Abstract
Non-structural proteins NS3 and NS5 of Japanese encephalitis virus (JEV) were expressed in Escherichia coli and purified by dialysis. Two monoclonal antibodies (MAbs) named 1H7 and 2D4 against NS3 protein and three MAbs named 3C4, 3H7, and 3F10 against NS5 protein were generated by fusing mouse myeloma cell line SP2/0 with spleen lymphocytes from NS3 or NS5 protein immunized mice. Then activity of MAbs was characterized by enzyme-linked immunosorbent assay (ELISA), Western blot analysis, and indirect immunofluorescent assays (IFA). Our results demonstrated that all the MAbs showed high specificity and sensitivity in IFA at 1:100 dilution and in Western blot analysis at 1:500 dilution, which indicated that these MAbs against NS3 and NS5 proteins of JEV may be used as valuable tools for analysis of the protein functions and pathogenesis of JEV.
Introduction
Japanese encephalitis (JE) is a viral disease caused by Japanese encephalitis virus (JEV). Since it emerged in Japan in the 1870s, Japanese encephalitis has spread across Asia and has become the most important cause of epidemic encephalitis worldwide.(1) Around 35,000–50,000 JE cases are reported each year, and approximately 25% of encephalitis patients die while about 50% of the survivors develop permanent neurologic and/or psychiatric sequelae, including memory loss, impaired cognition, behavioral disturbances, convulsions, motor weakness or paralysis, and abnormalities of tone and coordination.(2,3) Meanwhile, JE is also an important pig disease, mainly causing porcine reproductive failure. Outbreak of JE can result in great economic losses and restrict the development of animal husbandry.
JEV is a member of the genus Flavivirus, family Flaviviridae. It contains a single-stranded positive-sense RNA with approximately 11 kb in length and a single open reading frame (ORF) that encodes three structural proteins (C, PrM, and E) and seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5).(4,5) Among them, NS3 and NS5 are membrane-associated proteins and form the complex at the perinuclear site in the infected cells.(6) NS3 possesses enzymatic activities of serine protease, helicase, and nucleoside 5′-triphosphatase,(7) and NS5, as the largest and most conserved flavivirus protein, is homologous to methyltransferase and RNA-dependent RNA polymerase. Therefore, both NS3 and NS5 play important roles in viral replication and pathogenesis.(8)
Nowadays function of all the proteins of JEV is under ongoing study; however, the mechanism of JEV pathogenesis remains poorly understood.(9) As the geographical area affected by JEV is expanding,(10–14) effective treatment for JEV is urgently needed. In this study, we generated MAbs against NS3 and NS5 of JEV, which could be applied as useful tools for studying protein function of JEV, and pathogenic mechanism and treatment of JE.
Materials and Methods
Plasmid construction and protein expression
The NS3 and NS5 gene fragments were amplified from JEV P3 strain (GenBank: U47032)-infected BHK-21 cells by a one-step RT-PCR. NS3 forward primer: 5′-CGGAATTCATGGGGGGCGTGTTTTGGGACACGC-3′; NS3 reverse primer: 5′-CCCTCGAGCTATCTCTTCCCTGCTGCAAAGTC-3′; NS5 forward primer: 5′-CGGGATCCATGGGAAGGCCTGGGGGCAGGACGC-3′; NS5 reverse primer: 5′-CCCTCGAGCTAGATGACCCTGTCTTCCTGGATC-3′). Subsequently, the target fragments were cloned into the pET-28a vector. The recombinant plasmids, named pET-NS3 and pET-NS5, and the control plasmid (pET-28a) were then transformed into competent E. coli BL21 cells and induced with isopropyl-β-thio-galactopyranoside (IPTG). After centrifugation, the bacterial pellet was resuspended and sonicated until a clear lysate was obtained. The target proteins were then purified as previously described(15) and divided into small aliquots, with the concentration of 2 mg/mL and stored at −80°C.
Monoclonal antibody production
The MAbs against the NS3 and NS5 proteins were produced as previously described.(15) Briefly, 4-week-old female SPF BALB/c mice were immunized subcutaneously with 100 μg of the purified NS3 or NS5 protein at 2-week intervals. Four weeks after the last booster and 3 days before cell fusion, the mice were boosted with 40 μg of NS3 or NS5 protein. Three days later, mice splenocytes were harvested and fused with SP2/0 using 50% polyethylene glycol. Hybridoma culture supernatants were screened using ELISA. The positive hybridoma cells were cloned by a limiting dilution, and the stable hybridoma clones were injected into liquid paraffin-pretreated abdominal cavities of BALB/c mice. Subsequently, the MAbs were harvested and purified from the seroperitoneum with an antibody purification kit, according to the manufacturer's specifications (NAbTM Protein A/G Spin Kit, Thermo Scientific, Fremont, CA). Their activity was characterized by Western blot and indirect immunofluorescence assay (IFA).
Indirect enzyme-linked immunosorbent assay
Indirect ELISA was conducted in the following manner. ELISA plates were coated overnight at 4°C with 100 μL purified NS3 or NS5 protein diluted in bicarbonate coating buffer (pH 9.6) and then blocked with 5% bovine serum albumin (BSA) in PBS (PBSA) for 1 h at 37°C. The wells were drained and incubated with 100 μL/well 2-fold MAb dilutions in PBSA (from 1:200 to 1:12,800) for 30 min at 37°C. After three washes with PBS containing 0.05% Tween-20 (PBST), 100 μL horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG was added, and wells were incubated for 30 min at 37°C. After washing, 50 μL/well substrate solution A (0.1 M citrate/phosphate buffer [pH 5.0]) and 50 μL/well substrate solution B (0.04% OPD; 0.14% H2O2) were applied for 10 min at room temperature. Reactions were terminated by the addition of 50 μL/well 2 M H2SO4, and optical densities (OD) were measured at 630 nm using a microplate reader. Absorbance values two times higher than the background level reactivity were considered to be positive.
Immunofluorescence assay
For IFA, Hela cells were seeded into 24-well tissue culture plate (Costar Corning, Corning, NY) and inoculated with JEV genotype 3 strain (p3 strain) at 1 multiplicity of infection (MOI) when the cells reached approximately 70–80% confluence. At 72 h post-infection, the cells were fixed with absolute methanol and processed for indirect IFA using MAbs, followed by fluorescein isocyanate-conjugated goat anti-mouse IgG. Fluorescent images were examined under a fluorescent microscope.
Western blot analysis
Infected JEV (genotype 1 strain and genotype 3 strain, respectively) cells were collected, after separated by SDS-PAGE and then transferred after 1 h at 350 mA to nitrocellulose membranes. Then the membranes were blocked overnight with 1% bovine serum albumin (BSA) and 5% skimmed milk powder in TBST buffer (0.01 M Tris–HCl [pH 8.0], 150 mM NaCl, and 0.05% Tween-20) before being reacted with a 1:500 dilution of MAbs for 1 h. After washing with TBST buffer three times, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Southern Biotechnology, Birmingham, AL) secondary antibody (dilution of 1:1000 in blocking solution) for 1 h at 37°C. After washing with TBST buffer three times and TBS (TBST without 0.05% Tween-20) two times, the protein bands were visualized by 3,3′-diaminobenzidine tetrahydrochloride (DAB).
Identification of MAb subtype
The subtype identification kit (Pierce Rapid ELISA Mouse MAb Isotyping Kit, Thermo Scientific) was used to identify the MAb subtypes, according to the manual instructions.
Results
NS3 and NS5 expression in E. coli
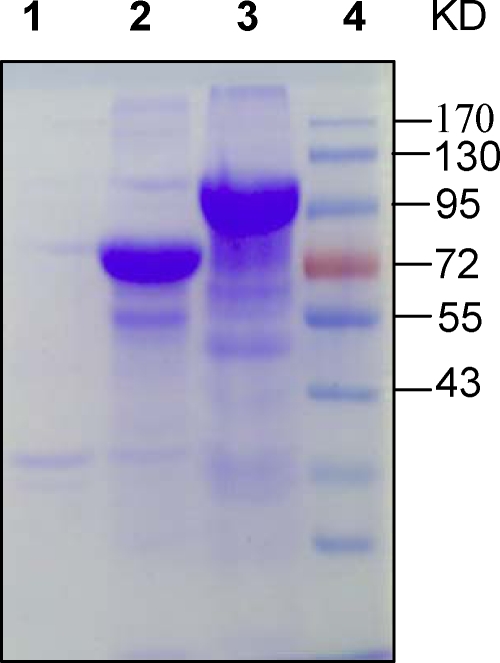
The recombinant vectors were transformed into competent E. coli BL21 cells and induced with 0.8 mmol/L IPTG at 37°C for 4 h to express the recombinant proteins. SDS-PAGE and Western bolt results showed that the molecular weight of recombinant proteins were about 73 kDa and 100 kDa, respectively (Fig. 1), which corresponded with the molecular weight of fusion proteins His-NS3 and His-NS5.
FIG. 1.
SDS-PAGE analysis of recombination proteins NS3 and NS5. The bacteria harboring the recombinant plasmids were induced with IPTG and the bacterial proteins were analyzed by SDS-PAGE. Then protein bands were visualized by staining the gel with Coomassie blue. Lane 1, bacilli precipitation of pET-28a; lane 2, bacilli precipitation of pET-NS3; lane 3, bacilli precipitation of pET-NS5; lane 4, protein marker.
Generation of MAbs against NS3 and NS5 proteins of JEV
After immunization of mice with purified proteins, two MAbs (1H7 and 2D4) against NS3 and three MAbs (3C4, 3H7, and 3F10) against NS5 were eventually isolated through hybridoma fusion and expanded for further characterization.
Indirect ELISA and MAb subtype
For indirect ELISA, 96-well plates were coated with 100 ng/100 μL purified NS3/NS5 proteins. The MAbs (preprocessed by E. coli lysate) and the normal mouse serum were diluted from 1:2000 to 1:1,024,000. The ELISA titers of 1H7, 2D4, 3C4, 3H7, and 3F10 were 1:256,000, 1:512,000, 1:512,000, 1:512,000, and 1:512,000, respectively (Table 1).
Table 1.
Detection and Characterization of Monoclonal Antibodies
| |
MAb against NS3 |
MAb against NS5 |
|||
|---|---|---|---|---|---|
| 1H7 | 2D4 | 3C4 | 3H7 | 3F10 | |
| ELISA titer | 1:256,000 | 1:512,000 | 1:512,000 | 1:512,000 | 1:512,000 |
| MAb subclass | IgG1 | IgG1 | IgG1 | IgG2a | IgG2a |
| Light chain | κ | κ | κ | κ | κ |
Immunofluorescence assay
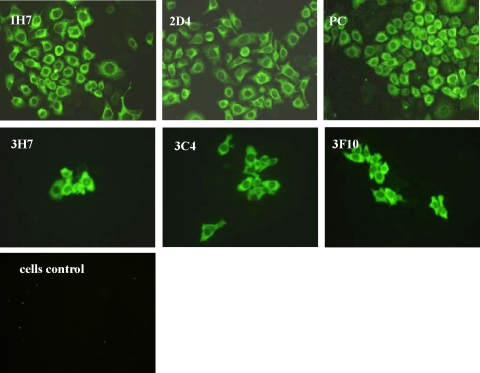
In IFA, four MAbs––1H7 and 2D4 against NS3 and 3H7 and 3F10 against NS5––showed positive reaction to JEV-infected Hela cells. Specific fluorescence signals were observed with MAbs 1H7 and 2D4 against NS3 protein throughout the cells (Fig. 2). Meanwhile, because cell infection percentages were different, fluorescence signals of MAbs 3C4, 3H7, and 3F10 against NS5 protein were more sporadic. Both positive control cells showed strong specific fluorescence signals, while negative control cells did not portray any fluorescence staining (Fig. 2).
FIG. 2.
Immunofluorescence staining (IFA) of JEV genotype 3 strain-infected Hela cells with different MAbs. At 48 h post-JEV infection, cells were washed with PBS three times and fixed with 100% paraformaldehyde for 10 min. After being blocked with PBSA for 30 min at 37°C, cells were washed with PBS, reacted with a 1:100 dilution of MAb for 1 h, and incubated with Alexa Fluor 488 goat anti-mouse IgG (H+L) for another 30 min. Cells were then washed three times with PBS and photographed using a fluorescence microscope. IH7 and 2D4 against NS3; 3H7, 3C4, and 3F10 against NS5; PC, JEV polyclone antibody control; cell control, uninfected Hela cells control (200x).
Western blot analysis
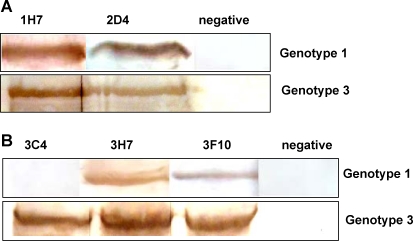
Western blot assay was applied to analyze the specificity of produced MAbs. The results showed that NS3 MAbs 1H7 and 2D4, and NS5 MAbs 3H7 and 3F10 specifically reacted with the proteins NS3 and NS5 of JEV, respectively, but 3C4 had no reaction with NS5 protein of JEV genotype 1 strain, while no reaction was observed with pET-28a protein (Fig. 3).
FIG. 3.
Western blot analysis of monoclonal antibodies against NS3 and NS5 proteins. Infected JEV cells (genotype 1 strain and genotype 3 strain, respectively) were collected after separating by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked overnight with blocking buffer, then reacted with a 1:500 dilution of MAbs against NS3 and NS5 proteins. Finally, protein bands were visualized by DAB followed by incubation with HRP-conjugated goat anti-mouse IgG secondary antibody (A) NS3; (B) NS5.
Subtype identification
A rapid ELISA mouse MAb isotyping kit was applied to identify the subtypes of produced MAbs. The result showed that both MAbs of NS3 (1H7 and 2D4) and one MAb against NS5 (3C4) belonged to the subtype of IgG1, and the other two NS5 MAbs (3H7 and 3F10) were attributed to IgG2a subtype. The light chains of these five MAbs were all kappa (Table 1).
Discussion
In this study, purified NS3 and NS5 proteins were used to immunize mice for the production of monoclonal antibodies. Supernatants of hybridoma cultures were screened by enzyme-linked immunosorbent assay, and two MAbs against NS3 protein and three MAbs against NS5 protein were obtained. All the MAbs showed high specificity in indirect immunofluorescent assays at 1:100 dilution and Western blot at 1:500 dilution.
NS3 is a non-structural protein containing 619 amino acids with the molecular weight of about 68 kDa, and NS5 is the largest non-structural protein (95 kDa) of JEV at approximately 900 amino acids in length. As mentioned earlier, both NS3 and NS5 of JEV play important roles in viral replication and pathogenesis. NS3 participates in viral replication and viral assembly by virtue of its RNA helicase and NTPase activity, while NS5, the largest among all the proteins of JEV, by the methyltransferase activity on its N-terminal region and RNA-dependent RNA polymerase (RdRp) activity on C-terminal motifs. The two proteins bind together and form a viral replication complex (RC), which is an important component during viral replication.(16) Therefore, the interaction of NS3/NS5 and their cellular factors may play important roles in viral RNA replication, protein synthesis, and viral assembly.(9) The characteristic and function of these host factors, once ascertained, may ultimately lead to the production of effective antiviral agents. However, the cellular proteins for JEV replication and the role of protein-protein interactions involved in viral replication have not been well defined. Here we produced MAbs against NS3 and NS5 of JEV, which will be used as a powerful tool for studying the functions of NS3 and NS5 proteins, and screening the cellular proteins involved in JEV replication, which will provide new insight into pathogenic mechanisms of JE.
It should be noticed that our Western blot results showed that the MAb 3C4 only reacted with NS5 of JEV serotype 3, rather than serotype 1, which was also confirmed by IFA (data not shown). This suggests that the epitope recognized by 3C4 is the non-homologous part of JEV serotypes 1 and 3. But which epitope the MAb exactly recognizes still needs to be further verified. Nevertheless, the MAb with such a high specificity will be useful material to make a distinction between JEV serotypes 1 and 3. We also identified the subtypes of the five monoclonal antibodies by subtype identification kit. Both 1H7 and 2D4 against NS3 belonged to the same subtype (IgG1), and two of NS5 MAbs, 3H7 and 310, belonged to the subtype of IgG2a. The light chains were all kappa.
In conclusion, five monoclonal antibodies against JEV NS3 and NS5 were produced and characterized in this study. These MAbs may be used as powerful tools for studying replication mechanism of JEV and may lay the foundation for future research on JEV pathogenic mechanism.
Acknowledgments
This research work was supported by the Natural Science Foundation of China (grant no. 31172325). We are deeply grateful to Dr. Ping Qian for providing the JEV genotype 1 strain.
Author Disclosure Statement
The authors declare that they have no financial conflict of interest.
References
- 1.Solomon T. Ni H. Beasley DWC. Origin and evolution of Japanese encephalitis virus in Southeast Asia. J Virol. 2003;77:3091–3098. doi: 10.1128/JVI.77.5.3091-3098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Hurk AF. Ritchie SA. Mackenzie JS. Ecology and geographical expansion of Japanese encephalitis virus. Annu Rev Entomol. 2009;54:17–35. doi: 10.1146/annurev.ento.54.110807.090510. [DOI] [PubMed] [Google Scholar]
- 3.Li M-H. Fu S-H. Chen W-X. Genotype V Japanese encephalitis virus is emerging. PLoS Negl Trop Dis. 2011;5:e1231. doi: 10.1371/journal.pntd.0001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumiyoshi H. Mori C. Fuke I. Morita K. Kuhara S. Kondou J. Kikuchi Y. Nagamatu H. Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 5.Chambers TJ. Hahn CS. Galler R. Rice CM. Flavivirus genome organization, expression and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 6.Edward Z. Takegami T. Localization and functions of Japanese encephalitis virus nonstructural proteins NS3 and NS5 for viral RNA synthesis in the infected cells. Microbiol Immunol. 1993;37:39–43. doi: 10.1111/j.1348-0421.1993.tb03206.x. [DOI] [PubMed] [Google Scholar]
- 7.Deng X. Shi Z. Li S. Characterization of nonstructural protein 3 of a neurovirulent Japanese encephalitis virus strain isolated from a pig. Virol J. 2011;8:209. doi: 10.1186/1743-422X-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi WB. Hua RH. Yan LP. Effective inhibition of Japanese encephalitis virus replication by small interfering RNAs targeting the NS5 gene. Virus Res. 2008;132:145–151. doi: 10.1016/j.virusres.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Ren J. Ding T. Zhang W. Does Japanese encephalitis virus share the same cellular receptor with other mosquito-borne flaviviruses on the C6/36 mosquito cells? Virol J. 2007;4:83. doi: 10.1186/1743-422X-4-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna JN. Ritchie SA. Phillips DA. Lee JM. Hills SL. van den Hurk AF. Pyke AT. Johansen CA. Mackenzie JS. Japanese encephalitis in north Queensland, Australia, 1998. Med J Aust. 1999;170:533–536. doi: 10.5694/j.1326-5377.1999.tb127878.x. [DOI] [PubMed] [Google Scholar]
- 11.Burdon JT. Stanley PJ. Lloyd G. Jones NC. A case of Japanese encephalitis. J Infect. 1994;28:175–179. doi: 10.1016/s0163-4453(94)95640-5. [DOI] [PubMed] [Google Scholar]
- 12.Saito M. Sunagawa T. Makino Y. Tadano M. Hasegawa H. Kanemura K. Zamami Y. Killenbeck BJ. Fukunaga T. Three Japanese encephalitis cases in Okinawa, Japan, 1991. Southeast Asian. J Trop Med Publ Health. 1999;30:277–279. [PubMed] [Google Scholar]
- 13.Paul WS. Moore PS. Karabatsos N. Flood SP. Yamada S. Jackson T. Tsai TF. Outbreak of Japanese encephalitis on the island of Saipan. J Infect Dis. 1993;167:1053–1058. doi: 10.1093/infdis/167.5.1053. [DOI] [PubMed] [Google Scholar]
- 14.Hanna JN. Ritchie SA. Phillips DA. Shield J. Bailey MC. Mackenzie JS. Poidinger M. McCall BJ. Mills PJ. An outbreak of Japanese encephalitis in the Torres Strait Australia, 1995. Med J Aust. 1996;165:256–260. doi: 10.5694/j.1326-5377.1996.tb124960.x. [DOI] [PubMed] [Google Scholar]
- 15.Li YM. Hou LD. Ye J. Liu XQ. Dan HB. Jin ML. Chen HC. Cao SB. Development of a convenient immunochromatographic strip for the diagnosis of infection with Japanese encephalitis virus in swine. J Virol Methods. 2010;168:51–56. doi: 10.1016/j.jviromet.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Unni SK. Růžek D. Chhatbar C. Mishra R. Johri MK. Singh SK. Japanese encephalitis virus: from genome to infectome. Microbes Infect. 2011;13:312–321. doi: 10.1016/j.micinf.2011.01.002. [DOI] [PubMed] [Google Scholar]