Abstract
Spontaneous abortion is a significant clinical problem of different etiologies. Certain thrombophilia gene mutations have been associated with an increased risk of spontaneous abortion. Also, mutations in folate-related genes can lead to abnormal chromosomal segregation during meiosis which is the most common cause of spontaneous abortion. We have developed a multiplex single-base extension reaction assay that allows simultaneous analysis of 10 different mutations in thrombophilia- and folate-related genes (Factor V Leiden G1691A, Factor V H1299R, Factor II G20210A, Factor XIII V34L, PAI-I -675 4G/5G, FGB -455G/A, MTHFR C677T, MTHFR A1298C, MTR A2756G, and MTRR A66G). Using this method we have studied 232 women who had a spontaneous abortion and 209 of their male partners. Prevalence of Factor II G20210A and Factor V H1299R mutations was significantly higher in the women than in their male partners (2.4% and 0.7%, respectively [p=0.0499] for the Factor II mutation and 9.3% and 5.7%, respectively [p=0.0485] for the Factor V mutation). The prevalence of MTHFR C677T, MTHFR A1298C, MTR A2756G, and MTRR A66G mutations did not differ between the studied groups. In conclusion, we have developed a rapid, simple, reliable, and inexpensive multiplex SNaPshot method for determination of 10 thrombophilic mutations that may result in spontaneous abortions.
Introduction
Spontaneous abortion is the most common complication of pregnancy, affecting approximately 10–15% of clinically recognized pregnancies. Recurrent spontaneous abortions, defined as the occurrence of three or more consecutive, clinically detectable pregnancy failures, are estimated to occur in 0.5–3% of all couples trying to conceive. After excluding anatomic, metabolic, endocrine, infectious, genetic, and environmental factors, the cause remains unknown in approximately 50% of women who experience spontaneous abortions (Frosst et al., 1995).
There is growing evidence of involvement of inherited thrombophilia in the etiology of spontaneous abortion and placenta-related complications in pregnancy, including intrauterine growth restriction, intrauterine fetal death, placental abruption, and preeclampsia (Ford and Schust, 2009; Rodger et al., 2010). Such thrombophilias augment the prothrombotic state of pregnancy and lead to inadequate fetomaternal circulation and affect the process of placentation in the developing embryo. The most commonly related to spontaneous abortions are the Factor V Leiden mutation G1691A, mutation in the promoter region of the prothrombin gene G20210A, and homozygosity of C677T mutation in the methylene tetrahydrofolate reductase (MTHFR) gene with general prevalence of 2–15%, 2–3%, and 11%, respectively. These mutations are associated with mild thrombotic risks, and their association with spontaneous abortion remains controversial (Brenner et al., 1999; Pickering et al., 2001; Rai et al., 2001; Hefler et al., 2004; Tranquilli et al., 2004; Coulam et al., 2006; Yenicesu et al., 2010). Mutations that affect genes for factors involved in the common pathway of the coagulation cascade and the fibrinolytic system, such as in Factor XIII, β Fibrinogen, and plasminogen activator inhibitor-1 (PAI-1), may also contribute to the thrombotic tendency (Coulam et al., 2006; Yenicesu et al., 2010).
Hyperhomocysteinemia leads to endothelial injury and vascular inflammation and is an independent risk factor for thrombosis. However, reduced regeneration of methionine caused by mutations in folate-related enzymes of one-carbon metabolism can lead to impaired segregation (nondisjunction) of chromatids during meiosis I or II. This nondisjunction produces aneuploid gametes that give rise of aneuploid embryos. Chromosomal aneuploidies are a major and well-defined cause of first trimester spontaneous abortion (Stephenson et al., 2002). Cytogenetic evaluation of spontaneous abortions has revealed chromosomal abnormalities in 50–70% (Hogge et al., 2003). The most common are de novo numerical abnormalities (94%), in particular autosomal trisomies for chromosomes 13, 14, 15, 16, 21, and 22, followed by monosomy X and polyploidy (Hassold et al., 1980; Strom et al., 1992; Stephenson et al., 2002).
Available genotyping methods for the thrombophilic mutations include polymerase chain reaction (PCR) with fragment length polymorphism (PCR-RFLP) analysis, real-time PCR with allele-specific Taqman probes (Ulvik and Ueland, 2001), allele-specific reverse hybridization (Yilmaz et al., 2006), and MALDI TOFF mass spectrometry (Jurinke et al., 2002; Jurinke et al., 2004). Here we describe a multiplex single-base extension reaction assay that allows simultaneous analysis of 10 different mutations in thrombophilia- and folate-related genes in one reaction.
Materials and Methods
In total, 441 DNA samples were studied using a multiplex single-base extension reaction assay, 232 samples from women experiencing spontaneous abortions and 209 from their male partners who were used as controls. Of the women, 137 were of Slovenian, 55 of Albanian, and 40 of Macedonian origin, while of the men, 127 were of Slovenian, 51 of Albanian, and 31 of Macedonian origin. Couples of Slovenian origin were selected in the Institute of Medical Genetics, University Medical Center-Ljubljana, Ljubljana, Slovenia, while those of Macedonian and Albanian origin were selected in the Research Centre for Genetic Engineering and Biotechnology “Georgi D. Efremov,” Macedonian Academy of Sciences and Arts, Skopje, Macedonia. The study was approved by the Ethical Committee of the Macedonian Academy of Sciences and Arts, and all subjects gave informed consent for participation in the study.
Multiplex SNaPshot method for detection of 10 mutations in five thrombophilia- and three folate-related genes
All individuals were analyzed for the presence of 10 different mutations: Factor V Leiden G1691A, Factor V H1299R, Factor II G20210A, Factor XIII V34L, PAI-I -675 4G/5G, FGB -455G/A, MTHFR C677T, MTHFR A1298C, MTR A2756G, and MTRR A66G. We have designed a multiplex single-base extension method for simultaneous detection of the 10 mutations using the Multiplex SNaPshot kit (Multiplex SNaP-shot™; Applied Biosystems, Warrington, WA) for the reaction, followed by capillary electrophoresis on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
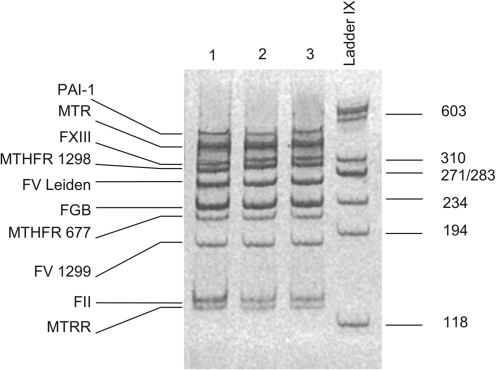
The PCR primers were designed to produce PCR fragments of 129–420 bp and to allow their separation by 6% polyacrylamide gel electrophoresis. Briefly the PCR amplification reaction contained 200 ng genomic DNA, 10 pmol of specific forward and reverse oligonucleotide primers (Table 1), 200 μL of each deoxyribonucleotide triphosphate (dNTP), 1.5 mM magnesium chloride, and 1 U Taq DNA polymerase (Ampli Tag Gold™; Applied Biosystems, Foster City, CA) in a total volume of 25 μL. The PCR conditions were 10 min initial denaturation at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 55°C, and 2.45 min at 72°C. A final extension was at 72°C for 30 min. After analysis of the PCR products on 6% polyacrylamide gels (Fig. 1), they were purified by the use of 1 μL of ExoSAP-IT® (USB, Cleveland, OH) per 5 μL of PCR product, with incubation at 37°C for 60 min and at 86°C for 15 min to deactivate the enzyme.
Table 1.
Sequences of PCR Primers and Primers Used for Single-Base Extension Reaction
| Mutation | Primer | Sequence (5'-3') |
|---|---|---|
| FV Leiden | PCR For | TGA TGC CCA GTG CTT AAC AA |
| PCR Rev | TCA CAC TGG TGC TAA AAA GGA | |
| SNaPshot | CCC CCC CCC CCC AGC AGA TCC CTG GAC AGG C | |
| FV H1299R | PCR For | GTC AGA TGC CCA TTT CTC CA |
| PCR Rev | AGA GGT TTG TCT GGC TGA GG | |
| SNaPshot | CCC CCC CCC CCC CCC CCC GCT GAA GTC TAG AGA AAG GGT TGT A | |
| F II G20210A | PCR For | GAG AGT AGG GGG CCA CTC AT |
| PCR Rev | CAT GAA TAG CAC TGG GAG CA | |
| SNaPshot | CCC CCC CCC CGT TCC CAA TAA AAG TGA CTC TCA GC | |
| FIII V34L | PCR For | TTT GGA GGC AGA AGA GCA GT |
| PCR Rev | CAA GGT CAG TAA GGG GCA GA | |
| SNaPshot | CAC AGT GGA GCT TCA GGG C | |
| PAI-1 4G/5G | PCR For | GGC AGC TCG AAG AAG TGA AA |
| PCR Rev | ACC TCC ATC AAA ACG TGG AA | |
| SNaPshot | CCC CCC CCC CCC CCC CCC ATG ATA CAC GGC TGA CTC CCC | |
| FGB -455G/A | PCR For | GGG TCT TTC TGA TGT GTA TTT TTC A |
| PCR Rev | TGA CCT ACT CAC AAG GCA ACC | |
| SNaPshot | ATT CTA TTT CAA AAG GGG C | |
| MTHFR C677T | PCR For | CCA GTC CCT GTG GTC TCT TC |
| PCR Rev | TCA CAA AGC GGA AGA ATG TG | |
| SNaPshot | CCC CCC CCC CCG CGG GAG CCG ATT TCA TCA T | |
| MTHFR A1298C | PCR For | TTT GGG GAG CTG AAG GAC TA |
| PCR Rev | GGA GGT CTC CCA ACT TAC CC | |
| SNaPshot | CCC CCC CCC CCC CCC CCC CCC CTG ACC AGT GAA GAA AGT GTC TTT GA | |
| MTRR A66G | PCR For | TCC CCC ATT TTT CAG TTT CA |
| PCR Rev | CCA TGT ACC ACA GCT TGC TC | |
| SNaPshot | AGG CCA TCG CAG AAG AAA T | |
| MTR A2756G | PCR For | CCA AGC CCA CTG AGT TTA CC |
| PCR Rev | TCC AAA GCC TTT TAC ACT CCT C | |
| SNaPshot | CCG AAT ATG AAG ATA TTA GAC AGG |
PCR, polymerase chain reaction.
FIG. 1.
Analysis of ten-plex polymerase chain reaction for detection of 10 thrombophilic mutations by 6% polyacrylamide gel electrophoresis.
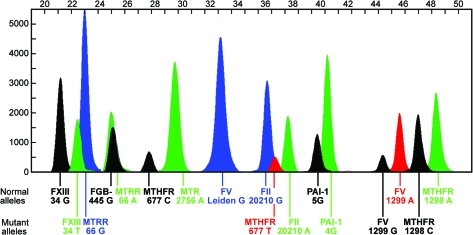
The purified PCR products formed the template for detection of the 10 mutations. In the following single-base extension reaction, the detection primers (SNaPshot primers) annealed adjacent to the single-nucleotide polymorphism position. Poly (dC) tails of different lengths were attached to the single-base extension primers (Table 1). The SNaPshot reaction contained 1 μL primer mix of all 10 single-base extension primers (final concentration of 1 pM each), 2 μL of purified PCR products, and 2 μL of SNaP-shot Multiplex kit in a final volume of 5 μL. The cycling profile consisted of 25 cycles of 95°C for 10 s, 55°C for 10 s, and 60°C for 30 s. After the reaction, 5'-phosphophoryl groups of unincorporated dideoxynucleotide triphosphates were removed by addition of 1.0 U of shrimp alkaline phosphatase (SAP; USB) and incubation at 37°C for 2 h and at 86°C for 20 min to inactivate the enzyme. Capillary electrophoresis was performed on an ABI PRISM 3130 Genetic Analyzer and the results were analyzed with Gene Mapper ID, Ver 4.0 (Applied Biosystems, Foster City, CA). Details of multiplex PCR and SNaPshot reaction products are given in Table 2. Due to the influence of the dye, size, and nucleotide composition on the mobility shift of the DNA fragments, the reported sizes differ by a few bases from the actual sizes, and this is particularly true with the shorter fragments as the relative contribution of the dye is greater. A representative electrophoreogram from a patient heterozygous for seven thrombophilic mutations is shown on Figure 2.
Table 2.
Details of Multiplex PCR and SNaPshot Reaction Products
| Mutation | Nucleotide change | PCR fragment (bp) | SNaPshot primer orientation | SNaPshot result (N/M) | SNaPshot fragment N (bp) | SNaPshot fragment M (bp) |
|---|---|---|---|---|---|---|
| FXIII V34L | G/T | 331 | R | C/A | 21 | 22 |
| MTRR A66G | A/G | 129 | F | A/G | 22.6 | 24.5 |
| FGB -455G/A | G/A | 222 | R | C/T | 24.4 | 26.6 |
| MTHFR C677T | C/T | 213 | F | C/T | 27 | 37 |
| MTR A2756G | A/G | 383 | F | A/G | 29 | 26 |
| FV Leiden | G/A | 265 | F | G/A | 33 | 34.5 |
| FII G20210A | G/A | 133 | F | G/A | 36 | 38 |
| PAI-1 4G/5G | T/G | 420 | R | A/C | 41 | 40 |
| FV H1299R | A/G | 177 | R | T/C | 46 | 45 |
| MTHFR A1298C | A/C | 314 | F | A/C | 49 | 48 |
FIG. 2.
Electrophoreogram from Gene Mapper ID, Ver 4.0, analysis of 10 mutations in five thrombophilia- and three folate-related genes in a patient with seven mutations in heterozygous state. Normal and mutant alleles are indicated on the left. Color images available online at www.liebertonline.com/gtmb
Ten DNA samples with different alleles of the 10 studied thrombophilic mutations, determined previously by DNA sequencing, were used to validate the multiplex SNaPshot method. Further, the SNaPshot genotyping results from 50 selected samples were confirmed by PCR-RFLP for FV Leiden, MTHFR C677T, and MTR A66G using the enzymes MnlI (New England BioLabs, Beverly, MA) (Bertina et al., 1994), HinfI, and HindIII (New England BioLabs) (Frosst et al., 1995), respectively. Digested segments of DNA were separated by electrophoresis on a 2% agarose gel stained with ethidium bromide, and examined under ultraviolet light. For the other mutations, confirmation was done by sequencing of the specific PCR product using ABI Prism® BigDye terminator cycle sequencing ready reaction kit v1.1 (Applied Biosystems, Foster City, CA) and forward or reverse primer (Table 1) according to the manufacturer's protocol. The reactions were run on an ABI Prism 3130 Genetic Analyzer and the results were analyzed with Sequencing Analysis Software 5.3.1.
Statistical analysis
Statistical analysis was performed using SISA (www.home.clara.net/sisa) statistics. Discrete variables were analyzed using Fisher's exact test or the χ2 test. p-Values less than 0.05 were taken as statistically significant. Relative risk and 95% confidence intervals were calculated where appropriate.
Results
The multiplex SNaPshot method for the testing of 10 thrombophilic mutations was validated by initial analysis of 10 DNA samples genotyped previously by DNA sequencing and representing normal, heterozygotes, and homozygotes for each studied mutation. Fifty samples representing normal and/or mutant alleles of the studied mutations, obtained by SNaPshot analysis, were genotyped by PCR-RFLP (for FV Leiden, MTHFR C677T, and MTR A66G mutations) and sequencing for evaluating the method. The positive and negative predictive values for the new multiplex SNaPshot method were 100%.
Allele and genotype frequencies were obtained for 232 female partners who had experienced spontaneous abortions and their male partners (n=209) using this multiplex single-base extension reaction assay. In all the studied groups, no significant deviation from the Hardy–Weinberg equilibrium was found (data not shown).
The allele frequencies of the mutations are shown in Table 3. Those of Factor II G20210A, A allele and Factor V H1299R (A→G), G allele were significantly higher in the female partners than in the male partners, 2.4% and 0.7% (p=0.0499), and 9.3% and 5.7% (p=0.0485), respectively. Those of Factor V Leiden mutation and PAI-1 5G/4G, 4G allele were slightly higher in the female partners (2.8% and 56.5%, respectively) than in the male partners (2.4% and 52.9%, respectively), but the difference was not statistically significant. There was a reverse trend for the alleles of Factor XIII V34L and FGB -455G/A, which were more frequent in the male partners (28.9% and 28.5%, respectively) than in the female partners (25% and 25.2%, respectively). Frequencies of MTHFR C677T, MTHFR A1298C, MTR A2756G, and MTRR A66G mutations were similar in both genders.
Table 3.
Distribution of Allele Frequencies of 10 Mutations in Thrombophilia- and Folate-Related Genes, Among Couples of Slovenian, Macedonian, and Albanian Ethnicity
| |
|
Slovenian couples |
Macedonian couples |
Albanian couples |
Total |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Allele | Female partner n=137 | Male partner n=127 | p | Female partner n=40 | Male partner n=31 | p | Female partner n=55 | Male partner n=51 | p | Female partner n=232 | Male partner n=209 | p |
| Factor V Leiden | G | 0.974 | 0.976 | 0.8875 | 0.962 | 0.952 | 0.7494 | 0.973 | 0.990 | 0.3504 | 0.972 | 0.976 | 0.7034 |
| A | 0.026 | 0.024 | 0.038 | 0.048 | 0.027 | 0.010 | 0.028 | 0.024 | |||||
| Prothrombin G20210A | G | 0.985 | 0.996 | 0.2063 | 0.962 | 0.984 | 0.4451 | 0.964 | 0.990 | 0.2030 | 0.976 | 0.993 | 0.0499 |
| A | 0.015 | 0.004 | 0.038 | 0.016 | 0.036 | 0.010 | 0.024 | 0.007 | |||||
| Factor V H1299R | A | 0.905 | 0.957 | 0.0203 | 0.938 | 0.952 | 0.7174 | 0.891 | 0.902 | 0.8602 | 0.907 | 0.943 | 0.0485 |
| G | 0.095 | 0.043 | 0.062 | 0.048 | 0.109 | 0.098 | 0.093 | 0.057 | |||||
| Factor XIII V34L | G | 0.737 | 0.693 | 0.2592 | 0.812 | 0.758 | 0.4307 | 0.736 | 0.725 | 0.8580 | 0.750 | 0.711 | 0.1866 |
| T | 0.263 | 0.307 | 0.188 | 0.242 | 0.264 | 0.275 | 0.250 | 0.289 | |||||
| FGB -455G/A | G | 0.745 | 0.717 | 0.4687 | 0.750 | 0.661 | 0.2474 | 0.755 | 0.745 | 0.8744 | 0.748 | 0.715 | 0.2757 |
| A | 0.255 | 0.283 | 0.250 | 0.339 | 0.245 | 0.255 | 0.252 | 0.285 | |||||
| PAI-1 5G/4G | 5G | 0.562 | 0.587 | 0.5686 | 0.575 | 0.452 | 0.1443 | 0.564 | 0.618 | 0.4244 | 0.435 | 0.471 | 0.3025 |
| 4G | 0.438 | 0.413 | 0.425 | 0.548 | 0.436 | 0.382 | 0.565 | 0.529 | |||||
| MTHFR C677T | C | 0.617 | 0.634 | 0.6855 | 0.538 | 0.565 | 0.7483 | 0.555 | 0.471 | 0.2218 | 0.588 | 0.584 | 0.8904 |
| T | 0.383 | 0.366 | 0.462 | 0.435 | 0.445 | 0.529 | 0.412 | 0.416 | |||||
| MTHFR A1298C | A | 0.686 | 0.681 | 0.9025 | 0.750 | 0.774 | 0.7379 | 0.736 | 0.794 | 0.3222 | 0.709 | 0.722 | 0.6588 |
| C | 0.314 | 0.319 | 0.250 | 0.226 | 0.264 | 0.206 | 0.291 | 0.278 | |||||
| MTR A2756G | A | 0.770 | 0.756 | 0.7024 | 0.762 | 0.742 | 0.7773 | 0.845 | 0.794 | 0.3302 | 0.787 | 0.763 | 0.4041 |
| G | 0.230 | 0.244 | 0.238 | 0.258 | 0.155 | 0.206 | 0.213 | 0.237 | |||||
| MTRR A66G | A | 0.431 | 0.417 | 0.7567 | 0.450 | 0.532 | 0.3307 | 0.400 | 0.441 | 0.5441 | 0.427 | 0.440 | 0.6873 |
| G | 0.569 | 0.583 | 0.550 | 0.468 | 0.600 | 0.559 | 0.573 | 0.560 | |||||
Values given in bold have statistical significance (p<0.05).
Frequencies of Factor V Leiden, Factor II G20210A, and MTHFR C677T mutations in Slovenian couples were slightly lower than in those of Macedonian or Albanian ethnicity (Table 3). In the Slovenian couples, there was a significantly higher frequency of Factor V H1299R mutation in the women than in their male partners (p=0.0203). A similar trend was obvious in the other ethnic groups, but their small number has restricted the power of the statistical tests.
Discussion
The SNaPshot genotyping method that we have designed detects 10 different mutations in thrombophilia- and folate-related genes that may be implicated in etiopathogenesis of spontaneous abortions and other vascular pathologies. This method efficiently and simultaneously detects the 10 mutations. Compared to other genotyping methods such as PCR-RFLP, reverse dot blot hybridization, sequencing, or mass spectrometry, this method is straightforward, accurate, easy to perform, and cost efficient.
With its use, we have estimated the prevalence of Factor V Leiden, prothrombin G20210A, Factor V H1299R, FXIII V34L, PAI-I -675 4G/5G, MTHFR C677T and A1298C, MTR A2756G, and MTRR A66G mutations in couples with spontaneous abortions of Slovenian, Macedonian, or Albanian ethnicity. We used the men as the control group, since thrombophilia is a maternal risk factor that leads to adverse pregnancy outcome. Several studies of couples with spontaneous abortions and fertile couples have found no difference in prevalence of thrombophilic mutations between studied groups (Jivraj et al., 2006; Jauniaux et al., 2006; Rodger et al., 2010).
The prothrombin G20210A and Factor V H1299R mutations in our study had a significantly higher prevalence among women than men. This implicates their possible involvement in pathogenesis of spontaneous abortions. These mutations were found only in the heterozygous state in both genders. We found no association between thrombophilia due to factor V Leiden, FXIII V34L, PAI-I −675 4G/5G, MTHFR C677T and A1298C, MTR A2756G and MTRR A66G and spontaneous abortions.
Prothrombin G20210A mutation, which results in increased levels of prothrombin and is a potential risk for thrombosis, has been identified as a risk factor for pregnancy loss in several studies and has been associated mainly with early spontaneous abortions (Finan et al., 2002). However, Factor V H1299R is associated with inherited mild-activated protein C resistance but mainly in homozygous state and predisposing to thrombosis. Factor V Leiden, responsible for more than 75% of inherited activated protein C resistance, is a common inherited thrombotic risk factor and there are conflicting results about but its association with spontaneous abortions especially early in pregnancy is controversial (Brenner, 1999; Brenner et al., 1999; Hatzis et al., 1999; Tranquilli et al., 2004; Coulam et al., 2006; Rodger et al., 2010). Its prevalence in our study was higher in women than in men (2.8% and 2.4%), but not statistically significant. Our results are in concordance with the one mentioned above and do not exclude the relevance of this mutation in second trimester abortions and late pregnancy complications.
Based on the association of the deficiency of coagulation factor XIII with high frequency of fetal wastage in mice, the polymorphic site within the coding region of coagulation factor XIII A subunit gene V34L that lowers plasma factor XIII–specific activity could be associated with spontaneous abortion (Koseki-Kuno et al., 2003). Mutation in -455 G→A promoter position of β-fibrinogen gene is associated with higher plasma β-fibrinogen level and increased clot formation. We found that the mutant genotypes of Factor XIII V34L (G/T+T/T) and FGB -455G/A (G/A+A/A) to be more frequent in the men than in the women (50.24% and 43.57% for F XIII V34L, and 48.8% and 42.66% for FGB -455G/A).
Factors that influence the fibrinolysis, particularly PAI-1, are active at implantation and placentation. They modify trophoblast migration and prevent additional hemorrhage at placentation. The -675 5G/4G polymorphism results in elevated endothelial expression of PAI-1 and reduced fibrinolytic activity. The difference between prevalence of mutant 4G allele in female and male partners (56.5% and 52.9%, respectively) that we have found was not statistically significant.
The lack of association between thrombophilia due to factor V Leiden, FXIII V34L, FGB -455G/A, and PAI-I -675 4G/5G mutations and spontaneous abortions that we have found accords with the published data of Brenner et al. (1999), Tranquilli et al. (2004), Coulam et al. (2006), and Rodger et al. (2010).
Mutations in enzymes involved in one-carbon metabolism lead to elevation of blood levels of homocysteine as a thrombotic risk factor. The MTHFR 677C/T mutation is the most prominent one giving reduced activity of MTHFR from 35% in heterozygous to 70% in homozygous state. The prevalence of MTHFR C677T and A1298C, MTR A2756G, and MTRR A66G mutations (Table 3) did not show statistical difference between the female and male partner groups. Hyperhomocysteinemia is a rare finding in pregnant woman today because of high vitamin supplementation during pregnancy. There are still conflicting results from meta-analysis elucidating the role of mutations in folate-related genes in adverse pregnancy outcome, and our results support those that did not find association (Zetterberg et al., 2002; Powers et al., 2003; Kujovich, 2004; Rodger et al., 2010).
We recognize the limitations of our study with respect to defining the association of thrombophilic mutations with spontaneous abortions and these are population-specific genotype effects, unequal representation of different ethnic groups, and lack of fertile couples as literally concordant controls. For valid conclusions about the role of the studied thrombophilic mutations in the Macedonian, Albanian, and Slovenian populations, analysis of ethnically matched couples with normal pregnancies as controls are warranted.
We conclude that this multiplex single-base extension reaction assay for simultaneous analysis of 10 different mutations in thrombophilia- and folate-related genes is an easy-to-perform and cost-effective genotyping technique. The mutations included in this assay have a substantial role in the pathogenesis of spontaneous abortion, which needs to be fully elucidated.
Acknowledgment
This study was supported by the grant 14-2414/1 from Ministry of Education and Science of the Republic of Macedonia (to D. Plaseska-Karanfilska).
Disclosure Statement
No competing financial interests exist.
References
- Bertina RM. Koeleman BP. Koster T, et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- Brenner B. Inherited thrombophilia and pregnancy loss. Thromb Haemost. 1999;82:634–640. [PubMed] [Google Scholar]
- Brenner B. Sarig G. Weiner Z, et al. Thrombophilic polymorphisms are common in women with fetal loss without apparent cause. Thromb Haemost. 1999;82:6–9. [PubMed] [Google Scholar]
- Coulam CB. Jeyendran RS. Fishel LA, et al. Multiple thrombophilic gene mutations rather than specific gene mutations are risk factors for recurrent miscarriage. Am J Reprod Immunol. 2006;55:360–368. doi: 10.1111/j.1600-0897.2006.00376.x. [DOI] [PubMed] [Google Scholar]
- Finan RR. Tamim H. Ameen G, et al. Prevalence of factor V G1691A (factor V-Leiden) and prothrombin G20210A gene mutations in a recurrent miscarriage population. Am J Hematol. 2002;71:300–305. doi: 10.1002/ajh.10223. [DOI] [PubMed] [Google Scholar]
- Ford HB. Schust DJ. Recurrent pregnancy loss: etiology, diagnosis, and therapy. Rev Obstet Gynecol. 2009;2:76–83. [PMC free article] [PubMed] [Google Scholar]
- Frosst P. Blom HJ. Milos R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Hassold T. Chen N. Funkhouser J, et al. A cytogenetic study of 1000 spontaneous abortions. Ann Hum Genet. 1980;44:151–178. doi: 10.1111/j.1469-1809.1980.tb00955.x. [DOI] [PubMed] [Google Scholar]
- Hatzis T. Cardamakis E. Drivalas E, et al. Increased resistance to activated protein C and factor V Leiden in recurrent abortions. Review of other hypercoagulability factors. Eur J Contracept Reprod Health Care. 1999;4:135–144. [PubMed] [Google Scholar]
- Hefler L. Jirecek S. Heim K, et al. Genetic polymorphisms associated with thrombophilia and vascular disease in women with unexplained late intrauterine fetal death: a multicenter study. J Soc Gynecol Investig. 2004;11:42–44. doi: 10.1016/j.jsgi.2003.07.008. [DOI] [PubMed] [Google Scholar]
- Hogge WA. Byrnes AL. Lanasa MC, et al. The clinical use of karyotyping spontaneous abortions. Am J Obstet Gynecol. 2003;189:397–400. doi: 10.1067/s0002-9378(03)00700-2. discussion 400–392. [DOI] [PubMed] [Google Scholar]
- Jauniaux E. Farquharson RG. Christiansen OB, et al. Evidence-based guidelines for the investigation and medical treatment of recurrent miscarriage. Hum Reprod. 2006;21:2216–2222. doi: 10.1093/humrep/del150. [DOI] [PubMed] [Google Scholar]
- Jivraj S. Rai R. Underwood J, et al. Genetic thrombophilic mutations among couples with recurrent miscarriage. Hum Reprod. 2006;21:1161–1165. doi: 10.1093/humrep/dei466. [DOI] [PubMed] [Google Scholar]
- Jurinke C. Oeth P. van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol. 2004;26:147–164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- Jurinke C. van den Boom D. Cantor CR, et al. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- Koseki-Kuno S. Yamakawa M. Dickneite G, et al. Factor XIII A subunit-deficient mice developed severe uterine bleeding events and subsequent spontaneous miscarriages. Blood. 2003;102:4410–4412. doi: 10.1182/blood-2003-05-1467. [DOI] [PubMed] [Google Scholar]
- Kujovich JL. Thrombophilia and pregnancy complications. Am J Obstet Gynecol. 2004;191:412–424. doi: 10.1016/j.ajog.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Pickering W. Marriott K. Regan L. G20210A prothrombin gene mutation: prevalence in a recurrent miscarriage population. Clin Appl Thromb Hemost. 2001;7:25–28. doi: 10.1177/107602960100700106. [DOI] [PubMed] [Google Scholar]
- Powers RW. Dunbar MS. Gallaher MJ, et al. The 677 C-T methylenetetrahydrofolate reductase mutation does not predict increased maternal homocysteine during pregnancy. Obstet Gynecol. 2003;101:762–766. doi: 10.1016/s0029-7844(02)03120-4. [DOI] [PubMed] [Google Scholar]
- Rai R. Shlebak A. Cohen H, et al. Factor V leiden and acquired activated protein C resistance among 1000 women with recurrent miscarriage. Hum Reprod. 2001;16:961–965. doi: 10.1093/humrep/16.5.961. [DOI] [PubMed] [Google Scholar]
- Rodger MA. Betancourt MT. Clark P, et al. The association of factor V leiden and prothrombin gene mutation and placenta-mediated pregnancy complications: a systematic review and meta-analysis of prospective cohort studies. PLoS Med. 2010;7:e1000292. doi: 10.1371/journal.pmed.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson MD. Awartani KA. Robinson WP. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: a case-control study. Hum Reprod. 2002;17:446–451. doi: 10.1093/humrep/17.2.446. [DOI] [PubMed] [Google Scholar]
- Strom CM. Ginsberg N. Applebaum M, et al. Analyses of 95 first-trimester spontaneous abortions by chorionic villus sampling and karyotype. J Assist Reprod Genet. 1992;9:458–461. doi: 10.1007/BF01204052. [DOI] [PubMed] [Google Scholar]
- Tranquilli AL. Giannubilo SR. Dell'Uomo B, et al. Adverse pregnancy outcomes are associated with multiple maternal thrombophilic factors. Eur J Obstet Gynecol Reprod Biol. 2004;117:144–147. doi: 10.1016/j.ejogrb.2004.01.044. [DOI] [PubMed] [Google Scholar]
- Ulvik A. Ueland PM. Single nucleotide polymorphism (SNP) genotyping in unprocessed whole blood and serum by real-time PCR: application to SNPs affecting homocysteine and folate metabolism. Clin Chem. 2001;47:2050–2053. [PubMed] [Google Scholar]
- Yenicesu GI. Cetin M. Ozdemir O, et al. A prospective case-control study analyzes 12 thrombophilic gene mutations in Turkish couples with recurrent pregnancy loss. Am J Reprod Immunol. 2010;63:126–136. doi: 10.1111/j.1600-0897.2009.00770.x. [DOI] [PubMed] [Google Scholar]
- Yilmaz S. Bayan K. Tuzun Y, et al. A comprehensive analysis of 12 thrombophilic mutations and related parameters in patients with inflammatory bowel disease: data from Turkey. J Thromb Thrombolysis. 2006;22:205–212. doi: 10.1007/s11239-006-9032-5. [DOI] [PubMed] [Google Scholar]
- Zetterberg H. Coppola A. D'Angelo A, et al. No association between the MTHFR A1298C and transcobalamin C776G genetic polymorphisms and hyperhomocysteinemia in thrombotic disease. Thromb Res. 2002;108:127–131. doi: 10.1016/s0049-3848(03)00004-5. [DOI] [PubMed] [Google Scholar]