Abstract
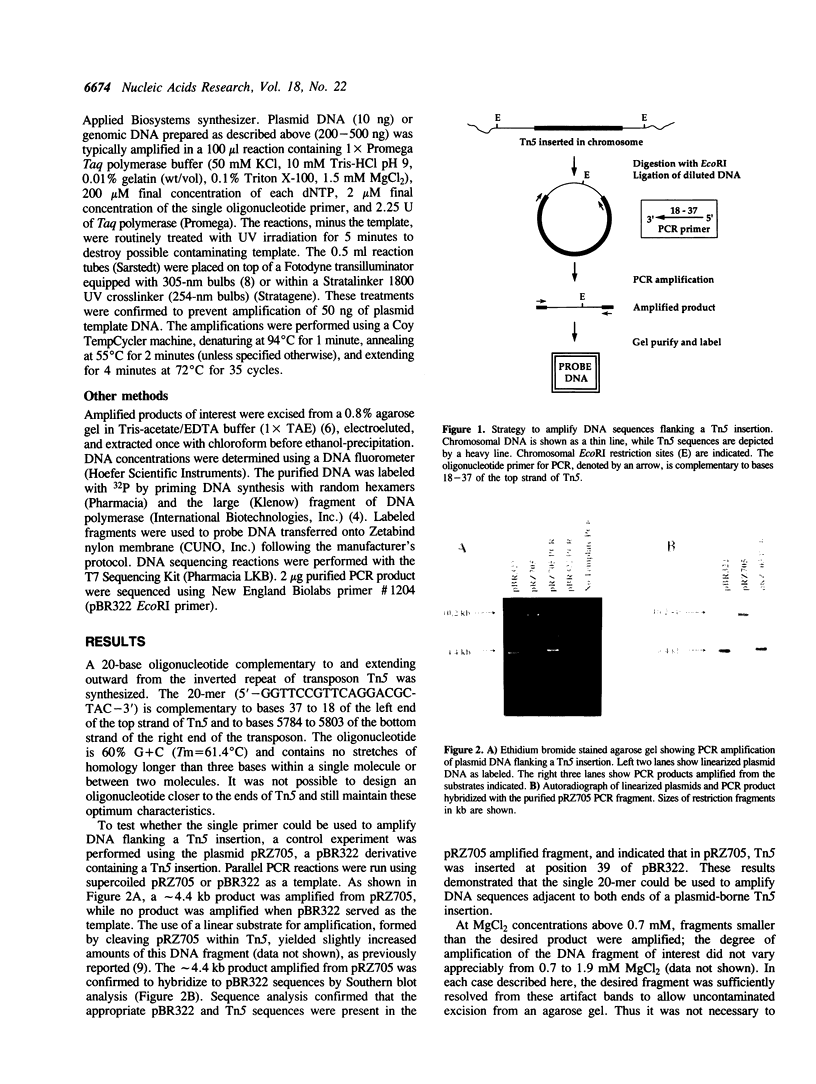
We have developed a strategy to rapidly construct DNA hybridization probes for the isolation of genes disrupted by transposon Tn5 insertions. A single oligonucleotide complementary to and extending outward from the ends of the inverted repeat of Tn5 was used to prime DNA synthesis in the polymerase chain reaction. The amplified product consisted of DNA sequences adjacent to both ends of the transposon insertion. The general feasibility of the approach was tested by amplifying pBR322 sequences from a derivative of pBR322 containing a Tn5 insertion. To amplify genomic DNA sequences flanking a Tn5 insertion in the chromosome of a Pseudomonas syringae strain, circular substrates were generated by ligating EcoRI-digested genomic DNA. Tn5 was contained intact within one such circular molecule, as the transposon does not contain sites for cleavage by EcoRI. The amplified product (approximately 2.5 kb) was used as a DNA hybridization probe to isolate the homologous fragment from a cosmid library of wild-type Pseudomonas syringae genomic DNA. This approach may be applied to the efficient isolation of sequences flanking any Tn5 insertion.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Neumann R., Keyte J. Amplification of human minisatellites by the polymerase chain reaction: towards DNA fingerprinting of single cells. Nucleic Acids Res. 1988 Dec 9;16(23):10953–10971. doi: 10.1093/nar/16.23.10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988 Nov;120(3):621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Sommer S. S. Shedding light on PCR contamination. Nature. 1990 Jan 4;343(6253):27–27. doi: 10.1038/343027a0. [DOI] [PubMed] [Google Scholar]
- Silver J., Keerikatte V. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J Virol. 1989 May;63(5):1924–1928. doi: 10.1128/jvi.63.5.1924-1928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T., Peterson M. G., Kemp D. J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988 Aug 25;16(16):8186–8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]