Abstract
Conventional cancer therapies are often limited in effectiveness and exhibit strong side effects. Therefore, alternative therapeutic strategies are demanded. The employment of tumor-colonizing bacteria that exert anticancer effects is such a novel approach that attracts increasing attention. For instance, Salmonella enterica serovar Typhimurium has been used in many animal tumor models as well as in first clinical studies. These bacteria exhibit inherent tumoricidal effects. In addition, they can be used to deliver therapeutic agents. However, bacterial expression has to be restricted to the tumor to prevent toxic substances from harming healthy tissue. Therefore, we screened an S. Typhimurium promoter-trap library to identify promoters that exclusively drive gene expression in the cancerous tissue. Twelve elements could be detected that show reporter gene expression in tumors but not in spleen and liver. In addition, a DNA motif was identified that appears to be necessary for tumor specificity. Now, such tumor-specific promoters can be used to safely express therapeutic proteins by tumor-colonizing S. Typhimurium directly in the neoplasia.
INTRODUCTION
Bacteria can cause devastating, sometimes fatal diseases. It is often overlooked that only a minority of bacteria is responsible for these severe illnesses. Most of them are not harmful and some are even beneficial for human health. For instance, the gut microflora counteracts the invasion of pathogenic microorganisms (1). Consequently, particular bacteria like the Escherichia coli Nissle 1917 strain are employed for medical treatment (2). An additional promising application of bacteria or bacterial products is the therapeutic treatment of cancer. Astonishingly, already >150 years ago first attempts to use bacteria for such therapeutic approaches have been made. They were based on the observation of several clinicians who correlated bacterial infection of cancer patients with regression of the tumor (3). The major drawback of the therapy at that time was the uncontrollable toxicity of the bacteria. Therefore, this treatment strategy was disregarded for many decades.
Today, as various possibilities exist to tailor bacteria for a particular purpose, bacteria-mediated tumor therapy undergoes a spectacular renaissance (4). For several bacterial strains, including not only obligate anaerobic bacteria like Clostridia or Bifidobacteria but also facultative anaerobic bacteria like E. coli or Salmonella, it could be shown that they preferentially colonize tumor tissue when administered systemically. This colonization can lead to the shrinkage or even the clearance of the neoplasia (5). Possible explanations for this growth preference can be found in the physiological composition of solid tumors. Most of such tumors contain central areas of low oxygen and scarce external nutrient supply resulting in the formation of necrotic regions. These areas provide optimal growth conditions for the bacteria as they represent niches with high amounts of nutrients from the dead cells and protection from the immune system.
Salmonella enterica serovar Typhimurium is among the bacteria that preferentially target and colonize tumors upon systemic administration (6). As consequence, the tumor usually shrinks but often regrows after a lag period. Different possibilities to improve Salmonella's inherent anticancer effects are under intensive investigation. For instance, dedicated mutant strains were established like A1-R, which was rendered auxotrophic for leucine and arginine and resisolated from tumors. It preferentially colonizes various human tumors and metastates in nude mice but is cleared rapidly from other organs (7–10). In addition, the therapeutic power of the bacteria is strengthened by enabling the bacteria to express recombinant therapeutic molecules (5). However, such bacteria would also colonize spleen and liver, the normal target organs of S. Typhimurium. Thus, to fully exploit an enhanced antitumor potential such bacteria need to be manipulated to express the therapeutic molecules exclusively in the tumor tissue and not in healthy organs. This restricted expression represents an absolutely necessary safety aspect.
In the present work, this aspect was tackled by searching for tumor-specific Salmonella regulatory sequences. A promoter-trap library was used to systematically screen the Salmonella genome for sequences that activate the expression of their corresponding gene only when the bacteria reside within a tumor and not in other organs. Here, we indentified 12 genomic fragments that exhibited the desired expression pattern in vivo. Fragmentation of four of such sequences, as examples, leads to a further confinement of the genomic sequence that is necessary for tumor specificity. In addition, a DNA sequence motif was identified that dominantly contributes to the specific pattern of expression.
MATERIALS AND METHODS
Ethics statement
Procedures involving animals and their care were fully in compliance with the German Animal Welfare Act (Tierschutzgesetz, 1998) and with the permission number 33.9.42502-04-050/09 of LAVES (Niedersaechsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit).
Bacterial strains and growth conditions
Salmonella Typhimurium strain SL7207 (hisG, ΔaroA) was kindly provided by Bruce Stocker (11) and was grown with vigorous shaking in LB medium containing 30 µg/ml streptomycin (Sigma-Aldrich, Germany) and 50 µg/ml ampicillin (Sigma-Aldrich, Germany) when needed for plasmid selection at 37°C or on respective LB agar plates. If needed, hypoxic conditions were established by sparging an aerobic liquid culture in early log phase with nitrogen before further incubation at 37°C under air exclusion.
Cell lines and animals
Six-week-old female BALB/c mice were purchased from Harlan (Germany). CT26 colon carcinoma cells (ATCC CRL-2638) were grown as monolayers in IMDM medium (Gibco BRL, Germany) supplemented with 10% (v/v) heat-inactivated fetal calf serum (Integro, Netherlands), 250 µmol/l β-mercaptoethanol (Serva, Germany) and 1% (v/v) penicillin/streptomycin (Sigma-Aldrich, Germany).
Promoter-trap library construction
Briefly, S. enterica serovar Typhimurium SL1344 genomic DNA had been sheared and fragments of 500–700 bp had been inserted upstream of a promoter less gfp_ova gene on a medium copy number plasmid. The resulting plasmid library contains about 1.1 × 106 independent transcriptional fusions (12). This DNA was used to transform S. Typhimurium SL7207 resulting in about 1 × 106 independent transformants.
Mouse experiments
Female BALB/c mice aged 6–8-weeks were subcutaneously (s.c.) inoculated at the abdomen with 5 × 105 CT26 cells. When tumors reached volumes of approximately 200 mm3, infection experiments were started. For the first sorting, an aliquot of the promoter-trap library was thawed on ice for 30 min. Subsequently, it was included in prewarmed Lysogeny broth (LB) medium and incubated for 30 min at 37°C. After washing with PBS three times, the inoculum was adjusted to obtain an infectious dose of 1 × 107 either in 10 µl for intratumoral (i.t.) infections or in 100 µl for intravenous (i.v.) infections. For infection with single strains, the respective strain was grown overnight on agar plates, resuspended in phosphate-buffered saline (PBS) and adjusted to an infectious dose of 5 × 106.
Sorting and flow cytometric analysis
Twenty-four hours after infection, respective organs and/or tumors were removed and homogenized in 2 ml ice-cold PBS. Then, the homogenates were diluted to 1 : 10 (spleen, liver) or 1 : 100 (tumors) in 0.1% Triton-X/PBS containing 2 mM EDTA, filtered through a 30 -μm CellTrics filter (Partec, Germany) and sorted or analyzed via two-color flow cytometry on a FACSAria or LSRII (Becton Dickinson, USA). Two-color flow cytometry is a method that allows distinguishing GFP-expressing bacteria from autofluorescent cellular debris since GFP-expressing Salmonella have a substantially lower orange/green emission ratio (13). Additionally, forward and side scatter were used to distinguish Salmonella from larger particles by setting an appropriate scatter gate.
Sequencing and analysis of sequence data
Plasmid DNA of sorted bacteria forming colonies on LB agar plates were prepared and inserts were sequenced on Genome Analyzer (Illumina Inc., USA) using primer SLE06 (5′ GTGATGTCGGCGATATAG 3′) and SLE07 (5′ GAATTGGGACAACTCCAG 3′) (12). Sequences were aligned using BLAST with Salmonella chromosome and plasmid sequence (NCBI database accession number AE006468 and AE006471). Annotation of fragments was done using self-developed Perl scripts and data provided on NCBI database.
For identification of regulatory motifs, program MEME and FIMO (14) was used with all default parameters. Graphical representation of the motifs was built using WebLogo (15).
RNA preparation
RNA of strain SL7207 was isolated from fixed samples with the RNeasy mini Kit (Qiagen, Germany) according to the manufacturer's instructions using double amounts of buffers when appropriate. For preparation of bacterial RNA from infected tumor tissue, the tumors were cut into small pieces and the necrotic tissue was squeezed through a cell strainer, 70 µm (BD Falcon, USA) in presence of 3 ml RNA protect bacteria reagent (Ambion, USA). For RNA isolation of bacterial RNA from infected spleen tissue, the organs were homogenized with a tissue homogenizer in presence of 2 ml RNA protect bacteria reagent on ice. The samples were centrifuged briefly at 1000 rpm to separate the bacteria from tissue debris following a centrifugation step at maximum speed for 5 min to pellet the bacteria. RNA quality was accessed using the Bioanalyzer (Agilent, USA).
qRT–PCR analysis
To validate the data of the promoter-trap library, qRT–PCR was performed. Complementary DNA (cDNA) was prepared from RNA using the RevertAid First Strand cDNA synthesis Kit (Fermentas, Canada) according to the instructions of the manufacturer. qRT–PCRs were carried out with the Power Sybr Green PCR Master Mix (Applied Biosystems, USA) and a 7500 Real-Time PCR System (Applied Biosystems, USA). The data were analyzed by the Sequence Detection Software, version 1.4 (Applied Biosystems, USA), according to manufacturer's specifications. cDNA levels were normalized to the cDNA levels of the housekeeping gene gyrB. Normalized values were used to calculate the ratios of the expression levels in the colonized tumor tissue relative to the expression during colonization of the spleen.
Construction of different insert fragments
To construct plasmids that contain fragments of the original library inserts, oligonucleotides of the desired sequence were ordered (Eurofins MWG Operon, Germany) containing a BamHI recognition site at the 5′ and an XbaI site at the 3′-end (Table 1). For longer sequences, primers were designed accordingly to amplify the fragment from the original plasmid (Table 1). As vector the original plasmid pMW82 was used (12), opened via restriction digestion with BamHI and XbaI followed by a ligation with the respective fragments. SL7207 was transformed with the plasmids and in parallel plasmid DNA was sequenced to confirm correct amplification products.
Table 1.
Oligonucleotides used in this study
| Fragment/gene | Sequence of sense primer | Sequence of the reverse primer |
|---|---|---|
| Construction of library insert fragments | ||
| 48a | GGATCCattgtttttcctcacagttcgtttt | TCTAGAattgtagcctccgtggcccat |
| 48b | GGATCCtatgttgagtatttttaaaccagcg | TCTAGAtcccgccgccgacgttatgc |
| 48c | GGATCCattgtttttcctcacagttcgtttt | TCTAGAtcccgccgccgacgttatgc |
| 134a | CGGGATCCcgttattcttaaaaatgagcgtaa | CGTCTAGAtcataacggcgaagttagcg |
| 134b | CGGGATCCccggcgaagcgg | CGTCTAGAaataacgggtaatgtcatttgttt |
| 134c | GATCtaacttcgccgttatgatcggtcgtcttttaagcaac tattgacacacac | CTAGgtgtgtgtcaatagttgcttaaaagacgaccgatca taacggcgaagtta |
| 134d | GATCggttaaggtcaaagaaagacacgttctgataattctt acttgtcattcgctaacttcgccgttatgatcggtcgtc ttttaagcaactattgacacacac | CTAGgtgtgtgtcaatagttgcttaaaagacgaccgatcat aacggcgaagttagcgaatgacaagtaagaattatcaga acgtgtctttctttgaccttaacc |
| 134e | CGGGATCCcgttattcttaaaaatgagcgtaa | CGTCTAGAgtgtgtgtcaatagttgcttaaaagacgaccg atcataacggcgaagttagcg |
| 212a | GATCctttcattcgaaagtaatttaatctttatatga aataagagaggccgttt | CTAGaaacggcctctcttatttcatataaagattaaattac tttcgaatgaaag |
| 212b | GATCtaacgcctctttgtcagaacctctccattcgttgac gcacatcaagatagctttcattcgaaagtaatt taatctttatatgaaataagagaggccgttt | CTAGaaacggcctctcttatttcatataaagattaaattac tttcgaatgaaagctatcttgatgtgcgtcaacgaat ggagaggttctgacaaagaggcgtta |
| 301a | GGATCCggcccatattttatttagaggtaaac | TCTAGAattgtttttcctcacagttcgttttt |
| 301b | GGATCCtgaaaactcgcgactcgcaaac | TCTAGAatccgttttaaaatctggttttcac |
| 301c | GGATCCggcccatattttatttagaggtaaac | TCTAGAatccgttttaaaatctggttttcac |
| qRT-PCR | ||
| mltD | TGCCAGACGTCTCAGAACAC | CATCGTGCCTGACTCGTAAA |
| PflE | ATTTTCTGCCCTACCACACG | GCGTACTTCTGGGCAAAATC |
| glpA | CTGGTACCTGGCGATACGAT | ACGCAGCAGGATATCGACTT |
| glpT | CAACGAAAAAGCGGAAGAAG | GCGCAGCAGATAAACAAACA |
Listed are oligonucleotides for the construction of library insert fragments and for qRT-PCR.
RESULTS
Screening the Salmonella genome for tumor-specific regulatory elements
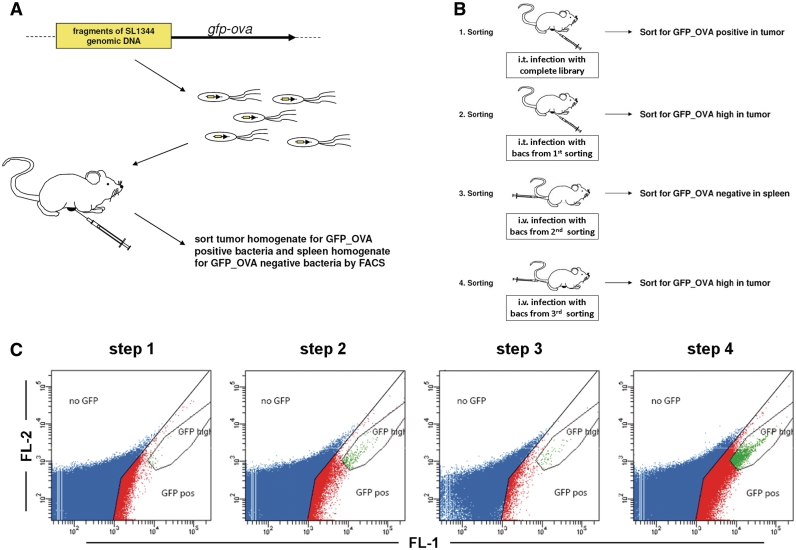
To define tumor-specific regulatory elements in S. Typhimurium a promoter-trap library was employed that has been described before (12). It consists of random, 500–700-bp long fragments of sheared genomic DNA from the S. Typhimurium strain SL1344 that were cloned upstream of a gfp_ovalbumin (ova) fusion reporter gene. The fusion of GFP with OVA renders the reporter protein unstable. Thus, the selection of clones carrying weakly active fusions that only accumulate GFP over time and clones with residual reporter activity that might have been induced in bacterial cultures or in locations other than the tumor can be prevented. The attenuated strain SL7207 was transformed with these library plasmids and used for selection of tumor-specific regulatory elements. Figure 1B displays the experimental schedule.
Figure 1.
Screening of a SL7207 promoter-trap library to identify tumor-specific clones. (A) Schematic depiction of the promoter-trap library. 500 to 700 bp fragments of sheared S. Typhimurium genomic DNA were inserted into a plasmid upstream of a promoterless gfp_ova reporter gene. SL7207 were transformed with these plasmids resulting in a promoter-trap library with >20-fold genome coverage. (B) Strategy to isolate tumor-specific regulatory elements. First, two steps of positive selection of reporter gene-expressing bacteria were carried out after intratumoral application of the bacteria (steps 1 and 2). The recovered bacteria where then negatively selected by infection of tumor-free mice and isolation of reporter negative bacteria (step 3). This should exclude elements that generally drive expression in vivo. Finally, bacteria exhibiting high expression of the reporter were enriched from tumors 24 h after i.v. application of the recovered bacteria (step 4). (C) Analyses of the different sorting steps. In the first step events in the red gate (GFP pos) were sorted and in the second step the green region (GFP high) was sorted. For the negative step the blue (no GFP) and the fourth step again the green region was taken as sorting gate.
First, 1 × 107 transformants were injected directly into the subcutaneous CT26 tumor of BALB/c mice (Figure 1A). This appeared to be necessary to preserve the library diversity. We had observed that the number of tumor-invading bacteria after systemic application is limited [(16) and Leschner, S. et al., manuscript in preparation]. Tumors were homogenized 24 h post-infection (p.i.) and green fluorescent bacteria were isolated by cell sorting using the two-color flow cytometry to distinguish autofluorescent cell debris from GFP signal (13). For gating, a broad region was employed to ensure that most positive bacteria were within the gate to enrich the library (Figure 1C). At this step, 1.7 × 105 positive events were sorted resulting in 1.2 × 104 colonies upon plating.
Bacteria were scraped off these plates and an aliquot of this sample was used to inject into CT26 tumors for a second enrichment cycle. Again, 24 h p.i. tumor homogenates were sorted with a more restricted setting to enrich transformants that strongly express the reporter gfp_ova. From the 8 × 103 positive-sorted events, 3.2 × 103 colonies were obtained.
To counterselect against transformants, with high expression elsewhere in the murine host, bacteria plated after the second sorting were used as an inoculum to intravenously infect mice not bearing a tumor. Spleens were homogenized 24 h p.i. and sorted for GFP negative events. From 1 × 107 sorted events, 3 × 104 colonies were obtained. The low fraction of viable Salmonella was due to the fact that in this sorting strategy, GFP-negative bacteria could not be separated from an overwhelming number of debris of the same size.
The colonies were scraped of the plates again and an aliquot was used to infect CT26 tumor bearing mice i.v. Again, 24 h p.i. tumor homogenates were sorted with restriction of the sorting to GFP-high expressing bacteria (Figure 1C). From the 1 × 104 sorted events, 3 × 103 colonies were formed. This strategy resulted in an efficient enrichment of GFP-positive and especially high GFP events in each step (Figure 1C).
Sequence analysis and classification of regulatory elements
Inserts of the library plasmids from all 3000 bacterial colonies that were obtained during the last sorting step were sequenced. This defined 314 unique fragments. The sequences of these fragments were aligned with the published genome sequence of S. Typhimurium LT2 using BLAST. Most of the fragments showed >95% sequence homology to the Salmonella chromosome. From the residual fragments, 24 were fused genomic fragments of which only the 3′ part was considered for further analysis. Five fragments matched the sequence of the Salmonella plasmid (pSLT) and 27 showed no match. The length of the fragments ranged from 64 to 1400 bp with an average of 583 bp.
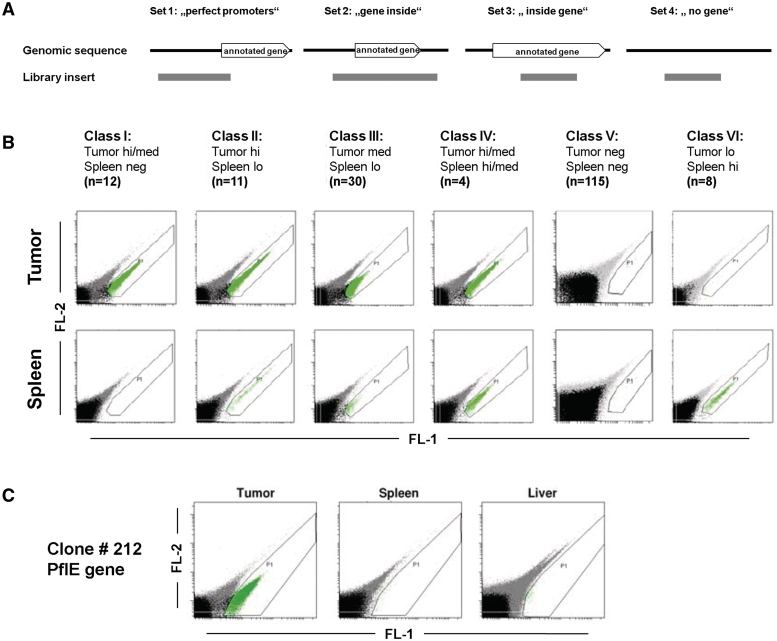
Annotation of the Salmonella genome was used to localize fragments within the genomic sequence and to identify genes for which a fragment may function as a promoter. According to the location of the fragments and in relation to associated genes, four different sets were defined (Figure 2A). A fragment was assigned to set 1 when its 3′-end resided within a gene sequence and the 5′-end was upstream of that gene. A total of 117 fragments fell into this category termed ‘perfect promoters’. Set 2 fragments were specified as containing the whole gene sequence and 24 fragments of that kind were identified (gene inside). In both the above cases, the gene closest to the 3′-end of the fragment was considered when a fragment overlapped with more than one gene. Set 3 contains fragments that were found to lie completely inside of a gene sequence. Thirty-eight of such fragments were found and termed ‘inside gene’. Fragments with sequences that showed no obvious association with a gene fell into set 4: ‘no gene’ (107 fragments).
Figure 2.
Classification of the obtained promoter sequences. (A) Definition of four different sets, according to the position of the respective sequence on the genome. (B) Classification of promoter sequences by expression pattern. After individual flow cytometric analysis, 180 insert sequences that matched the Salmonella genome in coding sequences or in close vicinity of it, were classified according to their expression level in tumor and spleen. Pictures show exemplary data of flow cytometric analysis, gated are GFP_OVA positive bacteria. (C) Exemplary depiction of results from tumor-specific clones in tumor versus spleen and liver tissue. Shown is the example of clone 212.
Using the enzyme commission (EC) nomenclature, these sets were further analyzed for overrepresented genes whose products might belong to the same enzymatic class. Two groups of enzymes could be identified: oxidoreductases (13 genes or 32.5%, expected 22.2%) and transferases (17 genes or 42.5%, expected 31.7%). This categorization should allow a reasonable order to verify promising tumor-specific regulatory sequences. This confirmation step was necessary as first random verifications showed that a relatively high number of sorted transformants did not show any expression in spleen and in tumor. Taking the overrepresentation as a sign of importance, transformants falling into these enzymatic classes and in addition belonging to set 1 (perfect promoter) were tested first for their expression. Subsequently, the residual clones from set 1 were queued for testing, then all clones from set 2, then from set 3 and finally from set 4.
Validation of individual regulatory sequences
Transformants representing individual regulatory sequences were intravenously injected into tumor-bearing mice and the expression of the GFP reporter was determined from homogenates of tumor and spleen. Depending on expression levels (high, medium, low and negative), each clone was classified into one of six classes (Figure 2B). Reporter expression of all Class I clones (tumorhi, spleenneg) were reanalyzed to confirm tumor specificity, including liver homogenates in addition. Strong reporter gene expression could again be found in tumor. No GFP signal could be detected for any of the clones in spleen and liver (Figure 2C).
Along with experimental validation of single clones, a positive discovery rate was monitored that appeared to degrade drastically when analyzing sets other than set 1 (Table 2). If estimated to be linear, using data for the entire set 1 and for sets 2 and 3 together, the expected number of tumor-specific clones to be found in set 4 would be about 1. On this basis, further experiments with clones that cannot be associated with a gene (set 4: ‘no gene’) were thought to be unreasonable and were not followed any further. Interestingly, after testing the two overrepresented EC classes (30 clones) 50% of all hits were discovered among those fragments.
Table 2.
Number of clones and positive discovery rate in data sets according to Figure 2A
| Data set | Number of clones tested | Number of Class I specific clones identified | Discovery rate per 10 experiments |
|---|---|---|---|
| Overrepresented classes from set 1 | 30 | 6 | 2.00 |
| Residual clones from set 1 | 87 | 4 | 0.46 |
| Set 2 and set 3 | 62 | 2 | 0.32 |
| Set 4 | 107 | ∼1a | ∼0.12a |
aExpected values.
Confirmation of exemplary clone sequences
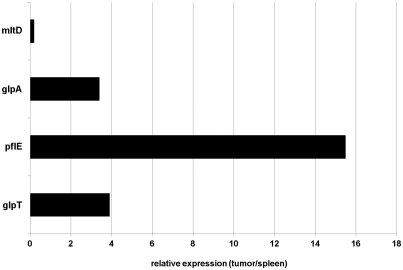
We made the assumption that each regulatory sequence we found can be assigned to the gene that is found downstream of it or that it comprises. This is represented in Table 3. To confirm that this assumption was correct, we carried out quantitative RT-PCR for four exemplary clones 48 (glpT), 134 (mltD), 212 (pflE) and 301 (glpA). Apart from gene mltD that did not show to be specifically upregulated in tumor tissue, tumor-specific expression could be confirmed for all the other genes (Figure 3). A complete correlation of the expression pattern given by the library plasmid in flow cytometric analyses and the result of the quantitative RT-PCR of the respective gene was not expected. This can be explained not only by the differences comparing the situation on the genome compared to the plasmid-like topology/context of the sequence but also by differential mRNA stabilities.
Table 3.
Clones of Class I with their respective gene and its function
| Clone no. | Gene | Function | Fold induction by hypoxia |
|---|---|---|---|
| 4 | ydiH | Putative cytoplasmic protein | 59.7 |
| 48 | glpT | sn-glycerol-3-phosphate transporter | 27.4 |
| 92 | bcsG | Membrane protein; endoglucanase | 0.3 |
| 134 | mltD | Membrane-bound lytic murein transglycosylase D | 1.1 |
| 154 | mdh | Malate dehydrogenase | 1.9 |
| 156 | mtfA | Mlc titration factor A | 15.4 |
| 172 | frdA | Fumarate reductase flavoprotein subunit | 1368.1 |
| 185 | pfkA | Similar to E.coli 6-phosphofructokinase I | 14.6 |
| 212 | pflE | Putative pyruvate formate lyase activating enzyme | 150.1 |
| 271 | nirB | Nitrite reducatase large subunit | 65.3 |
| 301 | glpA | sn-glycerol-3-phosphate dehydrogenase subunit A | 886.6 |
| 310 | ybaL | Putative cation::proton antiport protein | 0.4 |
Clones that belong to Class I (Figure 2B) show high or medium expression levels in tumor and no expression in spleen and are, therefore, introduced in this list with their corresponding gene and its function if known. Bacterial cultures of each clone were grown in vitro under aerobic and hypoxic conditions. Expression patterns were compared, dividing hypoxic by aerobic values resulting in fold induction during hypoxia. Exemplary flow cytometric results are shown in Supplementary Figure S1.
Figure 3.
Confirmation of tumor-specific expression four exemplary genes from Class I by qRT-PCR. The expression of the genes mltD, glpA, pflE and glpT in SL7207 colonizing tumors compared to spleens was tested.
Induction of reporter gene expression under hypoxic growth conditions
Due to their irregular vasculature, regions of low-oxygen partial pressure are a characteristic of many solid tumors. Therefore, promoters responding to a hypoxic environment should be found among the tumor-specific elements that we had defined. To test this possibility, liquid cultures of all Class I clones were grown under aerobic and hypoxic conditions and subsequently analyzed for their reporter gene expression by flow cytometry. Of the 12 clones, 8 showed strong differential GFP expression under oxygen-deprived conditions (Table 3). Downregulation by hypoxia could be observed in two clones whereas two showed equal expression under both conditions. Together, an unexpected complexity of microenvironments encountered by the bacteria in the tumor can be deduced from such findings.
Discovery of regulatory motifs
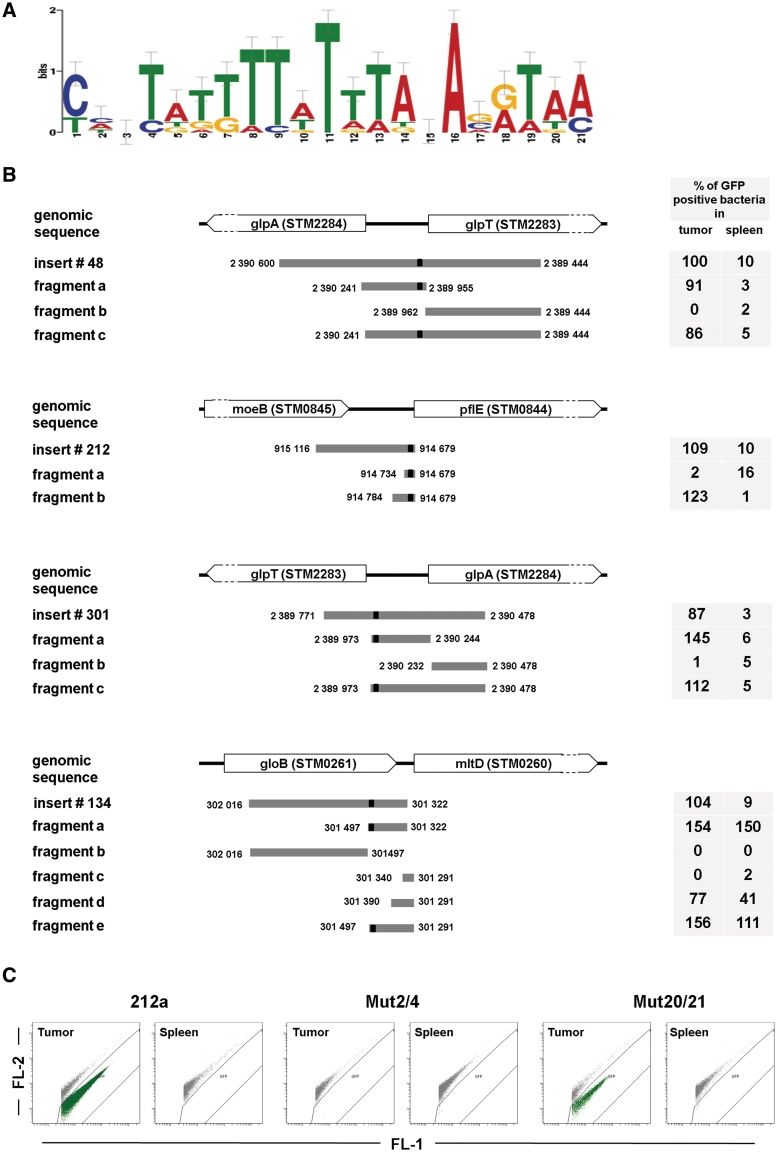
Proper gene expression is usually realized by specific binding of transcription factors (TFs) to their motifs on the DNA. To find such motifs, which in our case should be responsible for tumor-specific expression, the program MEME was applied for sequences of Class I clones. This resulted in three motifs. We expected, that true motif(s) should be specific only to Class I sequences, but not, for instance, to inserts of Class V exhibiting no expression. Using Class V sequences as a negative control, only one motif was identified, which showed the postulated specificity (Figure 4A). Each sequence (except clone 156) of Class I clones contains at least one copy of this motif (two have two copies), while only 6% (7 out of 113) of Class V sequences contain it. For convenience the motif (Figure 4A) will from now on be called tusp (tumor-specific) motif. Analysis of positional distribution of the tusp motif in Class I fragments did not revealed any localization preferences. The other two motifs either cover <80% (10 out of 12) of Class I sequences or more than 33% of Class V sequences.
Figure 4.
Tumor-specific motif and fragmentation of exemplary insert sequences. (A) Logo of the sequence motif found to be overrepresented in Class I clones. (B) Schematic depiction of four exemplary insert sequences, their respective genomic locus and how they were fragmented. Numbers at the fragment ends are genome position numbers. Right-hand tables show the percentage of Salmonella expressing GFP_OVA in tumor compared to spleen tissue for clone number 48, 134, 212, 301 and their respective fragments. Orientation is 5′–3′ direction of the insert according to its position on the library plasmid. The black box on the sequences shows the position of the 21 bp motif the sequence of which is shown in (A). Flow cytometric analysis of tumor and spleen homogenates from mice infected with SL7027 bearing plasmids that contained either the fragment 212a or its mutated forms (C).
Narrowing down the tumor-specific bacterial regulatory element
After the identification of 12 DNA fragments that drive tumor-specific expression, the aim was to define the shortest promoter possible, without losing functionality. This should help to identify key elements responsible for specificity. Since the Salmonella promoter-trap library was built from randomly generated genomic fragments, the potential for refinement appeared to be substantial. Therefore, four clones from Class I with strongest expression in tumors were selected. Depending on the location of the respective upstream and downstream genes, the four inserts were split into several fragments, such that the hypothesized promoter with the motif remained covered at least by one of the fragments and the discriminative potential was maximal (Figure 4B).
Tumor-bearing mice were i.v. infected with SL7207 transformed with each of the newly constructed plasmids, followed by a flow cytometric analysis of tumor and spleen homogenates after 24 h. To allow a quantitative statement about the percentage of bacteria that are GFP_OVA positive in the respective tissue, a defined volume of the homogenate was analyzed and plated in parallel. For the clones number 48 (glpT) and 301 (glpA), fragment ‘a’ representing the sequence directly upstream of the translation start (TLS) of the associated gene showed an expression pattern comparable to the complete original insert (Figure 4B). The sequences downstream of the TLS (fragments ‘b’) did not drive any expression. Fragments ‘c’ containing both sequences ‘a’ and ‘b’ again, were comparable in expression to the original insert sequence. For clone 134 and 212, where only the first 50 and 100 bp upstream of the TLS were tested, both 50 bp fragments did not show any expression, although the fragment from 212 contained the tusp motif. In additional experiments using 50 bp fragments we observed that none of them showed any expression (S. Leschner, unpublished data). In contrast, the 100 bp fragment 212 b showed an expression level comparable to the original insert.
For clone 134 (mltD) not only fragments of the original sequence were constructed but also the sequence was prolonged for 32 bp that included the region up to the TLS of the mltD gene (fragments c–e). Apart from the silent fragment c—which being 50 bp is obviously too short for expression—both prolonged fragments showed a stronger (S. Leschner, unpublished data) but less-specific expression.
Analysis of clones 48, 134, 212 and 301 and their fragments, provided evidence that the tusp motif identified above may contribute to gene expression in tumors (Figure 4A). Indeed, it exists in all subsequences of clones that showed similar expression patterns and does not exist in subsequences showing no expression. Only in one exception (clone 134e), expression was not restricted to tumor tissue, although the fragment contained the tusp motif (Figure 4B).
Finally, to prove the importance of the tusp motif, point mutation analysis was carried out. To this end, the DNA sequence of fragment 212b (Figure 4B) was studied in more detail since it is only about 100 bp long but still exerts the tumor-specific expression. The tusp motif is located on the negative strand at positions 399–419 (absolute genomic positions 915 183—915 203, TTTAATCTTTATATGAAATAA). To find positions within the tusp motif that most probably contribute to functionality, similarity to known regulatory motifs was analyzed using TOMTOM (17). In general, no hits with a reliable level of similarity could be identified that cover the entire motif. However, matches with lower similarity were preferably located on the edges of the motif. Hence, the following modification of the original motif was suggested: TGTCATCTTTATATGAAATAA (Mut2/4) and TTTAATCTTTATATGAAATGC (Mut20/21). (Bold letters indicate mutated positions).
Newly constructed plasmids bearing the mutated tusp motif within the fragment 212 b were tested in vivo in tumor-bearing mice. Results of flow cytometric analysis of tumor and spleen tissues revealed that single nucleotide substitutions at positions 2 and 4 of the original motif (Figure 4A) resulted in an abort of expression in the tumor while expression in spleen remained negative. Mutations at positions 20 and 21 had no effect (Figure 4C). This provides solid evidence for the functionality of the identified tusp motif. Further analysis of the motif against known motif libraries did not reveal significant matches. Thus, we speculate that the motif that drives tumor-specific expression is recognized by a transcription factor the binding pattern of which is not yet known.
DISCUSSION
For different types of bacteria it could be shown that they possess the ability to target and colonize tumors and even induce shrinkage of the cancerous tissue. This fact makes them an attractive tool for a novel therapeutic approach namely bacteria-mediated cancer therapy. For instance, the highly attenuated S. Typhimurium strain VNP20009 has been tested in the clinics with cancer patients. Although the results were rather disappointing, some of the patients that had received a high-titered bolus showed tumor colonization (18,19). In such cases, tumor-specific expression of therapeutic molecules should have been beneficial. Importantly, using the same bacterial strain to treat pet dogs with spontaneous tumors showed an incidence of 43% of tumor colonization and 15% of the dog patients responded favorably (20). Again, supportive expression of anticancer molecules might have dramatically increased the positive effects. This demonstrates the importance of appropriate expression systems for such molecules in the tumor-targeting bacteria.
Despite their therapeutic potential, the bacteria used are still pathogens and have to be rendered absolutely safe. In the present work, we demonstrate that the expression of therapeutic molecules by S. Typhimurium can be restricted to cancerous tissues using tumor-specific promoters. A similar concept using high-throughput chip technology has been already published by Arrach et al. (21). We screened a Salmonella promoter-trap library allowing the discovery of such tumor-specific regulatory elements without any prerequisite knowledge of the physiological conditions that induce them. The consecutive steps employed in the screening procedure resulted in the definition of 12 tumor-specific clones. Our findings demonstrate that tumor-specific promoters of S. Typhimurium exist and the strategy employed is suitable for the identification of such regulatory elements.
From the 184 clones analyzed, 23 clones were classified into either Class I or II, which were defined to have strong/medium tumor expression and no/negligible or weak expression in spleen, respectively. Here, the importance of a quantitative analysis became obvious. Only when a defined number of bacteria was analyzed, a reliable characterization of the performance of the control element could be revealed. The minor fraction of positive clones in spleen, especially of promoters that fall into Class I, can be explained by bacteria that had recently immigrated from the tumor tissue into the spleen. In addition, the number of GFP positive clones might be overestimated due to the discrepancy between flow cytometric measurement and plating results. Thus, such promoters should be considered truly tumor specific.
Many of the newly defined tumor-specific regulatory elements apparently respond to the anaerobic environment in the tumor (Supplementary Table S1). This was expected as most of the Salmonella reside within the necrotic core or at its border zone where oxygen supply is scarce (22–24).
On the other hand, we expected that among the tumor-specific clones several genes would be found that belong to the same metabolic pathways. However, this was not the case. The most ‘enriched’ pathways were the citrate cycle, glycolysis and the reductive carboxylate cycle, however, each pathway comprised not more than two clones. A trivial reason for that might be the low number of S. Typhimurium genes with assigned EC classes. From the genes associated to clones of Classes I and II, only 9 out of 23 are categorized in this system. Thus, the association of such uncharacterized genes with particular metabolic pathways might have been missed. Alternatively, some genes of such pathways might be preferentially expressed in cancerous tissue but not completely silent in healthy organs and thus excluded by our stringent selection.
Using bioinformatics tools, we searched for a common denominator within the Class I type of regulatory elements. We found a motif—now called tusp—which was present in all of such sequences except one and was represented in the genome all together only 42 times. Of such hits, 33 matches were located in potential promoters of genes, 21 of which were annotated (Supplementary Table S2). Using Gene Ontology (GO) classification, we found that protein products of five of such genes were associated with ‘structure-specific DNA binding’ with P-values of 3.4 × 10−5. Thus, we can speculate that tusp may have a role as a key regulator, which triggers a transcriptional cascade of downstream genes.
Investigation of the tusp motifs itself unfortunately gave no prominent results. No matches to established regulatory motifs could be detected using high stringency for alignment. Thus, we might have discovered a novel bacterial DNA element that is involved in the regulation of bacterial survival in a microenvironment that is simulated by the necrotic part of a tumor. By lowering the stringency of alignment, resemblance with known DNA motifs (e.g. cpxR, tyrR from E. coli or SygB from Bacillus subtilis) could be found at both borders of the tusp motif. In accordance, alteration of two nucleotides at the 3′ border of tusp (Mut2/4) completely silenced reporter expression. This clearly demonstrates that the tusp motif is important for the specific function of the regulatory element.
Furthermore, the question arises what the biological function of the tusp motif is and which environmental stimuli are linked to it. Different conditions are known to be present in solid tumors that should be considered. Hypoxia is a well-known state present in many cancers. In accordance, reporter gene expression in 8 of the 12 Class I promoters is induced after depletion of oxygen (Table 3). Low extracellular pH is also often observed in neopolastic tissues as a result from the high glycolytic flux (25). This might trigger the promoters not responding to hypoxia. Alternatively, we could recently show that the bacteria colonizing tumors are under severe stress most likely due to defense mechanisms of the host (26). This induces the formation of biofilms by the bacteria and could also provide key signals that activate some of the tumor-specific promoters.
Interestingly, an evaluation of the functions of genes comprising the tusp motif (Class I clones and matches according to genome blast – Supplementary Table S2) suggests a trend where Salmonella are preparing for relatively slow growth under anaerobic conditions. Furthermore, they appear to react to a deprivation of amino acids but a high supply of lipids like glycerol and fatty acids (27).
A similar approach to define tumor-specific promoters using high-throughput chip technology was reported recently (21). Screening was carried out in nude mice bearing the transplantable human prostate carcinoma PC3. When our list of tumor-specific promoters was compared with the list of Arrach et al., five fragments were found in both reports. If we consider such hits as real, a discrepancy in the discovery accuracy could be noted. While in our case an accuracy of 47% (5/12) can be noted, only 5.7% (5/87) can be deduced from Arrach et al. However, this is most likely due to methodological differences of the two screening procedures. Our approach is highly stringent and considers classified elements only when expression in spleen is completely absent. In contrast, Arrach et al. used arrays to test for spleen expression, which probably also includes some low-level expressers showing that this approach lacks the possibility for real quantification as it can be provided by FACS or qRT-PCR analyses. For example, 17 fragments (20% of hits) reported by Arrach et al. are found to have expression not restricted to tumor tissue. This might also explain why the tusp motif is not found in most of the regulatory elements described by Arrach et al.
Unexpectedly, in our work, a high number of unresponsive clones were present in the final positively sorted sample. This indicates that the sorting performance was suboptimal. Negative events should have been excluded. However, at least the redundancy of such clones was not as prominent as that for positive clones. Nevertheless, a second round of screening was carried out with experimental alterations (S. Leschner, unpublished data). First, the organ preparation included a centrifugation step to exclude most of the cellular debris and second, sorting parameters were adjusted using a GFP threshold. The analysis of 100 randomly selected clones of this altered sorting revealed that a strong enrichment had occurred. Almost 50% of these clones could be identified as clone 212 from the first screen. Another 16% exhibited the sequence of clone 301. The third most prominent clone (10%) was unknown so far but also found to be unspecific. The residual clones occurred only once or twice, and only two of them showed tumor-specific expression. They could be assigned to the genes spvD and arcA. Therefore, the initial screening strategy with a lower gating stringency was more efficient although more laborious.
Cooperative application of lab and bioinformatic techniques proved to be a fruitful and cost-effective approach. Used complementarily in the present work, tumor-specific clones were identified and a possible mechanism of their regulation via a novel DNA motif called tusp is suggested. In addition, some minor aspects regarding fine regulation and regulation in the absence of the tusp motif, could be made. The 100 bp part of clone 134 (fragment 134d) lost some of the tumor specificity, which was observed for the original clone 134. This fact suggests that the regulation of tumor-specific promoters might be more complex. Additional regulatory elements may be involved, which in general improve or repress transcriptional initiation. This also becomes clear when looking at fragment 134e that contain 32 additional bases spanning the region down to the TLS of the mltD gene. In this case, the expression is stronger but less specific compared to the original insert. We may speculate that other features of DNA, like melting or specific conformation may play a role, as well. The presented experimental data show that such putative elements are not common for all fragments, but introduce their minor effects in each individual fragment.
The sequences identified in this work allow the tumor-specific expression of potential therapeutic molecules by S. Typhimurium. This is a crucial safety aspect for the use of bacteria as vehicles in anticancer therapy and needs to be thoroughly investigated. Improving expressivity of the identified promoter sequences is a forthcoming activity and unraveling the complete regulatory logic underlying the promoters is the first step to this goal. In addition, this will allow computer generation of many artificial promoters targeting other types of cancers in many different model organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Tables 1 and 2 and Supplementary Figure 1.
FUNDING
Federal Ministry of Education and Research (BMBF) (in part); Deutsche Krebshilfe, the German Research Council (DFG); Helmholtz Gemeinschaft via HIRSIB; Hannover Biomedical Research School (HBRS). Funding for open access charge: Helmholtz Centre for Infection Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Regina Lesch, Susanne zur Lage, Martina Krey, Ansgar Conrad, Maren Scharfe and Gabriele Nordsiek for expert technical and scientific help. Sara Leschner designed, performed and analyzed experiments and wrote the manuscript. Igor V. Deyneko performed bioinformatic analyses and motif prediction and wrote the manuscript. Stefan Lienenklaus performed and analyzed experiments. Kathrin Wolf performed and analyzed experiments. Helmut Bloecker supervised the DNA sequence analysis. Dirk Bumann provided the promoter-trap library DNA and supervised the screening procedures. Holger Loessner supervised bacteria work. Siegfried Weiss designed, analyzed and supervised experiments and wrote the manuscript.
REFERENCES
- 1.Allen CA, Torres AG. Host-microbe communication within the GI tract. Adv. Exp. Med. Biol. 2008;635:93–101. doi: 10.1007/978-0-387-09550-9_8. [DOI] [PubMed] [Google Scholar]
- 2.Trebichavsky I, Splichal I, Rada V, Splichalova A. Modulation of natural immunity in the gut by Escherichia coli strain Nissle 1917. Nutr. Rev. 2010;68:459–464. doi: 10.1111/j.1753-4887.2010.00305.x. [DOI] [PubMed] [Google Scholar]
- 3.Barbe S, Van Mellaert L, Anne J. The use of clostridial spores for cancer treatment. J. Appl. Microbiol. 2006;101:571–578. doi: 10.1111/j.1365-2672.2006.02886.x. [DOI] [PubMed] [Google Scholar]
- 4.Dolgin E. From spinach scare to cancer care. Nat. Med. 2011;17:273–275. doi: 10.1038/nm0311-273. [DOI] [PubMed] [Google Scholar]
- 5.Leschner S, Weiss S. Salmonella-allies in the fight against cancer. J Mol. Med. 2010;88:763–773. doi: 10.1007/s00109-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 6.Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57:4537–4544. [PubMed] [Google Scholar]
- 7.Zhao M, Yang M, Li XM, Jiang P, Baranov E, Li S, Xu M, Penman S, Hoffman RM. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc. Natl Acad. Sci. USA. 2005;102:755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao M, Yang M, Ma H, Li X, Tan X, Li S, Yang Z, Hoffman RM. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;66:7647–7652. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
- 9.Zhao M, Geller J, Ma H, Yang M, Penman S, Hoffman RM. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc. Natl Acad. Sci. USA. 2007;104:10170–10174. doi: 10.1073/pnas.0703867104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu F, Zhang L, Hoffman RM, Zhao M. Vessel destruction by tumor-targeting Salmonella typhimurium A1-R is enhanced by high tumor vascularity. Cell Cycle. 2010;9:4518–4524. doi: 10.4161/cc.9.22.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 12.Bumann D, Valdivia RH. Identification of host-induced pathogen genes by differential fluorescence induction reporter systems. Nat. Protoc. 2007;2:770–777. doi: 10.1038/nprot.2007.78. [DOI] [PubMed] [Google Scholar]
- 13.Bumann D. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol. Microbiol. 2002;43:1269–1283. doi: 10.1046/j.1365-2958.2002.02821.x. [DOI] [PubMed] [Google Scholar]
- 14.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 15.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leschner S, Westphal K, Dietrich N, Viegas N, Jablonska J, Lyszkiewicz M, Lienenklaus S, Falk W, Gekara N, Loessner H, et al. Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-alpha. PLoS One. 2009;4:e6692. doi: 10.1371/journal.pone.0006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 2002;20: 142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heimann DM, Rosenberg SA. Continuous intravenous administration of live genetically modified salmonella typhimurium in patients with metastatic melanoma. J. Immunother. 2003;26:179–180. doi: 10.1097/00002371-200303000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thamm DH, Kurzman ID, King I, Li Z, Sznol M, Dubielzig RR, Vail DM, MacEwen EG. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin. Cancer Res. 2005;11:4827–4834. doi: 10.1158/1078-0432.CCR-04-2510. [DOI] [PubMed] [Google Scholar]
- 21.Arrach N, Zhao M, Porwollik S, Hoffman RM, McClelland M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008;68:4827–4832. doi: 10.1158/0008-5472.CAN-08-0552. [DOI] [PubMed] [Google Scholar]
- 22.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res. 2003;63:5188–5193. [PubMed] [Google Scholar]
- 23.Loessner H, Endmann A, Leschner S, Westphal K, Rohde M, Miloud T, Hammerling G, Neuhaus K, Weiss S. Remote control of tumour-targeted Salmonella enterica serovar Typhimurium by the use of l-arabinose as inducer of bacterial gene expression in vivo. Cell Microbiol. 2007;9:1529–1537. doi: 10.1111/j.1462-5822.2007.00890.x. [DOI] [PubMed] [Google Scholar]
- 24.Westphal K, Leschner S, Jablonska J, Loessner H, Weiss S. Containment of tumor-colonizing bacteria by host neutrophils. Cancer Res. 2008;68:2952–2960. doi: 10.1158/0008-5472.CAN-07-2984. [DOI] [PubMed] [Google Scholar]
- 25.Wojtkowiak JW, Verduzco D, Schramm K, Gillies RJ. Drug resistance and cellular adaptation to tumor acidic pH microenvironment. Mol. Pharm. 2011 doi: 10.1021/mp200292c. October 26 (doi:org/10.1021/mp200292c; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crull K, Rohde M, Westphal K, Loessner H, Wolf K, Felipe-Lopez A, Hensel M, Weiss S. Biofilm formation by Salmonella enterica serovar Typhimurium colonizing solid tumours. Cell Microbiol. 2011;13:1223–1233. doi: 10.1111/j.1462-5822.2011.01612.x. [DOI] [PubMed] [Google Scholar]
- 27.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.