Abstract
Circular RNA forms had been described in all domains of life. Such RNAs were shown to have diverse biological functions, including roles in the life cycle of viral and viroid genomes, and in maturation of permuted tRNA genes. Despite their potentially important biological roles, discovery of circular RNAs has so far been mostly serendipitous. We have developed circRNA-seq, a combined experimental/computational approach that enriches for circular RNAs and allows profiling their prevalence in a whole-genome, unbiased manner. Application of this approach to the archaeon Sulfolobus solfataricus P2 revealed multiple circular transcripts, a subset of which was further validated independently. The identified circular RNAs included expected forms, such as excised tRNA introns and rRNA processing intermediates, but were also enriched with non-coding RNAs, including C/D box RNAs and RNase P, as well as circular RNAs of unknown function. Many of the identified circles were conserved in Sulfolobus acidocaldarius, further supporting their functional significance. Our results suggest that circular RNAs, and particularly circular non-coding RNAs, are more prevalent in archaea than previously recognized, and might have yet unidentified biological roles. Our study establishes a specific and sensitive approach for identification of circular RNAs using RNA-seq, and can readily be applied to other organisms.
INTRODUCTION
Circular RNA forms (cRNAs) arise from a 3′–5′ ligation of both ends of linear RNA molecules. Instances of cRNAs have been observed in all domains of life, including eukaryotes, archaea, bacteria, and viruses, but overall cRNAs are considered extremely rare in nature. Examples for functional, naturally occurring cRNAs include the 1.75-kb circular single-stranded RNA genome of the hepatitis delta virus (1), and the RNA genomes in a family of plant pathogens called viroids (2,3). In the red algae Cyanidioschyzon merolae, RNA circularization followed by further processing is essential for maturation of permuted tRNAs, in which the 5′-end of the pre-tRNA occurs downstream to the 3′ end (4). Functional RNA circularity was also implied in the life cycle of eukaryal and bacterial group I introns. Following self-splicing from their precursor RNA, these introns may appear as circular RNA molecules, which can then reintegrate into other mRNAs, suggesting a potential role in intron mobility by reverse splicing (5–7). Additional rare cases of circular mRNA processing derivatives in eukaryotes have also been reported (8–12).
In archaea, cRNAs were mainly described in tRNA and rRNA introns, and rRNA processing intermediates. Once cleaved from their precursor RNA by the archaeal splicing endonuclease, archaeal introns undergo circularization by an RNA ligase (13,14). Although the cleaved-out circular introns are considered non-functional, unstable intermediates (15), some circular introns possess biological functions. For example, the 105-bp intron of tRNATrp in the euryarchaeote Haloferax volcanii contains a C/D box RNA (16). Once cleaved from its pre-tRNATrp and circularized, the C/D box located in the circular intron can guide chemical modifications on nucleotides 34 and 39 of the mature tRNATrp (16,17). Indeed, this circular tRNATrp intron is highly stable in Haloferax volcanii, as compared to other tRNA introns (14). Large, ORF-containing introns derived from rRNAs of the Crenarchaeotes Desulfurococcus mobilis (18) and Pyrobaculum species (19,20) were also found to be stable in their post-splicing circular forms, suggesting a functional role.
Circular 23S and 16S rRNAs can be found as processing intermediates in the course of rRNA maturation in archaea (21). Both these rRNA species are excised as circular pre-rRNA forms out of a single, long RNA precursor, and are then further processed to become mature rRNAs. Such processing intermediates were observed in representatives of the two major archaeal kingdoms, Euryarchaeota and Crenarchaeota, and are thus considered a widely conserved rRNA processing step across archaea (21).
Intriguingly, a single report by Starostina et al. (22) demonstrated that most, if not all, of the C/D box RNAs in the hyperthermophilic euryarchaeon Pyrococcus furiosus stably exist in both linear and circular forms. Archaeal C/D box RNAs, when complexed as an RNP with three specific proteins, guide 2′-O-methylation on target nucleotide positions in rRNAs and tRNAs (23). Starostina et al. have elegantly shown that the circular forms of Pyrococcus furiosus C/D box RNAs are found associated with the C/D box protein complex, and could thus function in guiding RNA modifications in vivo (22). This discovery points to the hypothesis that circular transcripts might have, at least in archaea, more biological roles than currently appreciated.
Deep transcriptome sequencing (RNA-seq) is an emerging method enabling the study of RNA-based regulatory mechanisms in a genome-wide manner (24). We have recently used RNA-seq to map the transcriptome of Sulfolobus solfataricus (25), a sulfur-metabolizing aerobic archaeon that grows optimally at 80°C and pH 2–3 and is one of the most widely studied model archaeal organisms (26). However, the standard mapping scheme of short RNA-seq reads has so far prevented identification of circular transcripts in our S. solfataricus RNA-seq data.
We now developed a directed computational scheme to pinpoint RNA-seq reads that have a permuted mapping to the genome, a hallmark of circular RNA (Figure 1). To enrich for circular transcripts in the sample and to overcome possible artifacts, we pre-treated the RNA sample by RNase R, an exoribonuclease that degrades linear RNA but leaves circular transcripts intact (27). This combined experimental and computational approach, which we coin circRNA-seq, has allowed us to map circular transcripts in S. solfataricus in an unbiased, genome-wide manner.
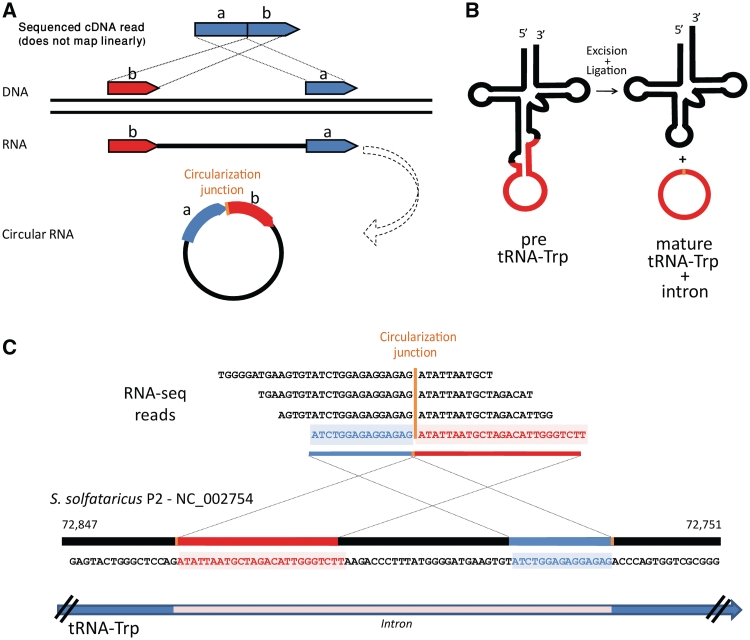
Figure 1.
Identification of circular RNA products in RNA-seq data. (A) An RNA-seq cDNA read that maps to the reference DNA in a non-linear, chiastic manner, is a hallmark of circular RNA. (B) Schematic representation of the Sulfolobus solfataricus tRNATrp, which contains a 65 b intron that is cleaved in the process of tRNA maturation and becomes a stable RNA circle. (C) Reads derived from the region around the circularization junction of the excised tRNATrp intron show chiastic mapping to the genome of S. solfataricus P2, exemplifying the power of RNA-seq in circular RNA discovery. Multiple different reads spanning the circularization junction confirm that the observed circular junction is not an amplification artifact. Numbers denote position of the S. solfataricus P2 genome.
MATERIALS AND METHODS
Archaeal cells growth
Sulfolobus solfataricus P2 and S. acidocaldarius cells were grown organotrophically in 300-l enameld bioreactor (Bioengineering AG, Switzerland) in 280-l MAL-medium and yeast extract (0.1% w/v, Difco) as substrate. The final pH of the solution was adjusted to pH 3.0. S. solfataricus cells were grown at 80°C and S. acidocaldarius cells were grown at 75°C. Both kinds of cells were grown at aerobic conditions and harvested at stationary phase.
Halobacterium sp. NRC-1 cells were grown in CM+complex media containing (final concentrations) 250 g/l NaCl, 20 g/l MgSO4·7H2O or 9 g/l MgSO4, 3 g/l Na citrate, 2 g/l KCl, 7.5 g/l Casamino acids, 10 g/l yeast extract, and 0.5 ml/l trace metals stock ×2000 containing 3.5 mg/ml FeSO4×7H2O, 0.88 mg/ml ZnSO4×7H2O, 0.66 mg/ml MnSO4×H2O, 0.02 mg/ml CuSO4×5H2O dissolved in 0.1 N HCl. The final pH of the solution was adjusted to pH 7.2 with NaOH. The cells were grown to either logarithmic phase or stationary phase at 42°C at 250 rpm at aerobic conditions as previously described (47)
Total RNA and genomic DNA isolation
Total RNA and DNA samples were isolated from each archaeal cells sample (∼107 cells) using TRI reagent according to the manufacturer's protocol (Molecular Research Center Inc.). Total RNA samples were treated with DNase I (Turbo DNA-free kit, Ambion) and nucAway spin columns (Ambion) according to the manufacturer's protocol to remove DNA contamination and salts. RNA and DNA quality was determined using either 2.0% TAE–agarose gel electrophoresis or 2100 Bioanalyser (Agilent).
RNase R digestion
RNase R digestion reaction was carried out at 37°C for 45 min in 20 μl 10× reaction buffer [20 mM Tris–HCl (pH 8.0), 0.1 M KCl and 0.1 mM MgCl2], with S. solfataricus P2 total RNA (20 μg), and E. coli RNase R (120 U) (Epicentre Biotechnologies). Ethanol precipitation was carried out in order to remove the enzyme and salts. The digestion and precipitation reactions were repeated two more times with a ratio of 3 U enzyme/1 μg RNA. The treated RNA sample was analyzed using either 2.0% TAE-agarose gel electrophoresis or 2100 Bioanalyser (Agilent).
Library preparation and Illumina sequencing
Sulfolobus solfataricus, S. acidocaldarius, H. salinarum (two samples) total RNA and S. solfataricus RNase R-treated samples were used as templates for cDNA libraries that were prepared according to the mRNA-seq Illumina protocol, omitting the polyA-based mRNA purification step. In brief, for each sample 100 ng of RNA were first fragmented by divalent cations at 94°C for 5 min. Double strand cDNA was generated using SuperScriptII and random primers following QIAquick PCR Purification kit. cDNA was then end-repaired, adenylated and ligated to adapters. In order to maintain small RNAs, a 120–240 nt fragment was gel-purified in the cDNA purification step of both S. solfataricus cDNA samples. Other samples were gel purified as indicated in the protocol. All five cDNA libraries were further amplified and sequenced using 40 single-read cycles on a Genome Analyser II (Illumina).
Reads mapping and analysis
Mapping of reads was performed as previously described (25) with the following changes. Briefly, sequencing reads were mapped to the reference genome (S. solfataricus P2: GenBank:NC_002754; S. acidocaldarius: GenBank:NC_007181; H. salinarum NRC1: GenBank:NC_002607, NC_002608, NC_001869) using blastn with an e-value of 0.0001 and the ‘–F F’ flag. Reads mapped by 33 bp and more to the genome with up to two mismatches were considered ‘high quality’ aligned, while reads showing partial mapping (of 20–32 bp) were considered ‘low quality’ reads. The alignment with the best bit-score was accepted as the correct mapping for each read. Reads having more than one best-scoring position on the genome were discarded. A transcript coverage map was calculated based on the alignment of high quality reads.
Detection and clustering of putative circularization junctions
Circularization junctions were predicted by first identifying uniquely aligned permuted reads. All partially mapped reads, defined as reads having between 20 and 32 bp of continuous alignment to the genome (with maximum two mismatches), were evaluated for evidence for being permuted reads. Permuted reads were defined as reads that their non-aligned part (‘short part’) was uniquely mapped to the genome in a non-sequential, chiastic manner (Figure 1). In the short part, only nucleotides with quality score above 20 were counted as alignable. To remove false positive artifacts, reads whose short part had less than 8 nt having quality score above 20 were discarded, as well as reads that half or more of the nucleotides in their short part were identical (homopolymeric). Based on the observation that G to T and A to C errors are common in the Illumina platform (48), reads whose short part aligned directly after the anchor with such substitutions and one additional mismatch were also discarded. Reads that their short part was aligned to Illumina adapters were also ignored.
Circular RNAs junctions were defined by reads for which both parts aligned uniquely to the genome within 10 000 nt in a chiastic order. It should be noted that circular RNAs sized <40 nt were defined by reads that were mapped in chiastic order and their short part overlapped their anchor part. These cRNAs could be detected only when the reverse transcriptase completed more than one round around the cRNA.
Next, reads showing identical circularization junctions were clustered. For each identified junction, further identification of additional reads supporting the junction was performed by adding reads that span the junction by at least four nucleotides on each side. This was performed to include supporting reads that were aligned by 33–36 bp to the genome and hence were not included in the initial analysis, as well as reads whose short parts aligned non-uniquely to the genome but supported a junction identified earlier by unique mapping. Junctions whose location on the genome differed by up to 3 nt were clustered into one junction, based on the observation that small junction shifts are common in cRNAs [in this study and (22)]. Identification of reliable cRNA candidates was performed by integration of total RNA (from both this study and Wurtzel 2010) and RNase R treated RNA-seq cRNA candidates. Matching candidates from both runs that differed by up to 3 nt were considered reliable cRNAs (Table 1).
Table 1.
Identified circular RNAs in S. solfataricus P2
| Genea | Circle startb | Circle endb | Circle size | No. of reads RNA- seqc | No. of reads RNase R-seqd | Norm. RNase R enrich.e | Other cRNAs in genef | Conserved in S. acido. | RT-PCR validated |
|---|---|---|---|---|---|---|---|---|---|
| (I) rRNAs and tRNA introns | |||||||||
| 5S rRNA/SSOr02 | 7 945 | 78 067 | 123 | 95 | 312 | 7.1 | 4 | Yes | No |
| 16S rRNA/SSOr03 | 871 658 | 873 216 | 1559 | 4124 | 4957 | 1.6 | Yes | ||
| 23S rRNA/SSOr04 | 873 334 | 876 429 | 3096 | 2629 | 1605 | 0.8 | Yes | ||
| tRNA-Trp/SSOt04 | 72 767 | 72 831 | 65 | 1148 | 1694 | 0.7 | 1 | Yes | Yes |
| tRNA-Lys/SSOt07 | 138 386 | 138 407 | 22 | 211 | 825 | 91.5 | |||
| tRNA-Met/SSOt11 | 184 817 | 184 841 | 25 | 25 | 240 | 20.9 | |||
| tRNA-Pro/SSOt42 | 898 313 | 898 333 | 21 | 550 | 1482 | 4.1 | |||
| tRNA-Ser/SSOt33* | 640 978 | 641 001 | 24 | 4 | NA | Yes | |||
| (II) Other non-coding RNAs | |||||||||
| C/D box sR106 | 285 707 | 285 760 | 54 | 2 | 17 | 271.5 | 1 | Yes | |
| C/D box Sso-180 | 362 308 | 362 369 | 62 | 4 | 1 | 46.2 | |||
| C/D box sR133 | 442 392 | 442 417 | 26 | 13 | 9 | 1.0 | |||
| C/D box sR102 | 563 241 | 563 296 | 56 | 2 | 3 | 31.1 | Yes | Yes | |
| C/D box Sso-sR8 | 647 783 | 647 833 | 51 | 10 | 9 | 0.9 | |||
| C/D box Sso-sR4 | 666 143 | 666 186 | 44 | 56 | 277 | 4.1 | 5 | Yes | |
| C/D box Sso-sR10 | 794 186 | 794 240 | 55 | 1 | 1 | 1223.5 | Yes | ||
| C/D box Sso-207 | 816 021 | 816 075 | 55 | 1 | 2 | 68.6 | |||
| C/D box SSOs02 | 829 352 | 829 405 | 54 | 2 | 2 | 52.8 | |||
| C/D box Sso-sR12 | 2 189 397 | 2 189 456 | 60 | 2 | 1 | 11.1 | Yes | ||
| C/D box sR105 | 2 237 915 | 2 237 962 | 48 | 2 | 1 | 7.4 | Yes | ||
| H/ACA box sR109 | 595 510 | 595 579 | 70 | 2 | 2 | 15.7 | 1 | Yes | Yes |
| ncRNA | 442 786 | 442 854 | 69 | 3 | 18 | 160.0 | 1 | Yes | |
| ncRNA (within ISC1476) | 722 538 | 722 578 | 41 | 6 | 8 | 1.4 | 2 | ||
| Sso-117 (antisense to ISC1225) | 1 576 633 | 1 576 671 | 39 | 8 | 1 | 0.0 | 1 | ||
| Sso-109 (antisense to ISC1225) | 1 927 228 | 1 927 258 | 31 | 5 | 1 | 0.1 | |||
| 7S rRNA/SSOr01 | 49 977 | 50 023 | 47 | 3 | 5 | 4.5 | Yes | ||
| Sso-214 | 105 148 | 105 181 | 34 | 13 | 2 | 0.2 | |||
| RNase P | 224 732 | 224 765 | 34 | 40 | 8 | 0.2 | 6 | Yes | Yes** |
| Sso-83 | 581 818 | 581 860 | 43 | 23 | 2 | 2.3 | 1 | ||
| ncRNA | 1 275 500 | 1 275 567 | 68 | 14 | 66 | 20.9 | 3 | Yes | |
| (III) Possible processing errors | |||||||||
| tRNA pseudouridine synthase/SSO0393 | 343 138 | 343 264 | 127 | 1 | 1 | 167.1 | 6 | ||
| Intergenic region upstream rRNA operon | 871 573 | 871 657 | 85 | 4 | 2 | 1.0 | |||
| Intergenic region between 16S and 23S rRNA operon | 873 215 | 873 331 | 117 | 7 | 8 | 8.9 | Yes | ||
| (IV) Inside ORFs | |||||||||
| hypothetical protein/SSO0389 | 335 563 | 335 635 | 73 | 7 | 2 | 128.8 | |||
| hypothetical protein/SSO0845 | 725 923 | 726 085 | 163 | 2 | 2 | 33.1 | |||
| succinate dehydrogenase subunit/SSO2359 | 2 154 297 | 2 154 322 | 26 | 4 | 1 | 2.5 | |||
| peptide ABC transporter/SSO2619 | 2 385 872 | 2 385 901 | 30 | 10 | 1 | 80.4 | |||
| rubrerythrin (rr)/SSO2642 | 2 404 146 | 2 404 203 | 58 | 1 | 1 | 749.7 |
aGene containing the identified cRNA. Annotation taken either from NCBI accession NC_002754 (51) and from the following refs (25,34,36).
bIn cases of several cRNAs within the same gene, coordinates for dominant cRNA (one supported by most reads) are shown.
cNumber of reads supporting the circularization junction in the total RNA sample.
dNumber of reads supporting the circularization junction in the RNase R treated sample.
eEnrichment in the RNase R sample (Methods).
fNumber of additional cRNAs overlapping the gene, in case of multiple cRNAs per gene.
*Detected in RNase R sample only.
**The cRNA taken for validation was a variant longer than the dominant one, due to the short size (34 bp) of the dominant cRNA (Supplementary Table S4).
Gene annotations for all organisms were downloaded from GenBank. Additional gene and ncRNA annotations for S. solfataricus P2 were taken from refs (25,34,36,49).
Calculation of RNase R normalized enrichment
For cRNAs detected both in total RNA and in RNase R-treated samples, RNase R normalized enrichment was calculated. For each cRNA, circLevel was defined as the total number of supporting reads spanning the circularization junction, divided by the mean coverage inside the cRNA locus. This parameter is calculated to reflect the ratio between circular form of the RNA and all RNA in the cRNA locus. Normalized enrichment per each cRNA was then calculated by dividing RNase R-treated circLevel and total RNA circLevel.
Conservation analysis
To identify conservation of circular RNAs between S. solfataricus and either S. acidocaldarius or H. salinarum, a sequence composed of cRNA sequence with ±100 nt in both ends was locally aligned using the Smith-Waterman algorithm [implemented in the ssearch35 program (50)] to the reference genome of S. acidocaldarius/H. salinarum [NC_007181/(NC_002607,NC_002608,NC_001869)] (E < 10−6) and a search of cRNA with similar length (±20%) in this homologous region was conducted. The match of the cRNAs region in both organisms was further manually examined for the cRNAs in Table 1.
RT–PCR and northern analyses
Sulfolobus solfataricus P2 total RNA and RNase R-treated samples were used as templates for the RT–PCR. To improve detection of structured RNAs Verso reverse transcriptase (Thermo-Fischer Scientific), which is active at high temperatures, was used. The samples (non-treated and RNase R treated) were reverse transcribed using random hexamers as primers. The RT reactions were carried out at 53°C for 30 min according to the manufacturer’s instructions.
Two sets of primers, an outward-facing primer set for detection of the non-linear form and an inward-facing primer set for detection of the linear and circular forms were designed for the cRNA candidates (Supplementary Tables S5 and S6). Equal amounts of both cDNA samples and additional genomic S. solfataricus DNA sample were used as templates for PCR using Reddy mix PCR kit (Thermo Scientific) following the manufacturer's instructions. RT–PCR products from the RNase R-treated sample with outward-facing primers were cloned (in case of products smaller than 100 nt) and sequenced to verify circularization junction existence. Products with insufficient amounts for cloning were reamplified using the same PCR procedure and the products thus were further cloned and sequenced.
For the northern blot analyses, membranes were produced using the NuPage TBE-Urea gels system (Invitrogen). RNA probes were produced with MAXIscript In Vitro Transcription Kit (Ambion). Hybridization conditions and buffers were according to NorthernMax kit (Ambion) manufacturer instructions.
RESULTS
To obtain transcriptome data for S. solfataricus P2, we sequenced a total RNA sample from this archaeon grown on organotrophic medium to a stationary phase (Methods). The sample was sequenced by the Illumina mRNA-seq approach, yielding 27.54 million reads sized 40 bp that mapped uniquely to the genome [3-fold the number of reads sequenced in our earlier study (25)]. Of these, 26.94 million (97.82%) reads mapped to the genome with at least 33 bp and no more than two mismatches, indicating a relatively high quality of the sequenced sample (Supplementary Table S1).
We reasoned that reads spanning an RNA circularization junction should align to the reference genome in a permuted, chiastic order, i.e. with a downstream stretch of the sequence aligning to an upstream position in the genome [Figure 1A; (22)]. Since such reads do not map linearly to the genome, most RNA-seq analysis software, including the one we used (Methods), will define them as ‘unmapped’ or ‘low quality.’ We therefore searched the fraction of such unmapped reads (2.17% of the data, 598 620 reads) for reads that unambiguously represent junctions of circular RNA, i.e. in which both sides of the permuted read were uniquely mapped to the genome (Methods). We were able to detect 7018 such reads (excluding reads that mapped to rRNAs), suggesting a prevalence of circular products within the sequenced RNA sample.
We next clustered the putative circular reads according to the circularization junctions they spanned (as exemplified in Figure 1C), and added supporting reads that were previously mapped only partially to the genome. We noticed that in many cases (over 300 junctions) the circularization point may be shifted by 1–3 nucleotides, and therefore clustered such junctions to avoid redundancy (see ‘Materials and Methods’ section). A total of 3933 junctions were recorded, with 897 of these (23%) supported by two or more reads (Supplementary Tables S1 and S2). The relatively small fraction of junctions supported by multiple reads may suggest that many of the junctions we observed represent artifacts. It was previously shown that template switching during reverse transcription can result in non-linear cDNA products [coined RT-facts; (28–30)]. Indeed, 2833 reads had a strong potential for being a result of template switching, having 5 or more repetitive bases flanking the beginning and end of the predicted cRNA (Supplementary Figure S1). Sequencing errors leading to erroneous read mapping, and chimeric amplification products (31) may be another possible cause for spurious predictions of circularization junctions (Supplementary Figure S1).
To attain a higher-confidence data on circular RNAs, we decided to use the exo-ribonucleolytic enzyme RNase R (27). This exonuclease degrades linear RNA molecules in a 3′–5′ direction, but leaves circular RNA molecules, as well as some structured RNAs, intact (27,32). On the basis of our hypothesis that RNase R will enrich for circular RNAs in the sample, we used a technical replicate of the RNA sample described above, and pre-exposed it to RNase R (Methods). The RNase R-treated sample was then sequenced using the same protocol as above, yielding 16.66 million uniquely mapped reads. Of these, 96.65% were mapped to rRNAs (versus 75.02% in the non-enriched data), both because of their highly structured nature, and because rRNA precursors are circular (see below), leaving 557 562 reads mapping to the non-rRNA portion of the genome (Supplementary Table S1). Only 4% of the genome was covered by multiple (five or more) reads in the RNase R-treated sample, suggesting that most linear RNA was indeed degraded by RNase R.
Since RNase R does not degrade all linear RNA [due to its requirement for five unstructured bases at the 3′-end and other sequence preferences (32,33)], the treated sample can, in principle, also contain cRNA artifacts. We therefore defined a set of reliable circular transcripts as those for which there was reproducible evidence for permuted reads both in the non-enriched, regular RNA-seq sample [either produced in this study or in Wurtzel et al. (25)], and in the sample pretreated by RNase R (Table 1). As expected, in most cases the RNase R sample showed enrichment in the amount of reads supporting circularization junctions as compared to non-circular reads covering the same area (as exemplified in Figure 2).
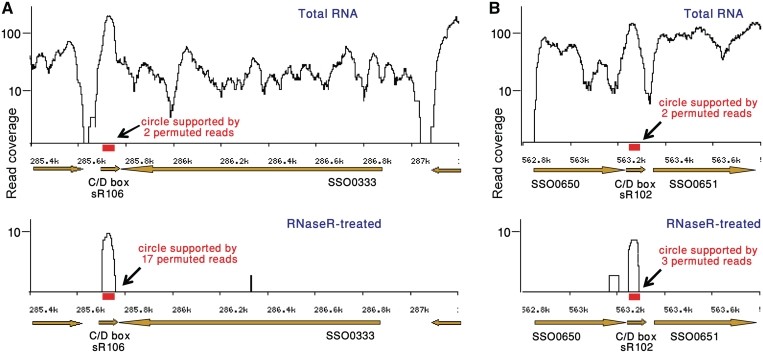
Figure 2.
Enrichment for circular transcripts in RNA-seq data from RNase R-treated sample. In both panels, top graph presents RNA-seq data from non-treated sample, and bottom graphs present data from RNase R-treated sample, where linear RNAs are depleted. X axis denotes the position on the S. solfataricus P2 genome, with brown arrows denoting genes (right- and left-pointing arrows correspond to genes encoded on the forward and reverse strands, respectively). Red box marks a cRNA region also supported by permuted reads. Y axis denotes RNA-seq coverage of reads fully mapping to the genome. (A) The C/D box RNA sR106 (34) is enriched in the RNase R-treated sample, and its circular formation is independently supported by 2 and 17 permuted reads in total RNA and RNase R-treated samples, respectively. (B) The C/D box RNA sR102 (34) is enriched in the RNase R-treated sample, and its circular formation is independently supported by two and three permuted reads in total RNA and RNase R-treated samples, respectively.
Functional categorization of cRNAs
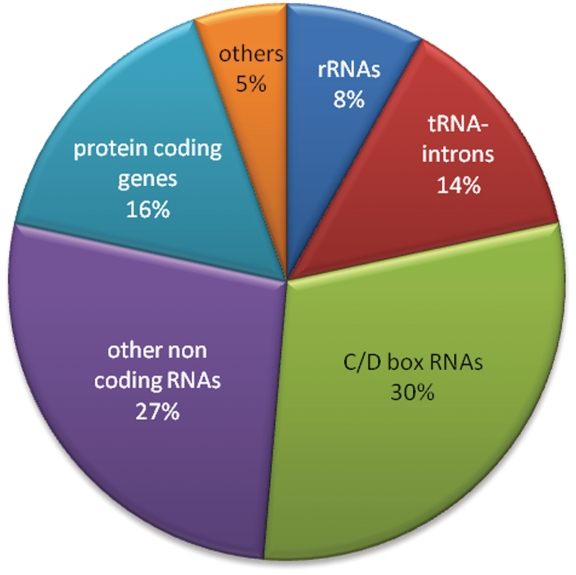
The circular transcripts we identified (Table 1) can be divided into several functional groups (Figure 3), as described below. This set contains cRNAs in 37 genes, seven of which were expected based on previous literature (i.e. tRNA introns and rRNA processing intermediates). The vast majority (78%) of the cRNAs we detected occurred in non-coding RNAs.
Figure 3.
Functional characteristics of genes encompassing circular transcripts identified in this study.
tRNA introns
Sulfolobus solfataricus P2 has 19 tRNAs that contain introns (35). Following excision from the pre-tRNA, these introns are predicted to become circular, relatively unstable products (14). We were able to detect four tRNA introns, of tRNATrp, tRNALys, tRNAMet, and tRNAPro. A fifth tRNA intron, tRNASer, was detected in the RNase R-treated sample only, probably reflecting instability of this intron leading to its rareness in the total RNA sample. Notably, our algorithm for search of permuted reads cannot detect circular transcripts shorter than 20 bp (see ‘Materials and Methods’ section). Indeed, most tRNA introns that we did not detect are shorter than this intrinsic threshold (see ‘Discussion’ section).
Remarkably, the 65-bp intron excised from tRNATrp was highly stable according to our data, with 1148 and 1694 supporting permuted reads in the non-treated and RNase R-treated samples, respectively (Figure 1B and C; Table 1). It was previously shown that an excised tRNATrp intron is highly stable in Haloferax volcanii as well (14). In Haloferax, this tRNATrp intron contains a functional C/D box RNA that is responsible for 2′-O-methylation at positions 34 and 39 in the tRNATrp (16). Although the consensus C and D boxes cannot be found in the tRNATrp intron of S. solfataricus (23), we did find two 7-bp stretches of sequence complementary between the S. solfataricus tRNATrp intron and positions 34 and 39 of the cognate tRNATrp (Supplementary Figure S2), implying that the tRNATrp intron might also have a role in guiding RNA modifications in S. solfataricus. If this is indeed the case, then what we observe here is a remarkable functional conservation of this intron across a phylum-level phylogenetic distance.
rRNA processing intermediates
Among the reads covering the rRNA operon we clearly observed high levels of the circular 16S and 23S processing intermediates previously described for S. solfataricus (21), with thousands of permuted reads supporting the expected circularization junctions (Table 1). However, we also observed evidence for circularization intermediates in the 5S rRNA, but to much lesser levels as compared to the 16S and 23S rRNAs. It is yet to be determined whether the 5S rRNAs are also pre-processed via a circular intermediate similar to the 16S and 23S. Alternatively, the circles we observe in the 5S rRNA might represent a specific step in its degradation. Notably, the 5S rRNA was the only non-coding cRNA we were unable to validate using RT–PCR (see below).
Non-coding RNAs
Circular forms were found in 11 C/D box RNAs, 8 of which encompass the full length of the C/D RNA, pointing to a general mechanism involving circularization of C/D box RNAs. In the hyperthermophilic archaeon Pyrococcus furiosus it was shown that most of the C/D box RNAs can exist in circular forms (22), and it was suggested that RNA circularization provides a better structural stability to the C/D box RNAs in high temperatures. Indeed, similar to P. furiosus, S. solfataricus is also a hyperthermophile, implying that C/D box circularity might have a stabilizing function in this organism as well (see ‘Discussion’ section). As opposed to Pyrococcus, where circular and linear forms of C/D box RNAs are found in roughly equal amounts in the cell, the circular forms of C/D box RNAs seem to be the minor form in S. solfataricus (Supplementary Table S1, Figure 2).
We also observed circular forms for additional ncRNAs, including in the H/ACA RNA sR109 that is predicted to guide pseudouridinylation in rRNA (34), and circles in two antisense-box ncRNAs that are predicted to target the third ORF of transposon ICS1225. A third circular ncRNA, found in this study (genomic location 442 786–442 857), has the potential of similarly targeting the second ORF of transposon ISC1904 (Table 1). Such ncRNAs were previously hypothesized to be involved in regulation of transposition activity (36). It is possible that additional cRNA forms of transposon-targeting ncRNAs have escaped detection, since reads mapped to repetitive regions were excluded from our analysis (see ‘Materials and Methods’ section).
Interestingly, seven circularization junctions, the dominant of which supported by 40 permuted reads, suggest that segments of RNase P can occur in circular forms, although these forms comprise a small minority as compared to the extensive abundance of the linear form (Supplementary Table S1). We also identified one circular segment of the 7S RNA which is the RNA component of the signal recognition particle. Notably, circular RNase P and 7S RNA transcripts were found to be conserved in Sulfolobus acidocaldarius (see below). Their functions, however, remain elusive.
Other circles
Two reproducible cRNAs occurred in the rRNA operon, one of them between the 16S and 23S rRNAs, and one directly upstream the 16S. A close examination revealed that these cRNAs begin exactly one nucleotide before the position from which the 16S or 23S were excised from the pre-rRNA transcript. A similar phenomenon was observed in the protein coding gene tRNA pseudouridine synthase (Cbf5), which contains a short intron (37). In that gene, seven different circularization junctions appeared one nucleotide away from the position where the intron was excised. These might represent errors in the excision/ligation procedure aimed to extract an intron, where the ligase accidentally circularizes the ends of the exon instead of, or in addition to, the excised intron.
Several additional cRNAs were observed within protein coding genes (Table 1). It is possible that these represent novel introns that were excised and circularized; however, we failed to detect spliced reads that support a mature form of the mRNA after intron excision, suggesting that these cRNAs are probably not intron excision products. Alternatively, based on our observation that over 75% of cRNAs occur in non-coding RNAs (Figure 3), it is possible that some of these ORF-encoded cRNAs actually represent non-coding RNAs expressed from within a protein. Perhaps the most likely possibility is that these cRNAs might represent degradation products, based on their high variability as observed from RT–PCR experiments (see below).
Detailed experimental verification of cRNAs
To independently verify the observed cRNAs in non-coding RNAs, we selected 11 candidates for RT–PCR analysis (Supplementary Table S4). We designed two sets of primers for each candidate, an outward-directed set that is expected to amplify only the circular form, and an inward-directed set that is expected to amplify both the linear and the circular forms (Figure 4). RT–PCR was then performed with S. solfataricus total RNA sample, RNase R-treated sample and a genomic DNA sample as a control (which is expected to be amplified only by the inward-directed primers).
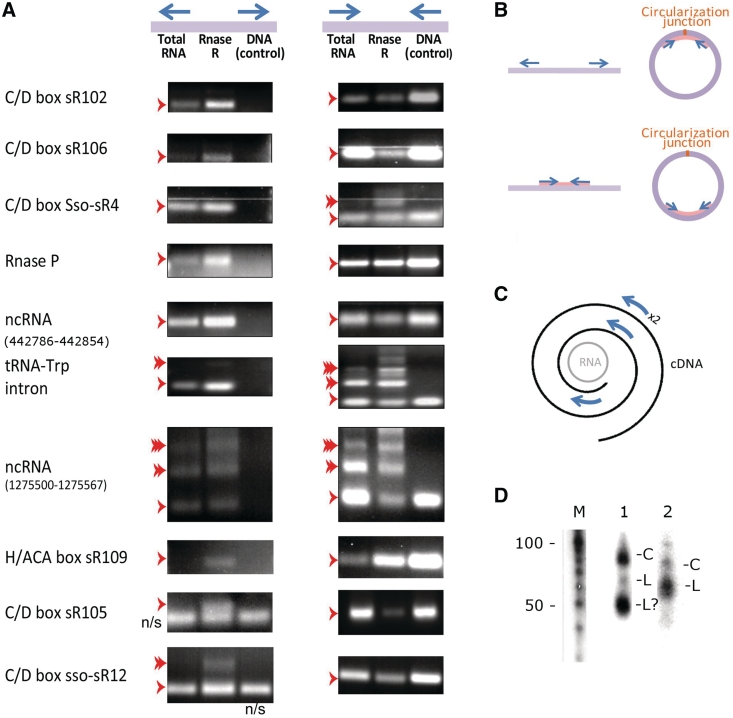
Figure 4.
Experimental verification of cRNAs by RT-PCR. (A) Left, RT–PCR results of amplification with outward-directed primers, designed to amplify cRNA; right, RT–PCR results of amplification with inward-directed primers, expected to amplify both linear and cRNA form. Single and multiple arrowheads represent single or double/triple size products, respectively. N/S, non-specific amplification (as verified by direct sequencing). RT–PCR with each primer set was performed on total RNA sample, RNase R-treated sample, and DNA sample, all extracted from S. solfataricus grown to stationary phase on organotrophic medium. (B) RT–PCR for verification of circular RNAs. Arrows indicate outward facing primers (top) and inward facing primers (bottom). Purple line/circle denotes the RNA template, pink line denotes expected PCR product. (C) Double and triple sized products can stem from multiple rounds of RT around a circular RNA template, followed by PCR amplification. Arrows mark illustrative PCR primers. (D) Northern blot analyses of two ncRNAs with circular forms: (M) Size marker, (1) ncRNA found in genomic location 1 275 500–1 275 567, (2) ncRNA found in genomic location 442 786–442 854. Circular forms are indicated by ‘C’, and linear forms by ‘L’.
For 10 out of the 11 tested cRNAs, clear amplification of the circular form was observed using the outward-facing primers (Figure 4). Moreover, in all 10 verified cases, the RNase R-treated sample showed higher band intensity with the outward-facing primers, providing further support that these are circular forms resistant for degradation by the RNase R exo-ribonucleolytic activity. In some of the cases, double and triple sized products were also observed; it was previously shown that such product multiplicity can stem from multiple rounds of RT around a circular RNA template [Figure 4B, (22)]. As expected, no amplification was observed using the outward facing primers on genomic DNA (except for two cases where directed sequencing showed non-specific amplification of a linear genomic sequence, which resulted from non-specific binding of one of the primers). Finally, RNase R bands usually had weaker intensity in the RT-PCR with the inwards-facing primers, probably implying that the total RNA sample is composed of a mixture of both circular and linear forms.
To further verify the composition of the amplified products we sequenced the RT–PCR products of the outward facing primers with the RNase R-treated sample. Five of the primary products included the exact predicted junction, while the remaining six showed variation of several nucleotides from the predicted junction (detailed in Supplementary Table S4). Minor length-heterogeneity and shifts in the circularization junction were previously observed also in circular C/D box RNAs of P. furiosus (22), and this was consistent with our transcriptome-wide sequencing results, where some genes were associated with several different circularization junctions (Table 1). It is therefore possible that several forms exist in vivo for some cRNAs.
It was previously shown that in northern blot assays, circular RNA forms have different electrophoretic mobility than the linear RNA of the same size (22). To provide further support for RNA circularity of novel ncRNAs, we selected the three non-coding RNAs corresponding to genomic positions 442 786–442 854 595 510–595 579 and 1 275 500–1 275 567 for northern blot analysis (Table 1). In two of the three cases, more than one distinct band was observed, suggestive of a circular form of different electrophoretic mobility (Figure 4D). Relative rarity of the circular form in total RNA (as also evident from the RT–PCR, Figure 4B) may explain the single case of ncRNA where no putative circular form was observed.
Next, we attempted to verify three additional circular RNA forms occurring within protein coding genes, as these might represent novel functions for cRNAs (Supplementary Table S4). However, distinct circular products were not observed upon PCR amplification with outward-facing primers. Instead, in two of the three cases we observed a ‘smear’ of multiple circular forms (Supplementary Figure S3). Cloning and direct sequencing of products from one of these smears showed a variety of distinct circular RNA forms occurring in various sizes (Supplementary Figure S3). This was in agreement with the observation of more than 50 different cRNAs that were identified inside this gene in the RNA-seq sequence data, most of which supported by a single read only (Supplementary Tables S1 and S2). These results imply that these ORF-residing cRNAs might be highly unstable, possibly representing degradation intermediates (see ‘Discussion’ section).
As described above, reliable cRNAs were defined as those supported both in the RNA-seq sample where total RNA was sequenced, and in the RNase R sample. To test whether this restriction is indeed warranted, we examined three putative cRNAs that were supported by multiple reads in the sequenced total RNA sample but were absent from the RNase R sample. Indeed, we were unable to amplify any of these products using outward-facing primers, further establishing our approach. The support of circular junction in these cases by multiple reads is therefore peculiar; it is tempting to speculate that these might represent permuted transcripts that were further processed and linearized, as in the case of the permuted tRNA genes in the red algae Cyanidioschyzon merolae (4).
Conservation of cRNAs in related species
We next set out to test whether the circles we detected in S. solfataricus are also present in the archaeon S. acidocaldarius. Although these two organisms belong to the same genus, they share only ∼40% sequence identity and thus represent a non-negligible genetic distance. To search for conservation, we first used the Smith–Waterman algorithm to locate the homologs of the S. solfataricus cRNA region on the S. acidocaldarius genome. Because the two genomes are relatively distant, we were able to find the homologs for only a subset of the cRNAs (20 regions with homology). We then produced RNA-seq data from reverse-transcribed total RNA of S. acidocaldarius grown in the same growth conditions as for the S. solfataricus, and searched for permuted reads indicating circularization, as described above (Supplementary Table S1). For 10 out of the 20 (50%) cRNAs in S. solfataricus, we were also able to detect a similarly sized cRNA in the homologous region in S. acidocaldarius (Table 1). The conserved circles included some of the ncRNAs, including RNase P and several C/D box RNAs, and also the three circular rRNA molecules. This observed conservation in circularity, found in these two relatively distant organisms, implies that the circular transcripts we detected represent products with possibly important function.
To gain further insights as to the extent of phylogenetic distance by which circular transcripts may be conserved, we also generated RNA-seq data from total RNA of the halophilic euryarchaeon Halobacterium salinarum NRC-1 (Methods). Among the highly represented circular transcripts in that organism we were able to identify the pre-16S and pre-23S circular intermediates, the 5S rRNA cRNA, two tRNA introns and also cRNAs inside the 7S RNA (but not in the same region as in S. solfataricus). However, the general lack of sequence similarity between the genomes of Halobacterium and Sulfolobus prevented further investigation into the presence of cRNAs in other, less conserved genes. Still, the observed circularity of pre-rRNAs and the tRNA introns further supports a general mechanism of rRNA/tRNA processing in archaea, as previously suggested (21).
DISCUSSION
We have described here a general approach aimed at genome-wide discovery of circular transcripts. This procedure, which we denote circRNA-seq, combines experimental enrichment for cRNAs using RNase R, whole-transcriptome sequencing, and a computational algorithm to detect short permuted reads that report on cRNAs. Using this approach in S. solfataricus, we found a set of cRNAs associated with 37 genes, most of which represent new cases of circular transcripts that were not described before.
The number of cRNAs we detected is probably an underestimate of the actual number of cRNAs in the S. solfataricus cell. First, very short circles might have escaped detection, as our algorithm has lower chances of detecting circles that are shorter than the sequencing read size, and cannot detect at all cRNAs that are 20 bp or shorter (Methods). In addition, circles that have low expression in the conditions of the experiment might have escaped sampling by the RNA-seq, as well as circles mapped to repeat regions that were excluded from our analysis (Methods). Therefore, more cRNAs probably exist in S. solfataricus than we reported here.
The functions of the new cRNAs we discovered might be diverse. In eukaryotes, the 5′- and 3′-termini of C/D box RNAs are usually base-paired (38). Starostina et al. (22) had hypothesized that circularization of C/D box RNAs in Pyrococcus might be essential to maintain the 5′- and 3′-ends in close vicinity in the hyperthermophilic growth conditions of this archaeaon. Our observation of many circular C/D box RNAs also in the S. solfataricus, which is evolutionarily very distant from Pyrococcus but shares the preference for hyperthermophilic conditions, might provide support for this stabilization hypothesis. Moreover, circularization of additional non-coding RNAs that we observed might serve for the same purpose, i.e. covalently maintaining a structure that is normally maintained by non-covalent base-pairing in mesophilic organisms. Notably, however, most of the C/D box ncRNA we detected had relatively low number of permuted reads as compared to linear reads. Because the previously identified archaeal circular RNAs (tRNA introns, rRNAs) are generated as intermediates of RNA processing, it is possible that circular forms of C/D box RNAs represent intermediates of a yet unidentified RNA processing mechanism as well.
In vitro, artificially circularized transcripts were shown to possess enhanced stability, being protected against degradation by exonucleases and by RNase H (39–41). One of the circularized ncRNAs we detected (in position 1 275 500–1 275 567) is genomically associated with a transposase, suggesting a function related to transposition. In this case, it could be hypothesized that circularization might provide protection against intrinsic cellular defense mechanisms, or might be involved in the process of transposition itself.
Many of the circular junctions that were detected in the non-treated, total RNA-seq data but were not reproducible in the RNase R treated sample occurred in transcripts in which high activity of RNA degradation was observed (25). For example, the beta subunit of the thermosome gene (SSO0282), coding for the archaeal chaperonin, contained 83 predicted cRNAs distributed all across its length. On the basis of our previous study (25) a high proportion of the RNA transcribed from this gene is fragmented, and it was shown that the half life of this gene is short (42), indicating rapid transcript turnover. It is therefore possible that the occurrence of seemingly ‘random’ cRNAs within protein coding genes stems from intermediate, short lived products of the RNA degradation process. This hypothesis is further corroborated by our attempts to verify ORF-residing cRNAs by RT–PCR, where amplification by outward-facing primers resulted in a ‘smear’ of multiple circular forms without a single distinct product.
In principle, our experimental approach can expose both perfectly circularized RNAs (with 3′–5′ end ligation), but also lariat circular formations. So far, to our knowledge no lariat formations were documented in archaea, although homologs of 2′–5′ ligases were found in species of this kingdom (43,44). Moreover, archaeal introns and C/D box RNAs were shown to form perfect circular RNAs, and the archaeal ligase was shown to have RNA circularization activity (14,18,22,45,46). Although we cannot exclude the possibility that some of the cRNAs we found have a lariat formation, we hypothesize that most of them represent 3′–5′ circularization.
Our circRNA-seq approach can be readily applied on any organism for which a sequenced reference genome is available. On the basis of our finding, in S. solfataricus, that circRNA-seq detects many more cRNAs than those that were previously known, we predict that application of circRNA-seq on other organisms might reveal many conserved and species-specific additional cRNAs. The biological functions of such putative cRNAs are yet to be determined.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S6, Supplementary Figures S1–S3.
FUNDING
ERC-StG program; Minerva foundation; EMBO YIP/IMOS program. Funding for open access charge: ERC-StG program.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Gil Amitai, Omri Wurtzel, Uri Gophna, Adi Stern, Eran Mick, Asaf Levy and Shulamit Michaeli for helpful discussions, Gadi Schuster and Victoria Portnoy for contribution of Halobacterium cells, and Shirley Horn-Saban and Daniella Amann-Zalcenstein for Illumina sequencing.
REFERENCES
- 1.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 2.Flores R, Hernandez C, Martinez de Alba AE, Daros JA, Di Serio F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005;43:117–139. doi: 10.1146/annurev.phyto.43.040204.140243. [DOI] [PubMed] [Google Scholar]
- 3.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen H, Fiskaa T, Birgisdottir AB, Haugen P, Einvik C, Johansen S. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA. 2003;9:1464–1475. doi: 10.1261/rna.5290903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen H, Johansen SD. Group I introns: moving in new directions. RNA Biol. 2009;6:375–383. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- 7.Vicens Q, Cech TR. A natural ribozyme with 3′,5′ RNA ligase activity. Nat. Chem. Biol. 2009;5:97–99. doi: 10.1038/nchembio.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hensgens LA, Arnberg AC, Roosendaal E, van der Horst G, van der Veen R, van Ommen GJ, Grivell LA. Variation, transcription and circular RNAs of the mitochondrial gene for subunit I of cytochrome c oxidase. J. Mol. Biol. 1983;164:35–58. doi: 10.1016/0022-2836(83)90086-4. [DOI] [PubMed] [Google Scholar]
- 9.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 11.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 12.Halbreich A, Pajot P, Foucher M, Grandchamp C, Slonimski P. A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA. Cell. 1980;19:321–329. doi: 10.1016/0092-8674(80)90506-1. [DOI] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett RA. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci. 1997;22:326–331. doi: 10.1016/s0968-0004(97)01113-4. [DOI] [PubMed] [Google Scholar]
- 14.Salgia SR, Singh SK, Gurha P, Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA. 2003;9:319–330. doi: 10.1261/rna.2118203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjems J, Garrett RA. Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc. Natl Acad. Sci. USA. 1991;88:439–443. doi: 10.1073/pnas.88.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J. Biol. Chem. 2004;279:47661–47671. doi: 10.1074/jbc.M408868200. [DOI] [PubMed] [Google Scholar]
- 17.Clouet d'Orval B, Bortolin ML, Gaspin C, Bachellerie JP. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 2001;29:4518–4529. doi: 10.1093/nar/29.22.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54:693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- 19.Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalgaard JZ, Garrett RA. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene. 1992;121:103–110. doi: 10.1016/0378-1119(92)90167-n. [DOI] [PubMed] [Google Scholar]
- 21.Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B, Branlant C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002;30:921–930. doi: 10.1093/nar/30.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starostina NG, Marshburn S, Johnson LS, Eddy SR, Terns RM, Terns MP. Circular box C/D RNAs in Pyrococcus furiosus. Proc. Natl Acad. Sci. USA. 2004;101:14097–14101. doi: 10.1073/pnas.0403520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clouet-d'Orval B, Gaspin C, Mougin A. Two different mechanisms for tRNA ribose methylation in Archaea: a short survey. Biochimie. 2005;87:889–895. doi: 10.1016/j.biochi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 25.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 30.Roy SW, Irimia M. When good transcripts go bad: artifactual RT-PCR ‘splicing’ and genome analysis. Bioessays. 2008;30:601–605. doi: 10.1002/bies.20749. [DOI] [PubMed] [Google Scholar]
- 31.Haas BJ, Gevers D, Earl A, Feldgarden M, Ward DV, Giannokous G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent HA, Deutscher MP. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J. Mol. Biol. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 34.Zago MA, Dennis PP, Omer AD. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:1812–1828. doi: 10.1111/j.1365-2958.2005.04505.x. [DOI] [PubMed] [Google Scholar]
- 35.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 36.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe Y, Yokobori S, Inaba T, Yamagishi A, Oshima T, Kawarabayasi Y, Kikuchi H, Kita K. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- 38.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 40.Mackie GA. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 2000;275:25069–25072. doi: 10.1074/jbc.C000363200. [DOI] [PubMed] [Google Scholar]
- 41.Puttaraju M, Been MD. Generation of nuclease resistant circular RNA decoys for HIV-Tat and HIV-Rev by autocatalytic splicing. Nucleic Acids Symp. Ser. 1995;33:152–155. [PubMed] [Google Scholar]
- 42.Andersson AF, Lundgren M, Eriksson S, Rosenlund M, Bernander R, Nilsson P. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 2006;7:R99. doi: 10.1186/gb-2006-7-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arn EA, Abelson JN. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J. Biol. Chem. 1996;271:31145–31153. doi: 10.1074/jbc.271.49.31145. [DOI] [PubMed] [Google Scholar]
- 44.Kato M, Shirouzu M, Terada T, Yamaguchi H, Murayama K, Sakai H, Kuramitsu S, Yokoyama S. Crystal structure of the 2′-5′ RNA ligase from Thermus thermophilus HB8. J. Mol. Biol. 2003;329:903–911. doi: 10.1016/s0022-2836(03)00448-0. [DOI] [PubMed] [Google Scholar]
- 45.Brooks MA, Meslet-Cladiere L, Graille M, Kuhn J, Blondeau K, Myllykallio H, van Tilbeurgh H. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008;17:1336–1345. doi: 10.1110/ps.035493.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Englert M, Sheppard K, Aslanian A, Yates JR, III, Soll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl Acad. Sci. USA. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berquist BR, Müller JA, DasSarma S. 27 Genetic Systems for Halophilic Archaea. In: Rainey FA, Aharon O, editors. Methods Microbiology. Jerusalem: Academic Press; 2006. pp. 649–680. [Google Scholar]
- 48.Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat. Methods. 2008;5: 679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 50.Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 51.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.