Abstract
Studies addressing the role of somatic copy number variation (CNV) in the genesis of congenital heart defects (CHDs) are scarce, as cardiac tissue is difficult to obtain, especially in non-affected individuals. We explored the occurrence of copy number differences in monozygotic (MZ) twins discordant for the presence of a CHD, as an illustrative model for chromosomal mosaicism in CHDs. Array comparative genomic hybridization was performed on peripheral blood-derived DNA obtained from 6 discordant MZ twin pairs and on sex-matched reference samples. To identify CNV differences between both twin members as well as potential CNVs in both twins contributing to the phenotype, DNA from each twin was hybridized against its co-twin, and against a normal control. Three copy number differences in 1 out of 6 MZ twin pairs were detected, confirming the occurrence of somatic CNV events in MZ twins. Further investigation by copy number and (epi)genome sequencing analyses in MZ twins, discordant for the presence of CHDs, is required to improve our knowledge on how postzygotic genetic, environmental and stochastic factors can affect human heart development.
Key Words: Array comparative genomic hybridization, Congenital heart defects, Copy number variation, Monozygotic twins
Introduction
Somatic mosaicism is defined by the presence of genetically distinct populations of somatic cells in a single organism. Somatic mutations are implicated in an increasing variety of genetic disorders, and may induce a milder phenotype, hereby complicating diagnosis and genetic counseling. The mutation size ranges from base pair level to megabase-sized chromosomal segments [Erickson, 2010]. Studies investigating the role of mosaicism in the genesis of congenital heart defects (CHDs) have evoked conflicting results. CHDs arise from an abnormal heart development, induced either by environmental influences [Jenkins et al., 2007], by an altered gene dosage or function [Bruneau, 2008], by stochastic factors [Kurnit et al., 1987] or by combinations thereof. Human genetic studies have implicated mutations of numerous genes and chromosomal regions in CHD. A high prevalence of somatic point mutations in specific cardiac genes have been reported by a single research group in DNA extracted from formalin-fixed affected cardiovascular tissue of patients with non-syndromic septal defects [Reamon-Buettner and Borlak 2004a, b, 2005, 2006; Reamon-Buettner et al., 2007, 2008, 2009]. These mutations were not present in unaffected tissue of the same patients, suggesting a role of somatic mutations in the pathogenesis of CHD. However, these findings could not be replicated by other research groups using fresh-frozen cardiac tissues rather than formalin-fixed tissues [Majumdar et al., 2006; Draus et al., 2009; Salazar et al., 2011]. Poor DNA quality from formalin-fixed tissue presumably resulted in artifacts [Draus et al., 2009].
Studies addressing the role of somatic copy number variation (CNV) in the genesis of CHDs are scarce [Notini et al., 2008; Erickson, 2010]. Recently, using array comparative genomic hybridization (aCGH) experiments on DNA derived from fully differentiated tissues, the presence of CNV differences between unaffected cardiac tissue and the cerebellar cortex of a single individual was evidenced by Piotrowski et al. [2008]. However, to date, no causal somatic CNVs in CHD were detected. Since germline deletions on 22q11.2 are a common chromosomal cause of syndromic conotruncal heart defects, Rauch et al. [2004] searched for somatic 22q11.2 copy number changes in fresh-frozen cardiac tissues from 14 patients with conotruncal heart defects without a germline deletion on this locus. However, no somatic 22q11.2 deletions were found.
We investigated whether CNV differences in monozygotic (MZ) twins discordant for CHDs may improve our insight into chromosomal mosaicism in CHDs. Discordant MZ twins represent an intriguing observation, as they are derived from a single fertilization event, and are considered to be genetically identical. Any CNV identified in 1 twin but not in the other would have a somatic origin, and would be an example of mosaicism [Bruder et al., 2008]. In this study, we aimed to investigate the role of somatic CNV events in 6 MZ twin pairs discordant for the presence of CHDs.
Patients and Methods
Patients
Six MZ twin pairs were recruited from the pediatric cardiology unit and Center for Human Genetics of the University Hospitals Leuven. The twins were discordant for a CHD. The cardiac phenotype and extracardiac anomalies of the affected twins are depicted in table 1. CHDs were excluded in all non-affected twin members by means of echocardiography. Medical history of the non-affected twin members was unremarkable with respect to congenital defects, dysmorphism and developmental delay, with the exception of the ‘non-affected’ member of twin pair 6, who died neonatally due to severe multicystic renal disease and aplasia of the bladder, which resulted in oligohydramnion and a Potter sequence. Prenatal ultrasound was not suggestive of twin-to-twin transfusion syndrome (TTTS), except for twin pair 1, undergoing laser ablation at 24 weeks of gestation. Twin pair 3 was of Asian origin, twin pair 1 of African origin. All other twin pairs originated from 2 Caucasian parents. Familial history was negative with respect to congenital anomalies and developmental delay. Peripheral blood for DNA extraction was taken within the first week of life, except for twin pair 4, from whom DNA samples were taken at 4 years of age. Conventional karyotyping (G-banding analysis at least at ISCN +550 bands) was normal in all patients.
Table 1.
Phenotypes of the affected twins
| Twin | Gender | Chorionicity | Weeks of | Birth weight and length | CHD type | Associated anomalies | ||
|---|---|---|---|---|---|---|---|---|
| pair | gestation | |||||||
| number | at birth | affected | unaffected | |||||
| 1 | f | MCDAa | 32 | 1,050 g (<p3) | 1,640 g (p25) | 6 mm pm. VSD, | none | |
| 36 cm (<p3) | 36 cm (<p3) | CoAo | ||||||
| 2 | f | MCDA | 28 6/7 | 1,165 g (p50) | 1,160 g (p50) | 3 mm pm. VSD, | none | |
| 36 cm (p25) | 35 cm (p10–25) | ASD II, PDA | ||||||
| 3 | m | MCDA | 30 | 1,320 g (p25) | 1,560 g (p50) | ToF, DCRV, left | facial dysmorphism; cryptorchidism; webbed | |
| 37 cm (p10–25) | 41 cm (p50–75) | SCV, PDA | neck, dry skin, widely spaced nipples, low-set | |||||
| ears, developmental delay | ||||||||
| 4 | f | MCDA | 34 | 720 g (<p3) | 2,000 g (p25) | VSD, CoAo | short stature (<3rd centile), microcephaly (3rd | |
| NA | NA | centile), neonatal hypoglycemia, pointed chin, | ||||||
| narrow palpebral fissures, mild limb | ||||||||
| asymmetry, asymmetric nipples | ||||||||
| 5 | m | MCDA | 32 | 1,580 g (p25) | 1,970 g (p50–75) | musc. VSD, | none | |
| 41 cm (p25) | 44 cm (p50–75) | CoAo | ||||||
| 6 | m | DCDAb | 36 | 2,770 g (p25–50) | 2,040 g (<p10) | TGA, coronary | none | |
| 46 cm (p25) | 46 cm (p25) | anomaly | ||||||
aLaser ablation at 24 weeks of gestation for twin-to-twin transfusion syndrome (TTTS).
b Unaffected twin died neonatally due to severe multicystic renal disease.
ASD II = Type 2 atrial septal defect; CoAo = aortic coarctation; DCDA = dichorionic diamniotic; DCRV = double chambered right ventricle; left SCV = left-sided superior caval vein; MCDA = monochorionic diamniotic; musc. = muscular; NA = not available; PDA = persistent ductus arteriosus; pm. = perimembranous; TGA = transposition of the great arteries; ToF = tetralogy of Fallot; VSD = ventricular septal defect.
The study was approved by the Ethics Committee at the University Hospitals Leuven. Appropriate informed consent was obtained from all participating patients or their legal representatives.
Methods
Array Comparative Genomic Hybridization
Peripheral blood-derived DNA from both twins and sex-matched reference samples was hybridized to a 180-k oligo array platform (OGT CytoSure Syndrome Plus array, OGT Oxford, UK) according to the manufacturer's instructions with minor modifications as described in Srisupundit et al. [2010]. To identify inter-twin CNV differences as well as potential intra-twin CNVs contributing to the phenotype, DNA from each twin was hybridized against its co-twin, and against a genetically well-characterized normal control. All hybridizations were done in duplicate, with the Cy3 and Cy5 dyes being swapped between the twin members of each pair in the second hybridization to eliminate a possible dye-specific bias.
Data Analysis
The CytoSure Interpret software (OGT) was used to visualize the data and perform the analysis. Thresholds for CNV calling were set at ±0.36 from a log2 ratio of 0, and at least 5 consecutive probes within a segment, achieving a theoretical resolution of 80 kb for targeted regions and of 16 kb for targeted genes [Srisupundit et al., 2010]. For a region to be called as discrepant between both twin members, it had to be present in both datasets within the dye-swap experiment.
An internal database of ‘benign’ CNVs at ∼200 kb resolution could be constructed by applying array experiments in healthy control individuals by means of the CytoSure Syndrome Plus V2 105K array (OGT). CNVs were also considered phenotypically indifferent when within regions reported in >1% of healthy controls by at least 2 independent research groups in the Database of Genomic Variants (http://projects.tcag.ca/variation/).
Confirmatory Tests
To confirm the presence of CNV differences, quantitative polymerase chain reaction (qPCR) was performed on DNA from both twin members and 3 unrelated controls (2 males and 1 female) using the LightCycler® 480 Real-Time PCR system (Roche Applied Science, Belgium, https://www.roche-applied-science.com/lightcycler-online/), following the manufacturer's instructions. Quantitative PCR was performed as described by Menten et al. [2006]. Reference primers were selected within the TP53 gene, whereas reference primers within the MID1 gene were selected for CNVs on the X chromosome. For every potential inter-twin CNV difference 2 PCR fragments were analyzed. Every qPCR experiment was performed in duplicate. The average of both analyses per PCR fragment was calculated. Oligonucleotide primer sequences are available upon request. All genome coordinates were determined according to NCBI human genome build 36 (hg18, March 2006).
Results
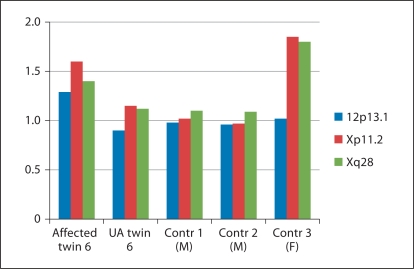
Array comparative genomic hybridization was performed on peripheral blood-derived DNA from 6 MZ twin pairs. Nine potential inter-twin CNVs were detected, representing 7 gains and 2 losses in 3 twin pairs (no. 1, 3 and 6) (online suppl. table 1, for all online suppl. material see www.karger.com/doi/10.1159/000335284). The average size was 36 kb (median 6 kb), and average probe count was 22 (median 9 probes). Six of these copy number differences were excluded by means of qPCR: 4 CNVs were not confirmed (false-positive), and 2 CNVs were present in both twin members (false-negative in 1 twin member). The 3 remaining CNV differences involved copy number gains in the affected member of twin pair 6, and were confirmed independently by qPCR. The average values for 1 PCR fragment of the confirmed inter-twin differences were depicted in figure 1. The average log2 ratio of one of these duplications was 1.25, whereas a ratio of 1.50 should be expected (fig. 1, table 2).
Fig. 1.
Quantitative PCR results of the 3 confirmed CNV differences between the members of twin pair 6. Quantitative PCR results are shown for the affected and unaffected (UA) member of twin pair 6, as well as for 3 unrelated normal controls (2 males (M), 1 female (F)). The boxes represent the average log2 ratios for 1 PCR fragment within the corresponding region (indicated as a red box in online suppl. fig. 1). The affected twin member harbors a discordant duplication on 12p13.31, Xp11.23 and Xq28. The log2 ratio was at 1.25 for the region on 12p13.31, which may indicate mosaicism of this CNV in the blood of the CHD-positive twin.
Table 2.
Characteristics of inter-twin CNV differences confirmed by qPCR
| Twin | CNS | Chr | Start, bp | End, bp | Size, kb | Probe count | Genes |
|---|---|---|---|---|---|---|---|
| affected twin 6 | gain | 12p13.31 | 6,922,387 | 6,926,060 | 3.7 | 14 | C12orf57 |
| affected twin 6 | gain | Xp11.23 | 47,390,509 | 47,397,970 | 7.5 | 8 | ELK1, UXT |
| affected twin 6 | gain | Xq28 | 153,245,504 | 153,282,881 | 37.4 | 101 | EMD, FLNA, RPL10 |
Copy number variants can cause CHDs with a reduced penetrance (e.g. 22q11 duplication). As a consequence, CNVs present in both twins, might affect heart development of only 1 member. Therefore, CNVs present in both twins were evaluated using an algorithm for CNV interpretation in patients with CHD, which was reported recently by our group [Breckpot et al., 2011]. In summary, copy number variants which are not recurrently found in >1% of normal individuals can be considered causal when they comprise known dosage-sensitive heart genes or known chromosomal syndromes. Rare CNVs not affecting genes or regions known to be involved in CHDs should be regarded as unclassified. Following this algorithm, none of the CNVs present in both members of a twin pair were considered to be causal for CHD.
Discussion
Monochorionic twins are at increased risk for CHDs, occurring in 5–7% of monochorionic diamniotic twins [Manning and Archer, 2006; Bahtiyar et al., 2007]. A proportion of these may result from altered hemodynamics during development due to placental vascular anastomoses [Manning and Archer, 2006; Bahtiyar et al., 2007]. Virtually all monochorionic twin placentae have vascular anastomoses, which can be located superficially on the placenta (i.e. arterioarterial and venovenous), or form capillary connections in the placental parenchyma (i.e. arteriovenous or oppositely directed arteriovenous connections). Depending on the size, type and direction, these anastomoses may provoke large uncompensated fetofetal transfusions, which results in a discordant hemoglobin concentration between the twins or TTTS [Manning and Archer, 2006; Bahtiyar et al., 2007]. Recently, hemodynamic analyses unveiled the presence of additional ‘deep-hidden’ anastomoses, which cannot be coagulated by laser therapy as done for TTTS. Although the incidence of deep-hidden anastomoses is significant (in 50–70% of monochorionic twin placentae), the hematological (hemoglobin differences) and clinical consequences are considered to be limited [Wee et al., 2005; Lewi et al., 2006; van den Wijngaard et al., 2007].
Approximately 15–35% of monochorionic diamniotic twin pregnancies are complicated by TTTS, 6.9% of which are associated with CHD. Cardiovascular changes, such as ventricular hypertrophy and right ventricle outflow tract obstruction, occur predominantly in recipient twins of TTTS pregnancies [Karatza et al., 2002; Bahtiyar et al., 2007; Divanovic et al., 2011]. In our study, prenatal ultrasound was not suggestive of TTTS, except for twin 1, of which the donor was diagnosed with a ventricular septal defect.
TTTS is typically diagnosed between 15 and 26 weeks of gestation, based on inter-twin differences in amniotic volume on prenatal ultrasound and flow abnormalities on Doppler evaluation. First-trimester diagnosis of TTTS is challenging, as no reproducible correlations between first-trimester measurements (e.g. inter-twin differences in crown-rump length or in nuchal translucency thickness) and the presence of TTTS could be established [El Kateb et al., 2007]. Studies addressing the role of altered hemodynamics during early heart development, before human embryonic day 32, are scarce. Four-dimensional Doppler optical coherence tomography, recently introduced for hemodynamic measurements during avian heart tube development [Jenkins et al., 2010], might enable the study of hemodynamic changes at early stages of heart development in MZ twin models.
The challenge remains how to explain the discordance in cardiac development observed in monochorionic twins without obvious fetofetal transfusions and in dichorionic MZ twins where altered hemodynamics does not play a role. One possibility is that the MZ twinning process in itself increases the incidence of CHD, with postzygotic unequal division of the inner cell mass or disturbance of laterality being responsible for discordant cardiovascular anatomy. Other possible explanations are postzygotic genetic, epigenetic mutations and stochastic factors [Gringras and Chen, 2001; Singh and O'Reilly, 2009].
Previously, multiple CNV differences were detected by means of a 32-k BAC array on peripheral blood-derived DNA from 9 MZ twin pairs discordant for Parkinson's disease [Bruder et al., 2008]. There were 31 loci that deviated in 3 discordant MZ pairs and 4 loci that deviated in 4 such pairs. Similarly, Sasaki et al. [2011] identified a discordant loss of the SHC2 gene in the affected member of 1 MZ twin, discordant for multiple system atrophy. Subsequently, this SHC2 deletion was found in 10 out of 31 unrelated patients with multiple system atrophy, but not in normal controls, and, therefore, was deemed causal for the multiple system atrophy phenotype in the affected twin member. Conversely, 2 recent aCGH studies on peripheral blood-derived DNA from 1 MZ twin pair discordant for breast cancer and from 3 MZ twin pairs discordant for schizophrenia failed to reproduce the presence of inter-twin CNV differences [Lasa et al., 2010; Ono et al., 2010].
We aimed to investigate the role of postzygotic CNV events in MZ twin pairs discordant for CHDs. One twin pair was found to carry 3 copy number differences, involving 3 duplications in the CHD-positive member. The copy number gain on chromosome 12 reached a log2 ratio of about 1.25 by qPCR, whereas an expected ratio of 1.50 was obtained for a positive (female) control and for other duplications occurring in both members of twin pair 6, suggesting that this duplication was present as a mosaic in the blood of the CHD-positive twin, compatible with the hypothesis of a postzygotic somatic event. Unfortunately, no other tissue of this twin was available to reproduce the results.
The Xq28 duplication partially affects the FLNA gene. Loss-of-function mutations in FLNA have been described in families with X-linked cardiac valvular dysplasia [Kyndt et al., 2007] and in boys with CHDs associated with nodular periventricular heterotopias [Jefferies et al., 2010] or otopalatodigital syndrome [Kondoh et al., 2007]. A similar duplication partially comprising FLNA and EMD was detected once, in a normal control from the HAPMAP population [Perry et al., 2008]. The ELK1 gene on Xp11.23 encodes a ternary complex factor that acts as a repressor of smooth muscle proliferation [Wang et al., 2004], but its potential role in heart development remains unexplored. The other genes within the duplicated segments are not known to be involved in the genesis of CHDs either. Further studies are required to determine whether the detected CNV differences are contributive to the patient's cardiac phenotype.
Tissue sampling is an important element when studying somatic events in twins, as the genotype of 1 tissue analyzed (usually blood) might not always be relevant to the genotype of the tissue responsible for the disease [Piotrowski et al., 2008]. Moreover, in many cases, the hematopoietic lineages of MZ twins are chimeric due to twin-to-twin exchange of hematopoietic stem cells during embryogenesis [Erlich, 2011]. Therefore, CNV profiles on DNA derived from a buccal smear, fibroblasts or cardiac tissue may differ from those obtained on peripheral blood-derived DNA, as was shown by Rohrer et al. in MZ twins discordant for Turner syndrome [Rohrer et al., 2004]. Cardiac tissue is however difficult to obtain, especially in non-affected individuals, and saliva samples were not available. Furthermore, the age at which (blood) samples are taken might theoretically influence somatic CNV profiles as well, since cellular subpopulations carrying a low-grade mosaic CNV can show impaired cell growth, and therefore gradually disappear in blood over time. Conversely, at an older age, inter-twin CNV differences could be induced by chromosomal rearrangements in hematologic malignancies, as shown by Bruder et al. [2008]. A longitudinal study in larger cohorts of MZ twins of different age is required to assess the impact of age on somatic CNV profiles. In our study, DNA samples were taken within the first week of life in 5 out of 6 MZ twin pairs.
In the absence of causal CNV differences, the question remains whether other genetic or epigenetic factors underlie the discordance of CHD in MZ twins. With the advent of (epi)genome-wide sequencing technologies, the answer to this is within our reach. Although exome sequencing data on MZ twins are still scarce, epigenetic differences in MZ twins have been identified in various diseases and tissues, either at selected genes [Petronis et al., 2003; Oates et al., 2006] or epigenome-wide [Fraga et al., 2005; Kaminsky et al., 2009]. In a cross-sectional study, Fraga et al. [2005] showed that MZ twins were epigenomically similar during their early years of life, indicative of high epigenetic heritability. However, epigenomes significantly diverged with age. The role of the environment was indicated by the fact that higher epigenetic variability was found in twins who differed more in lifestyle [Fraga et al., 2005; Wong et al., 2010]. Age-related epigenomic changes will definitely improve our understanding of complex traits, such as autoimmune disorders [Baranzini et al., 2010] or arterial hypertension, rather than that of inborn disorders, such as CHDs. Nevertheless, the broad application of genome and epigenome sequencing on MZ twins discordant for the presence of CHD will improve our knowledge on how genetic, environmental and stochastic factors can affect heart development.
In conclusion, we detected 3 CNV differences in 1 out of 6 MZ twin pairs discordant for the presence of CHD, confirming the occurrence of somatic CNV events in MZ twins. However, further studies are required to causally relate these CNVs to the cardiac phenotype. Copy number, genome and epigenome analyses in a larger MZ twin cohort are required to explore the role of somatic CNVs and mutations in the genesis of CHD.
Supplementary Material
Screenshots of the Cytosure Software output for the inter-twin CNV differences, confirmed by means of quantitative PCR. CNV profiles of the CHD-positive twin member and CHD-negative member of pair 6 are depicted as red or blue lines, respectively. The red and black boxes represent the two sets of PCR fragments in the discordant regions. The qPCR results for the red PCR fragments are depicted in figure 1.
Acknowledgements
J.B. is an aspirant investigator and K.D. a senior clinical investigator of the FWO (Fonds voor Wetenschappelijk Onderzoek) – Flanders. This work was made possible in part by grants from the IWT (SBO-60848) and GOA/2012/015, and the SymBioSys Center of Excellence (KUL PFV/10/016 SymBioSys) to J.R.V. and K.D. The authors would like to thank the patients and their parents for their cooperation. Many thanks to Nooshin Ardeshirdavani for performing the OGT array analysis.
References
- Bahtiyar MO, Dulay AT, Weeks BP, Friedman AH, Copel JA. Prevalence of congenital heart defects in monochorionic/diamniotic twin gestations: A systematic literature review. J Ultrasound Med. 2007;26:1491–1498. doi: 10.7863/jum.2007.26.11.1491. [DOI] [PubMed] [Google Scholar]
- Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckpot J, Thienpont B, Arens Y, Tranchevent LC, Vermeesch JR, et al. Challenges of interpreting copy number variation in syndromic and non-syndromic congenital heart defects. Cytogenet Genome Res. 2011;135:251–259. doi: 10.1159/000331272. [DOI] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Divanovic A, Cnota J, Ittenbach R, Tan X, Border W, et al. Characterization of diastolic dysfunction in twin-twin transfusion syndrome: Association between Doppler findings and ventricular hypertrophy. J Am Soc Echocardiogr. 2011;24:834–840. doi: 10.1016/j.echo.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Draus JM, Jr, Hauck MA, Goetsch M, Austin EH, 3rd, Tomita-Mitchell A, Mitchell ME. Investigation of somatic NKX2–5 mutations in congenital heart disease. J Med Genet. 2009;46:115–122. doi: 10.1136/jmg.2008.060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kateb A, Nasr B, Nassar M, Bernard JP, Ville Y. First-trimester ultrasound examination and the outcome of monochorionic twin pregnancies. Prenat Diagn. 2007;27:922–925. doi: 10.1002/pd.1802. [DOI] [PubMed] [Google Scholar]
- Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res. 2010;705:96–106. doi: 10.1016/j.mrrev.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Erlich Y. Blood ties: Chimerism can mask twin discordance in high-throughput sequencing. Twin Res Hum Genet. 2011;14:137–143. doi: 10.1375/twin.14.2.137. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringras P, Chen W. Mechanisms for differences in monozygous twins. Early Hum Dev. 2001;64:105–117. doi: 10.1016/s0378-3782(01)00171-2. [DOI] [PubMed] [Google Scholar]
- Jefferies JL, Taylor MD, Rossano J, Belmont JW, Craigen WJ. Novel cardiac findings in periventricular nodular heterotopia. Am J Med Genet A. 2010;152A:165–168. doi: 10.1002/ajmg.a.33110. [DOI] [PubMed] [Google Scholar]
- Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- Jenkins MW, Peterson L, Gu S, Gargesha M, Wilson DL, et al. Measuring hemodynamics in the developing heart tube with four-dimensional gated Doppler optical coherence tomography. J Biomed Opt. 2010;15:066022. doi: 10.1117/1.3509382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Karatza AA, Wolfenden JL, Taylor MJ, Wee L, Fisk NM, Gardiner HM. Influence of twin-twin transfusion syndrome on fetal cardiovascular structure and function: prospective case-control study of 136 monochorionic twin pregnancies. Heart. 2002;88:271–277. doi: 10.1136/heart.88.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh T, Okamoto N, Norimatsu N, Uetani M, Nishimura G, Moriuchi H. A Japanese case of oto-palato-digital syndrome type II: an apparent lack of phenotype-genotype correlation. J Hum Genet. 2007;52:370–373. doi: 10.1007/s10038-007-0108-7. [DOI] [PubMed] [Google Scholar]
- Kurnit DM, Layton WM, Matthysse S. Genetics, chance, and morphogenesis. Am J Hum Genet. 1987;41:979–995. [PMC free article] [PubMed] [Google Scholar]
- Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- Lasa A, Ramon y Cajal T, Llort G, Suela J, Cigudosa JC, et al. Copy number variations are not modifiers of phenotypic expression in a pair of identical twins carrying a BRCA1 mutation. Breast Cancer Res Treat. 2010;123:901–905. doi: 10.1007/s10549-010-0877-0. [DOI] [PubMed] [Google Scholar]
- Lewi L, Jani J, Cannie M, Robyr R, Ville Y, et al. Intertwin anastomoses in monochorionic placentas after fetoscopic laser coagulation for twin-to-twin transfusion syndrome: is there more than meets the eye? Am J Obstet Gynecol. 2006;194:790–795. doi: 10.1016/j.ajog.2005.08.062. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Yagubyan M, Sarkar G, Bolander ME, Sundt TM., 3rd Bicuspid aortic valve and ascending aortic aneurysm are not associated with germline or somatic homeobox NKX2–5 gene polymorphism in 19 patients. J Thorac Cardiovasc Surg. 2006;131:1301–1305. doi: 10.1016/j.jtcvs.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Manning N, Archer N. A study to determine the incidence of structural congenital heart disease in monochorionic twins. Prenat Diagn. 2006;26:1062–1064. doi: 10.1002/pd.1556. [DOI] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notini AJ, Craig JM, White SJ. Copy number variation and mosaicism. Cytogenet Genome Res. 2008;123:270–277. doi: 10.1159/000184717. [DOI] [PubMed] [Google Scholar]
- Oates NA, van Vliet J, Duffy DL, Kroes HY, Martin NG, et al. Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet. 2006;79:155–162. doi: 10.1086/505031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Imamura A, Tasaki S, Kurotaki N, Ozawa H, et al. Failure to confirm CNVs as of aetiological significance in twin pairs discordant for schizophrenia. Twin Res Hum Genet. 2010;13:455–460. doi: 10.1375/twin.13.5.455. [DOI] [PubMed] [Google Scholar]
- Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, et al. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29:169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- Piotrowski A, Bruder CE, Andersson R, Diaz de Stahl T, Menzel U, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- Rauch A, Hofbeck M, Cesnjevar R, Koch A, Rauch R, et al. Search for somatic 22q11.2 deletions in patients with conotruncal heart defects. Am J Med Genet A. 2004;124A:165–169. doi: 10.1002/ajmg.a.20323. [DOI] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Borlak J. Somatic NKX2–5 mutations as a novel mechanism of disease in complex congenital heart disease. J Med Genet. 2004a;41:684–690. doi: 10.1136/jmg.2003.017483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Borlak J. TBX5 mutations in non-Holt-Oram syndrome (HOS) malformed hearts. Hum Mutat. 2004b;24:104. doi: 10.1002/humu.9255. [DOI] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Borlak J. GATA4 zinc finger mutations as a molecular rationale for septation defects of the human heart. J Med Genet. 2005;42:e32. doi: 10.1136/jmg.2004.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Borlak J. HEY2 mutations in malformed hearts. Hum Mutat. 2006;27:118. doi: 10.1002/humu.9390. [DOI] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Cho SH, Borlak J. Mutations in the 3′-untranslated region of GATA4 as molecular hotspots for congenital heart disease (CHD) BMC Med Genet. 2007;8:38. doi: 10.1186/1471-2350-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Ciribilli Y, Inga A, Borlak J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet. 2008;17:1397–1405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Ciribilli Y, Traverso I, Kuhls B, Inga A, Borlak J. A functional genetic study identifies HAND1 mutations in septation defects of the human heart. Hum Mol Genet. 2009;18:3567–3578. doi: 10.1093/hmg/ddp305. [DOI] [PubMed] [Google Scholar]
- Rohrer TR, Gassmann KF, Rauch A, Pfeiffer RA, Doerr HG. Growth of heterokaryotic monozygotic twins discordant for Ullrich-Turner syndrome during the first years of life. Am J Med Genet A. 2004;126A:78–83. doi: 10.1002/ajmg.a.20446. [DOI] [PubMed] [Google Scholar]
- Salazar M, Consoli F, Villegas V, Caicedo V, Maddaloni V, et al. Search of somatic GATA4 and NKX2.5 gene mutations in sporadic septal heart defects. Eur J Med Genet. 2011;54:306–309. doi: 10.1016/j.ejmg.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Emi M, Iijima H, Ito N, Sato H, et al. Copy number loss of (src homology 2 domain containing)-transforming protein 2 (SHC2) gene: discordant loss in monozygotic twins and frequent loss in patients with multiple system atrophy. Mol Brain. 2011;4:24. doi: 10.1186/1756-6606-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SM, O'Reilly R. (Epi)genomics and neurodevelopment in schizophrenia: monozygotic twins discordant for schizophrenia augment the search for disease-related (epi)genomic alterations. Genome. 2009;52:8–19. doi: 10.1139/G08-095. [DOI] [PubMed] [Google Scholar]
- Srisupundit K, Brady PD, Devriendt K, Fryns JP, Cruz-Martinez R, et al. Targeted array comparative genomic hybridisation (array CGH) identifies genomic imbalances associated with isolated congenital diaphragmatic hernia (CDH) Prenat Diagn. 2010;30:1198–1206. doi: 10.1002/pd.2651. [DOI] [PubMed] [Google Scholar]
- van den Wijngaard JP, Lopriore E, van der Salm SM, Schaap AH, Vandenbussche FP, et al. Deep-hidden anastomoses in monochorionic twin placentae are harmless. Prenat Diagn. 2007;27:233–239. doi: 10.1002/pd.1652. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wee LY, Taylor M, Watkins N, Franke V, Parker K, Fisk NM. Characterisation of deep arterio-venous anastomoses within monochorionic placentae by vascular casting. Placenta. 2005;26:19–24. doi: 10.1016/j.placenta.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, et al. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Screenshots of the Cytosure Software output for the inter-twin CNV differences, confirmed by means of quantitative PCR. CNV profiles of the CHD-positive twin member and CHD-negative member of pair 6 are depicted as red or blue lines, respectively. The red and black boxes represent the two sets of PCR fragments in the discordant regions. The qPCR results for the red PCR fragments are depicted in figure 1.