited the human HSJ1a–mutant huntingtin association (Fig. 6A and B). Furthermore, human HSJ1a co-localized with K63 ubiquitin immunoreactivity in nuclear inclusions in double transgenic brain (Fig. 6C), thereby confirming that the human HSJ1a–mutant huntingtin complexes are K63 ubiquitylated in vivo. Interestingly, as was observed for human HSJ1a, the K63 ubiquitin signal was found only to decorate nuclear inclusions.
Figure 6.
HSJ1a associates with K63 ubiquitylated mutant huntingtin inclusions. Western blots probed with S830, S653 or α-Ub antibodies after HSJ1 (S653 or α-myc) immunoprecipitation of material from 15-week R6/2 brain lysates incubated with (A) purified HSJ1a or (B) lysates from SK-N-SH cells expressing wild-type or UIM inactive (ΔUIM) HSJ1a in the presence of UbK48 chains or UbK63 chains (A and B) (asterisks denote IgG bands). (C) Immunofluorescence images from the cortex of 15-week-old double transgenic mice stained with anti-huntingtin (S830), anti-HSJ1a (16321) and anti-UbK63 antibodies. Scale bars: 10 µm, arrowheads show the position of cytoplasmic inclusions; arrows show the position of nuclear inclusions.
HSJ1a associates with mutant huntingtin aggregates via client binding and ubiquitin-interacting motif domains
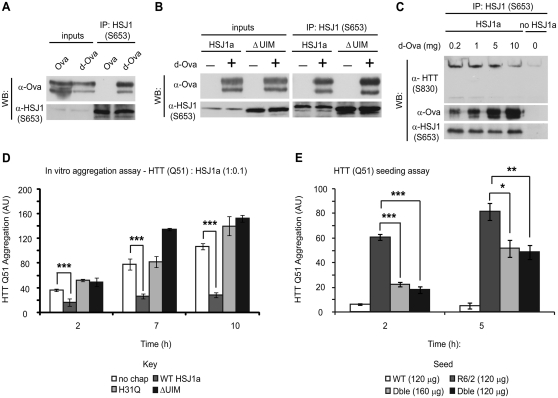
To confirm that the inability of ΔUIM HSJ1a to immunoprecipitate aggregated mutant huntingtin is not a consequence of a general impairment in HSJ1a client recognition and binding, we incubated HSJ1a with native or denatured ovalbumin (denatured ovalbumin, a classic chaperone substrate) and performed immunoprecipitation with S653. Immunoblotting demonstrated that HSJ1a interacted specifically with denatured ovalbumin, a finding that is consistent with HSJ1a’s role as a chaperone (Fig. 7A). Despite mutation in both UIMs, ΔUIM HSJ1a was able to recognize denatured ovalbumin as effectively as wild-type HSJ1a (Fig. 7B). Moreover, denatured ovalbumin was able to compete with mutant huntingtin complexes for human HSJ1a binding in a concentration-dependent manner (Fig. 7C). Taken together, these data show that the ability of HSJ1a to associate with high molecular weight mutant huntingtin from R6/2 brain tissue requires both chaperone client binding and UIMs.
Figure 7.
HSJ1a reduces huntingtin aggregation in a J-domain and UIM-dependent manner. Purified wild-type (A and B) or ΔUIM (B) HSJ1a was incubated with native (Ova) or denatured ovalbumin (d-Ova) for 30 min at room temperature, after which, anti-HSJ1 antibody S653 was used to immunoprecipitate material from incubations. Western blotting was performed to determine levels of HSJ1a and ovalbumin after immunoprecipitation. (C) Lysates from SK-N-SH cells expressing HSJ1a were incubated with 15-week R6/2 brain lysates in the presence of 0.2, 1, 5 or 10 mg of denatured ovalbumin. Levels of denatured ovalbumin, huntingtin and HSJ1a were determined by immunoblotting after immunoprecipitation with anti-HSJ1 (S653). (D) GST-HTT Q51 in vitro aggregation assays were performed in the presence of SK-N-SH cell lysates and ATP regenerating system containing no additional chaperone (white bars), wild-type HSJ1a (dark grey bars), J-domain inactive HSJ1a (light grey bars) or ΔUIM HSJ1a (black bars). HSJ1a was added to GST-HTT Q51 at a ratio of 1:10. Reactions were stopped 2, 7 and 10 h after removal of GST tag from GST-HTT Q51 by TEV protease cleavage. Levels of detergent insoluble mutant huntingtin were determined by filter trap, immunoblotting and densitometry. Mean signal was plotted for each group ± SEM (n = 4). (E) GST-HTT Q51 in vitro nucleation assays were performed using 120 ng of detergent insoluble material purified from brains of wild-type (white bars), R6/2 (light grey bars) or double transgenic (black bars) or 160 ng from double transgenic (dark grey bars). Mean signal was plotted for each group ± SEM (n = 6). Statistical analysis was performed by Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.005).
The J-domain and ubiquitin-interacting motifs are both important for HSJ1a-mediated suppression of mutant huntingtin aggregation
To assess the impact of the J-domain and UIM domains on the suppression of mutant huntingtin aggregation by human HSJ1a, we modified a previously published in vitro assay (Scherzinger et al., 1999; Tam et al., 2009) to quantify GST-HTT Q51 aggregation in the presence of purified HSJ1a or different SK-N-SH cell lysates and an ATP regenerating system. Consistent with previously published data (Tam et al., 2009), mutant huntingtin aggregation proceeded rapidly and reached saturation 10 h after initiation of AcTEV™ protease cleavage of the GST tag (Supplementary Fig. 8A). While addition of bovine serum albumin had no effect on aggregation kinetics, the addition of HSJ1a reduced mutant huntingtin aggregation in a substoichiometric manner (Fig. 7D and Supplementary Fig. 8B). Furthermore, while HSJ1a was unable to reverse aggregation after 7 h, the addition of HSJ1a to preformed aggregates at 2 h did significantly suppress further aggregation (Supplementary Fig. 8C). In contrast, both H31Q and ΔUIM HSJ1a were unable to suppress mutant huntingtin aggregation, suggesting that both the J and UIM domains are important for HSJ1a-mediated suppression of aggregation in the presence of other proteostasis factors (Fig. 7D).
The in vitro aggregation of mutant huntingtin can be accelerated by the addition of seeds of pre-aggregated material as nucleation factors (Scherzinger et al., 1999; Tam et al., 2009). We hypothesized that HSJ1a would modify the ability of mutant huntingtin aggregates to seed further aggregation. To test this hypothesis, we reduced the GST-HTT Q51 concentration so that it would only aggregate rapidly with nucleation from preformed mutant huntingtin aggregates and added detergent insoluble mutant huntingtin purified from R6/2 or double transgenic brains according to the method of Nekooki-Machida et al. (2009). The R6/2-derived mutant huntingtin aggregates efficiently nucleated GST-HTT Q51 aggregation (Fig. 7E). Importantly, double transgenic derived mutant huntingtin aggregates were significantly less able to nucleate aggregation, even when 30% more material was added to compensate for the reduction in aggregation observed in the Seprion assay (Fig. 2A). Therefore, human HSJ1a expression can modify the ability of aggregated mutant huntingtin to seed further aggregation, thereby reducing the recruitment of soluble mutant huntingtin and reducing aggregation.
Human HSJ1a modestly improves neurological phenotypes at late stage disease in R6/2 mice
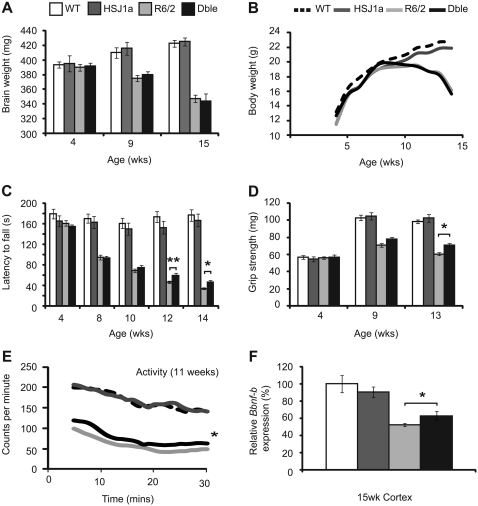
Finally, we wanted to determine whether the effects of HSJ1a on mutant huntingtin aggregation coincided with an amelioration of R6/2 disease progression. To address this we used a set of established quantitative tests to assess behavioural and physiological phenotypes in mice (Hockly et al., 2003a, b, 2006). Mice were well matched for CAG repeat size (Supplementary Table 1) and phenotypic parameters were measured from 4- to 14-weeks of age in wild-type, HSJ1a, R6/2 and double transgenic mice. As expected, R6/2 mice weighed less than wild-type mice by 11 weeks of age and gained weight at a slower rate. In keeping with previously published data, R6/2 mice also showed decreased brain weight with disease progression (Davies et al., 1997). Human HSJ1a over-expression had no significant effect on the brain or body weight of wild-type or R6/2 mice at any age tested (Fig. 8A and B).
Figure 8.
HSJ1a over-expression modestly improves neurological phenotypes in R6/2 mice. Wild-type (WT, white bars and dotted lines), HSJ1a (dark grey bars/lines), R6/2 (light grey bars/lines) and double transgenic (Dble, black bars/lines) mice were subjected to (A) body weight, (B) brain weight, (C) RotaRod, (D) grip strength or (E) activity assessment from 4- to 15 weeks of age. Mean values for each parameter were plotted for each group ± SEM (n ≥ 16 per group). Statistical analysis was performed by GLM-ANOVA in SPSS (*P < 0.05, **P < 0.01). (F) Relative levels of Bdnf-B (coding exon) messenger RNA in 15 week cortex as measured by TaqMan® quantitative polymerase chain reaction. Levels of Bdnf-B were normalized to the reference genes Atp5b and Canx. Statistical analysis was performed by Student's t-test (*P < 0.05, **P < 0.01).
RotaRod performance is a sensitive indicator of balance and motor coordination, which has been reliably shown to decline in R6/2 mice (Hockly et al., 2003a). Consistent with previous results, R6/2 RotaRod performance was impaired by 8 weeks and deteriorated with age. Human HSJ1a expression was found to significantly improve the RotaRod performance of R6/2 mice at 12- and 14-weeks of age (Fig. 8C). In addition, forelimb grip strength was assessed at 4-, 11- and 13-weeks of age. Consistent with previous data, the grip strength of R6/2 and double transgenic mice deteriorated with age. Human HSJ1a expression improved R6/2 grip strength at 13-weeks of age (Fig. 8D).
Exploratory activity was assessed fortnightly from 5- to 13-weeks of age as described previously (Hockly et al., 2006). Mice were assessed for a period of 30 min for total activity and mobility (Supplementary Fig. 9), and the statistical analysis is summarized in Supplementary Table 3. R6/2 mice exhibited significant hypo-activity with disease progression compared with wild-type mice. Both overall activity and mobility were improved at 11 weeks in double transgenic mice (Fig. 8E and Supplementary Table 2). Taken together, these findings show that over-expression of HSJ1a modestly improves several phenotypic parameters in R6/2 mice.
Finally, reduced levels of Bdnf promoter transcripts are a hallmark of disease progression in Huntington’s disease (Zuccato et al., 2001). TaqMan® real-time quantitative polymerase chain reaction and enzyme-linked immunosorbent assay were used to determine levels of BDNF messenger RNA and protein, respectively. We found that R6/2 mice had significantly reduced levels of BDNF messenger RNA and protein in cortex at 15-weeks of age. Human HSJ1a expression did not alter BDNF levels in wild-type mice but did significantly preserve levels of BDNF messenger RNA and protein in double transgenic mice (Fig. 8F and Supplementary Fig. 10). Taken together, these data show that HSJ1a reduces aggregate load with a concomitant improvement in disease progression in R6/2 mice.
Discussion
For over 10 years, DNAJ (HSP40) chaperones have been recognized as potent modifiers of polyglutamine aggregation and toxicity (Muchowski and Wacker, 2005). However, the effect of DNAJ over-expression on polyglutamine disease progression in the mouse brain has remained unknown. Over-expression of human HSJ1a (human HSJ1a/DNAJB2a) reduced mutant huntingtin exon 1 aggregate load in the cortex and striatum of R6/2 mice, but not cerebellum. This is most likely due to the fact that the cerebellar expression of human HSJ1a was much lower than in other brain tissues and suggests that the ability of HSJ1a to modify mutant huntingtin aggregation is dependent on its relative expression. These findings confirm that HSJ1a is a potent modifier of polyglutamine aggregation and, importantly, demonstrate that previous observations in cell culture translate to the mouse brain (Westhoff et al., 2005; Borrell-Pages et al., 2006; Howarth et al., 2007; Hageman et al., 2010).
HSJ1a was previously shown to suppress aggregation in cells by associating with soluble mutant huntingtin, thereby promoting its degradation via the proteasome (Westhoff et al., 2005). Surprisingly, we found no evidence of an association between HSJ1a and detergent soluble forms of mutant huntingtin in R6/2 brain tissue. It is possible that differences in polyglutamine length and the environment of the mouse brain result in a conformation of soluble mutant huntingtin that is not readily recognized by HSJ1a (Jana et al., 2000; Helmlinger et al., 2004). Alternatively, soluble mutant huntingtin–HSJ1a complexes may be rapidly degraded in mouse brain tissue making them difficult to detect by co-immunoprecipitation. However, we were also unable to detect soluble HSJ1a–mutant huntingtin complexes from ex vivo immunoprecipitations, or an effect of HSJ1a on soluble mutant huntingtin degradation ex vivo, which argues against this. While we cannot completely exclude the possibility that soluble HSJ1a:mutant huntingtin complexes are too transient for detection, these data suggest that in R6/2 mice human HSJ1a over-expression reduces aggregate load in a manner independent of soluble mutant huntingtin recognition and proteasomal targeting. This is intriguing given that other mammalian chaperones such as HSP70, HDJ-1 and TRiC have been proposed to suppress aggregation by acting at the level of soluble mutant huntingtin (Tam et al., 2009; Lotz et al., 2010).
In contrast, we found direct evidence that HSJ1a is able to associate with detergent insoluble high molecular weight complexes of mutant huntingtin. Consistent with previous data from cell culture (Westhoff et al., 2005; Howarth et al., 2007), we found that HSJ1a co-localized with mutant huntingtin inclusions when over-expressed in R6/2 brain tissue. Interestingly, despite the presence of HSJ1a in both the nucleus and cytoplasm, co-localization occurred almost exclusively with nuclear inclusions. This has been observed previously with the chaperones HDJ-1 (DNAJB1), HDJ-2 (DNAJA1), HSC70 (HSPA8), α-SGT and β-SGT (Hay et al., 2004) and raises the intriguing possibility that cytoplasmic inclusions adopt a conformation or composition that is unfavourable for recognition and binding by chaperones, or that chaperone decorated cytoplasmic inclusions are rapidly removed by autophagy. Furthermore, HSJ1a was found to co-immunoprecipitate insoluble material from R6/2 brain tissue more effectively at 9- and 15-weeks than at 4-weeks of age. While the temporal increase in HSJ1a expression may contribute to this, ex vivo immunoprecipitations demonstrated that human HSJ1a–mutant huntingtin complexes were preferentially formed at 9- and 15-weeks, even when equivalent levels of human HSJ1a were added to R6/2 brain lysates. These findings suggest that HSJ1a displays an age-dependent conformational and/or compositional selectivity when binding high molecular weight mutant huntingtin complexes and, coupled with increased HSJ1a expression, most likely explains why aggregate load only shows pronounced reduction in R6/2 mice at 15-weeks of age.
HSJ1 is distinguished by its N-terminal J-domain and UIMs (Chapple et al., 2004; Kampinga and Craig, 2010). The accumulation of ubiquitin chains and ubiquitylation of mutant huntingtin inclusions have been reported in Huntington’s disease models (Bence et al., 2001; Bennett et al., 2005, 2007). Interestingly, we found that HSJ1a co-localizes with K63 ubiquitylated inclusions in R6/2 brain tissue and that UIM mutants of HSJ1a exhibit a reduced ability to associate with mutant huntingtin. Conversely, a J-domain mutant (H31Q) that is unable to functionally co-operate with HSP70 could still form human HSJ1a–mutant huntingtin complexes. In addition, human HSJ1a–mutant huntingtin complex formation was inhibited in ex vivo immunoprecipitations by the addition of K63 but not K48 linked ubiquitin chains, and by the addition of an excess of denatured ovalbumin (a client for HSJ1a binding). Furthermore, human HSJ1a and K63 ubiquitin were both found to co-localize exclusively with nuclear inclusions in double transgenic mouse brains. Taken together, these data suggest that human HSJ1a–mutant huntingtin association requires HSJ1a chaperoning ability and the recognition of K63-ubiquitin chains. Sequential immunoprecipitation demonstrated that the ubiquitylated factor necessary for HSJ1a complex formation is tightly associated with mutant huntingtin. However, whether that factor is mutant huntingtin itself or a tightly sequestered protein is unclear. K63 linked ubiquitin chains have been implicated in the cellular formation of tau and SOD1 inclusions, with K63 positive inclusions preferentially targeted to lysosomes for degradation (Tan et al., 2008). No change in levels of p62 or LC3II were observed upon human HSJ1a over-expression and while p62 was found to associate with human HSJ1a in the presence of mutant huntingtin, this is most likely due to the fact that p62 is present in mutant huntingtin inclusions (Bjorkoy et al., 2005). These data suggest that despite human HSJ1a–mutant huntingtin complex formation occurring in the presence of factors that promote autophagy, HSJ1a-mediated reduction of mutant huntingtin aggregation does not occur primarily through stimulating lysosomal targeting.
To elucidate how HSJ1a reduces aggregate load in R6/2 mice, we subjected human HSJ1a:mutant huntingtin complexes to closer scrutiny. Human HSJ1a-associated material was found to comprise a high molecular weight complex subpopulation enriched for HSP70 and HSP90. HSP70 has been proposed to associate dynamically with polyglutamine inclusions (Kim et al., 2002) although its exact role in doing so is unclear. We found that in vitro, HSJ1a-mediated suppression of aggregation requires functional co-operation with HSP70. As such, it is likely that recruitment of HSP70 to mutant huntingtin aggregates represents an important step in the mechanism by which HSJ1a reduces protein aggregation in vivo.
Examination of high molecular weight mutant huntingtin size distribution by agarose gel electrophoresis resolution of aggregates revealed that HSJ1a over-expression predominantly reduced levels of high molecular weight material with a concomitant reduction in nuclear inclusion size. Given that levels of detergent soluble mutant huntingtin are significantly increased upon HSJ1a over-expression, it is unlikely that these observations are simply caused by HSJ1a restructuring aggregates into a more compact conformation. In vitro assays revealed that, as with other DNAJB chaperones (Hageman et al., 2010), HSJ1a effectively suppressed mutant huntingtin aggregation. In addition, while HSJ1a was unable to reverse aggregation, we found that addition of HSJ1a to preformed aggregates in vitro did significantly reduce further aggregation. Strikingly, the ability of HSJ1a to suppress mutant huntingtin aggregation was dependent on J-domain and UIM function and suggests that while non-essential for binding, a functional association with HSP70 is necessary to suppress aggregate formation. Interestingly, UIM mutants of HSJ1a significantly increased mutant huntingtin aggregation in vitro. This is perhaps not surprising given that these mutants retain their ability to interact with HSP70 molecules and may therefore impede HSP70-mediated suppression of aggregation in these reactions. We propose that HSJ1a selectively recruits HSP70 to a subpopulation of mutant huntingtin oligomers through K63 ubiquitin chain recognition and client protein binding. HSJ1a and HSP70 can then act to inhibit inclusion growth by physically blocking sites of polymerization and altering the on and off rates for polymerization. Alternatively, HSJ1a and HSP70 could act to refold mutant huntingtin oligomers into subtly different conformations (Muchowski et al., 2000; Wacker et al., 2004). However, if this is the case, the rearrangement of mutant huntingtin must be relatively subtle, as no clear shift in aggregate morphology was detectable by transmission electron microscopy.
In addition to reducing aggregate load, DNAJ chaperones have been shown to reduce polyglutamine toxicity in cells and flies (Fayazi et al., 2006; Hageman et al., 2010). We found that CNS specific over-expression of human HSJ1a improved activity, grip strength and RotaRod ability in R6/2 mice but did not improve body or brain weight at any age tested. This suggests that HSJ1a modifies disease progression independently of non-neurological symptoms. In addition, phenotypic improvements in R6/2 mice occurred late in disease progression, a time that coincides with optimal human HSJ1a:mutant huntingtin binding and correlates with a pronounced reduction in aggregate load.
Previous studies have shown that a cystamine-induced increase in levels of HSJ1 transcripts correlates with an improvement in disease progression in Huntington’s disease mice (Karpuj et al., 2002). Upregulation of HSJ1b rather than HSJ1a was proposed to be responsible for these beneficial effects by increasing BDNF levels (Borrell-Pages et al., 2006). We did not observe any change in BDNF transcript or protein levels upon over-expression of human HSJ1a in wild-type mice. However, levels of BDNF messenger RNA and protein were partly preserved in R6/2 mice over-expressing HSJ1a. This is most likely a reflection of reduced aggregate load (Zuccato et al., 2001) but could also contribute to the behavioural improvements observed in R6/2 mice.
In summary, our data provide the first evidence to our knowledge that a J-domain chaperone can reduce aggregate load and ameliorate disease progression, albeit modestly, in a mouse model of polyglutamine disease. HSJ1a acts through a previously undescribed chaperone mechanism that is focused at the level of ubiquitylated high molecular weight mutant huntingtin and works to prevent aggregate growth in a manner similar to that described for HSP104-mediated suppression of Sup35 prionogenesis in yeast (Shorter and Lindquist, 2008). This is an important proof of principle and shows that increasing levels of DNAJ chaperones can be beneficial in the context of the mammalian brain. As such we propose that efforts to increase HSJ1a expression pharmacologically may be a valid therapeutic strategy in Huntington’s disease and other polyglutamine diseases, particularly with regards to improving late stage disease outcomes.
Funding
MRC studentship to J.L.; MRC (0401350, 0700412 and 801314); Wellcome Trust; CHDI Foundation and the Huntington’s Disease Society of America Coalition for the Cure.
Supplementary Material
Acknowledgements
GST-TEV-huntingtin exon 1 -Q51-Stag (GST-HTT Q51) construct was kindly provided by Professor Ron Kopito (Stanford University, USA) and the bovine prion protein promoter by Dr Catherine Lemaire-Vieille (CNRS Grenoble, France). We thank Donna Groom and members of the Neurogenetics lab for R6/2 mice and CAG repeat sizing and Dr Jose' Gonzalez (EMBL Mouse Biology Unit, Italy) for human HSJ1a transgenic blastocyst production.
Glossary
Abbreviations
- BDNF
brain-derived neurotrophic factor
- UIM
ubiquitin-interacting motif
References
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–55. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Benn CL, Landles C, Li H, Strand AD, Woodman B, Sathasivam K, Li SH, Ghazi-Noori S, Hockly E, Faruque SM, Cha JH, Sharpe PT, Olson JM, Li XJ, Bates GP. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington's disease. Hum Mol Genet. 2005;4:3065–78. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- Benn CL, Fox H, Bates GP. Optimisation of region-specific reference gene selection and relative gene expression analysis methods for pre-clinical trials of Huntington's disease. Mol Neurodegener. 2008;3:17. doi: 10.1186/1750-1326-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn CL, Butler R, Mariner L, Nixon J, Moffitt H, Mielcarek M, et al. Genetic knock-down of HDAC7 does not ameliorate disease pathogenesis in the R6/2 mouse model of Huntington's disease. PLoS One. 2009;4:e5747. doi: 10.1371/journal.pone.0005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–65. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–8. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell-Pages M, Canals JM, Cordelieres FP, Parker JA, Pineda JR, Grange G, et al. Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase. J Clin Invest. 2006;116:1410–24. doi: 10.1172/JCI27607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9:2811–20. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- Chapple JP, Cheetham ME. The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J Biol Chem. 2003;278:19087–94. doi: 10.1074/jbc.M212349200. [DOI] [PubMed] [Google Scholar]
- Chapple JP, van der Spuy J, Poopalasundaram S, Cheetham ME. Neuronal DnaJ proteins HSJ1a and HSJ1b: a role in linking the Hsp70 chaperone machine to the ubiquitin-proteasome system? Biochem Soc Trans. 2004;32(Pt 4):640–2. doi: 10.1042/BST0320640. [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Brion JP, Anderton BH. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem J. 1992;284(Pt 2):469–76. doi: 10.1042/bj2840469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–54. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Sun Y, Opal P, Antalffy B, Mestril R, Orr HT, et al. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet. 2001;10:1511–8. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–48. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–3. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Fayazi Z, Ghosh S, Marion S, Bao X, Shero M, Kazemi-Esfarjani P. A Drosophila ortholog of the human MRJ modulates polyglutamine toxicity and aggregation. Neurobiol Dis. 2006;24:226–44. doi: 10.1016/j.nbd.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Hageman J, Rujano MA, van Waarde MA, Kakkar V, Dirks RP, Govorukhina N, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37:355–69. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Hansson O, Nylandsted J, Castilho RF, Leist M, Jaattela M, Brundin P. Overexpression of heat shock protein 70 in R6/2 Huntington's disease mice has only modest effects on disease progression. Brain Res. 2003;970:47–57. doi: 10.1016/s0006-8993(02)04275-0. [DOI] [PubMed] [Google Scholar]
- Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13:1389–405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123(Pt 22):3849–55. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Bonnet J, Mandel JL, Trottier Y, Devys D. Hsp70 and Hsp40 chaperones do not modulate retinal phenotype in SCA7 mice. J Biol Chem. 2004;279:55969–77. doi: 10.1074/jbc.M409062200. [DOI] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci USA. 2003a;100:2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E, Woodman B, Mahal A, Lewis CM, Bates G. Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res Bull. 2003b;61:469–79. doi: 10.1016/s0361-9230(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Hockly E, Tse J, Barker AL, Moolman DL, Beunard JL, Revington AP, et al. Evaluation of the benzothiazole aggregation inhibitors riluzole and PGL-135 as therapeutics for Huntington's disease. Neurobiol Dis. 2006;21:228–36. doi: 10.1016/j.nbd.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Howarth JL, Kelly S, Keasey MP, Glover CP, Lee YB, Mitrophanous K, et al. Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol Ther. 2007;15:1100–5. doi: 10.1038/sj.mt.6300163. [DOI] [PubMed] [Google Scholar]
- Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–18. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- Karpuj MV, Becher MW, Springer JE, Chabas D, Youssef S, Pedotti R, et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med. 2002;8:143–9. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–31. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- Ko J, Ou S, Patterson PH. New anti-huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins. Brain Res Bull. 2001;56:319–29. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
- Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–63. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H, Finkbeiner S, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington's disease. J Biol Chem. 2010;285:8808–23. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Vieille C, Schulze T, Podevin-Dimster V, Follet J, Bailly Y, Blanquet-Grossard F, et al. Epithelial and endothelial expression of the green fluorescent protein reporter gene under the control of bovine prion protein (PrP) gene regulatory sequences in transgenic mice. Proc Natl Acad Sci USA. 2000;97:5422–7. doi: 10.1073/pnas.080081197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz GP, Legleiter J, Aron R, Mitchell EJ, Huang SY, Ng C, et al. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J Biol Chem. 2010;285:38183–93. doi: 10.1074/jbc.M110.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci USA. 2010;107:7927–32. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt H, McPhail GD, Woodman B, Hobbs C, Bates GP. Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington's disease. PLoS One. 2009;4:e8025. doi: 10.1371/journal.pone.0008025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–6. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Nekooki-Machida Y, Kurosawa M, Nukina N, Ito K, Oda T, Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc Natl Acad Sci USA. 2009;106:9679–84. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak MJ, Tabrizi SJ. Huntington's disease. BMJ. 2010;340:c3109. doi: 10.1136/bmj.c3109. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Woodman B, Mahal A, Bertaux F, Wanker EE, Shima DT, et al. Centrosome disorganization in fibroblast cultures derived from R6/2 Huntington's disease (HD) transgenic mice and HD patients. Hum Mol Genet. 2001;10:2425–35. doi: 10.1093/hmg/10.21.2425. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, Finkbeiner S, et al. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington's disease. Hum Mol Genet. 2010;19:65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, et al. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc Natl Acad Sci USA. 1999;96:4604–9. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–24. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szapacs ME, Mathews TA, Tessarollo L, Ernest Lyons W, Mamounas LA, Andrews AM. Exploring the relationship between serotonin and brain-derived neurotrophic factor: analysis of BDNF protein and extraneuronal 5-HT in mice with reduced serotonin transporter or BDNF expression. J Neurosci Methods. 2004;140:81–92. doi: 10.1016/j.jneumeth.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16:1279–85. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–9. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- The Huntington's disease collaborative research group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11:1215–22. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–8. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Westhoff B, Chapple JP, van der Spuy J, Hohfeld J, Cheetham ME. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol. 2005;15:1058–64. doi: 10.1016/j.cub.2005.04.058. [DOI] [PubMed] [Google Scholar]
- Weiss A, Klein C, Woodman B, Sathasivam K, Bibel M, Regulier E, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington's disease. J Neurochem. 2008;104:846–58. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- Weiss A, Abramowski D, Bibel M, Bodner R, Chopra V, DiFiglia M, et al. Single-step detection of mutant huntingtin in animal and human tissues: a bioassay for Huntington's disease. Anal Biochem. 2009;395:8–15. doi: 10.1016/j.ab.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–8. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman B, Butler R, Landles C, Lupton MK, Tse J, Hockly E, et al. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res Bull. 2007;72:83–97. doi: 10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–8. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.