Abstract
Methyl-CpG-binding protein 2 (MeCP2) is a chromatin-binding protein that mediates transcriptional regulation, and is highly abundant in brain. The nature of its binding to reconstituted templates has been well characterized in vitro. However, its interactions with native chromatin are less understood. Here we show that MeCP2 displays a distinct distribution within fractionated chromatin from various tissues and cell types. Artificially induced global changes in DNA methylation by 3-aminobenzamide or 5-aza-2′-deoxycytidine, do not significantly affect the distribution or amount of MeCP2 in HeLa S3 or 3T3 cells. Most MeCP2 in brain is chromatin-bound and localized within highly nuclease-accessible regions. We also show that, while in most tissues and cell lines, MeCP2 forms stable complexes with nucleosome, in brain, a fraction of it is loosely bound to chromatin, likely to nucleosome-depleted regions. Finally, we provide evidence for novel associations of MeCP2 with mononucleosomes containing histone H2A.X, H3K9me2 and H3K27me3 in different chromatin fractions from brain cortex and in vitro. We postulate that the functional compartmentalization and tissue-specific distribution of MeCP2 within different chromatin types may be directed by its association with nucleosomes containing specific histone variants, and post-translational modifications.
INTRODUCTION
The precise and dynamic modulation of chromatin structure is essential for context-specific transcriptional, or replication-dependent processes. Chromatin alterations resulting from the disruption of such processes are traceable and provide potentially useful disease biomarkers (1). Mutations in the chromatin-binding protein, methyl–CpG-binding protein 2 (MeCP2), have been linked to the neurodevelopmental disorder Rett syndrome (RTT) and other neurological abnormalities (2,3).
MeCP2 is a member of the methyl–CpG-binding domain (MBD) family of proteins (4). MeCP2 can bind symmetrically methylated 5′-CpG dinucleotides that are located proximal to at least four A/T nucleotides within the surrounding 11 bp (5). This binding is dependent on the recognition of the hydrated methylated CpG by the wedge-shaped MBD (6,7).
Most work characterizing MeCP2 interactions with chromatin has been done with reconstituted templates. Under such conditions, MeCP2 has been shown to bind both methylated and unmethylated templates, with a preference for the former, particularly in the presence of competitor DNA (8,9). Under non-physiological ionic conditions, MeCP2 was demonstrated to condense unmethylated chromatin templates (10). Upon binding to nucleosomes, MeCP2 forms chromatosome-like structures and may even facilitate inter-nucleosomal fiber interactions in vitro (11). Early studies indicated that MeCP2 is able to compete with histone H1 for binding linker DNA, and is able to displace ∼40% of H1 (12). In vitro reconstitution of nucleosomes containing preferential binding sites for MeCP2 indicated that it preferred binding within 10 bp of the dyad axis (8,11). By contrast, the interactions of MeCP2 with native chromatin are less extensively characterized.
Much of the early work done on MeCP2 characterized it as a transcriptional repressor due to its association with histone deacetylases (HDACs, HDAC 1 and 2) and SIN3A (4,13). As well, MeCP2 was shown to associate with SUV39H1, the histone methyltransferase responsible for methylating lysine 9 of histone H3 (14), an activity also linked to the DNA methyltransferase, DNMT3A (15). Furthermore, involvement of MeCP2 in long-range silencing was shown through its ability to form chromatin loops containing the gene Dlx5 (16). Nevertheless, the perspective began to change with recent publications demonstrating that the majority of MeCP2-bound promoters were positively regulated (17,18). MeCP2 has been shown to localize to a broad range of chromatin types, supporting observations of a potential plurality to MeCP2 function (8). However, besides the requirement for sequence-specificity and DNA methylation, very little is known about the histone determinants [histone variants and post-translational modifications (PTMs)] associated with this multifaceted functionality.
In this paper, we look at the chromatin distribution of MeCP2 in several tissues, and characterize the histone composition variability of nucleosomes with which MeCP2 is associated in the brain.
MATERIALS AND METHODS
Biotin-conjugated peptides pull-down and immunoblotting
Biotinylated histone peptides were purchased from Anaspec (Anaspec, Fremont, CA). The following peptides were used: 1. Histone H3 (1–21)—GGK(Biotin), H3K9/14(Ac); 2. Histone H3 (1–21)—GGK (Biotin), H3K4(Me2); 3. Histone H3 (1–21)—GGK(Biotin), H3K4(Me3); 4 [Lys(Me3)9]—Histone H3 (1–21)—GGK(Biotin), H3K9(Me3); 5. [Lys (Me3)27]—Histone H3 (21–44)—GK(Biotin), H3K27(Me3). Each peptide was re-suspended in TGD (20 mM Tris pH 8.0, 1 mM EDTA, 0.1% Triton X-100) at a concentration of 1 µg/µl, flash frozen and stored at −80°C until use. For each pull-down experiment, 5 µg of biotinylated histone H3 tail peptides bound to streptavidin beads [Dynabeads M-280 Streptavidin, (Invitrogen, Carsbad, CA)] were incubated with 0.5 µg wild-type MeCP2 (previously dialyzed against TGD), in the presence of 5 µg of purified BSA, for 2 h at 4°C with rotation. After three TGD washes to reduce non-specific binding, MeCP2-bound peptides were acid-eluted (100 mM glycine pH 2.8) from the beads. The eluates and supernatant representing 40% total input were subjected to SDS–PAGE, transferred to nitrocellulose membranes, probed for the presence of MeCP2 using an N-terminal specific antiMeCP2 antibody [Abcam (Cambridge, UK), ab2828], and visualized with ECL (GE Health Care Life Sciences, Piscataway, NJ) according to the manufacturer's protocol.
Cytoplasmic and nuclear fractionation
HeLa cell cytoplasmic and nuclear fractions were prepared using an Active Motif kit (Active motif, Carlsbad, CA). Western blots for analysis of the MeCP2 content were normalized based on the volumes of each fraction that corresponded to the same amount of cells.
Preparation of nuclei and chromatin fractionation
Nuclei and chromatin from different cell lines and tissues were prepared as described in the Supplementary Data section.
Salt extraction of MeCP2 from rat brain and liver nuclei
Nuclei were prepared as described above, with inclusion of 20 mM EDTA in all buffers in the case of liver. Salt extracts were prepared by extracting equal aliquots of nuclei with 10 mM Tris pH 7.5 supplemented with concentrations of NaCl ranging from 0.10 to 0.50 M, in 0.05 M steps as, previously described (19), and in the case of liver nuclei, 10 mM EDTA. Preparations were vortexed vigorously and left on ice to incubate for 20 min, followed by centrifugation at 16 000 g. All extractions were carried out in the presence of Roche Complete Protease Cocktail Inhibitor.
Sucrose gradient fractionation
S1 and SE supernatants were run on 5–20% sucrose gradients in 25 mM NaCl, 10 mM Tris pH 7.5 and 5 mM EDTA buffer for 21 h at 96 000 g and 4°C, and collected thereafter in 0.5 ml aliquots.
Gel electrophoresis
SDS–(15%) PAGE (20), acetic acid-urea (AU)- PAGE (21), acetic acid-urea-triton X-100 (AUT)—(10%) PAGE (22), Native– (4%) PAGE (23) and 0.7% agarose gels were performed as described earlier (24,25).
Western blotting
Western analysis was carried out as in (26). The antibodies and the dilutions used were as follows: MeCP2 [Sigma (St. Louis, MO), M9317] 1:2500, H3K27me3 [Millipore (Billerica MA, 07-449] 1:5000, H3K9me2 [Upstate/Millipore (Billerica, MA), 07-212] 1:2000, H2A.X [abm (Vancouver, BC), Y021260] 1:3333, H2A.Xγ [Millipore (Billerica, MA), 07-164] 1:1000, H2A.Z [Abcam (Cambridge, UK), ab4174] 1:1000, H2A.Z acetylated (generous gift from Dr. Crane-Robinson) 1:1000, H4 (made in-house) 1:10 000, H4 pan acetylated [Upstate/Millipore (Billerica, MA), 06-866] 1:4000, H3K4me3 [Millipore (Billerica, MA), 04-745) 1:5000.
DNA methylation quantification
The relative amount of 5′-methylcytosine was analyzed by HPLC using a strong cation exchange (SCX) column (27) and the base preparation conditions described in (28). In brief, digestion buffer from a 10× stock (200 mM acetic acid, 200 mM glycine, 50 mM MgCl2, 5 mM ZnCl2 and 2 mM CaCl2, adjusted to pH 5.3 with NaOH), was added to 10–20 μg of DNA, which had been purified with a Qiagen (Qiagen, Germantown, MD) PCR kit. This was then treated with 1:50 of nuclease mix [2.5 U/ml nuclease P1 (Sigma-Aldrich, Saint Louis, MO) and 500 U/ml DNase I in 50% glycerol]. The mixture was incubated overnight at 37°C, and followed by an additional overnight incubation with one unit of Antarctic phosphatase (New England Biolabs) at 37°C. Immediately prior to injection, the sample was diluted 2× with 12 mM HCl, and resolved on a Luna SCX (Phenomenex, Torrance, CA) column at a flow rate of 1 ml/min, with isocratic gradient of 40 mM acetic acid and 15% acetonitrile, pH 4.8.
Real-time quantitative RT–PCR
RT–PCR experiments were performed as described elsewhere (29). The comparative Ct (2−ΔΔCt) method was used to quantify the relative fold changes in MECP2 expression levels between different treatment conditions or tissues (30–32). A serial dilution validation assay was performed to assess the appropriateness of the target and reference gene primers for RT–PCR. For all primer sets, when plotting the log cDNA concentration dilution to the ΔCt value, the slope of the line was <0.1, and efficiencies of the primer sets were >99%.
Two sets of primers were used to amplify MECP2: 1; [forward: 5′-AGCCATCAGCCCACCACT, reverse: 5′-CGCAATCAACTCCACTTTAGA (∼281-bp amplicon)] (HeLa cells), 2: [forward 5′-GCCTCTGCTTCTCCCAAAC and reverse 5′-CCACCTCCCTCACCCTTAC (485-bp amplicon)] (rat tissues). These primers were designed to amplify both MECP2 isoforms. After it was found that GAPDH (glyceraldehyde-3-phosphate dehydrogenase) exhibited a significant extent of variation across different tissues, rat GC-rich promoter-binding protein 1 gene (GPBP1) was instead used as a reference to normalize the target gene (MECP2) and to account for any variations that may be present in reference gene levels between the different tissues (33). The primers used for [GPBP1 were: [forward 5′-CTTAGTCCCCAAACCTGCTG and reverse 5′-TGAGAAATCACCGAGGCATT (462-bp amplicon)].
The Invitrogen Platinum SYBR Green qPCR SuperMix-UDG with Rox was used in the experiments described. For each reaction, 7.5 μl of the SYBR mix was added to 0.5 μl 10 μM primers, 2 μl 1/20 diluted cDNA and 5 μl of RNase- and DNase-free dH2O. For each cDNA dilution prepared, the reaction was run in quadruplicate per primer set. A non-template control was run to test the primers independently as well. The RT–PCR was done using an Agilent Technologies Stratagene Mx3005P instrument and MxPro – Mx3005P v4.10 Build 389, Schema 85 software (Stratagene). The run parameters were as follows. Segment 1 (one cycle): denaturation at 94°C for 9 min; Segment 2 (40 cycles): denaturation at 95°C for 15 s, annealing at 62°C for 30 s, and extension at 72°C for 45 s; Segment 3 (one cycle—for dissociation profile): 95°C for 1 min, 55°C for 30 s, 95°C for 30 s. Data analysis was done using the 2−ΔΔCt method, with the untreated (native) HeLa S3 culture used as the comparator for 3-ABA -treated cultures, in order to determine the relative fold changes in transcript levels. In the tissue analysis, a mixed cDNA comparator was prepared in which 2 μl of each 1/20 cDNA dilution for every tissue was combined together and run as a separate sample (31). Every tissue cDNA 1/20 dilution was run in triplicate, as were non-template (primer) controls.
Native coimmunoprecipitation of MeCP2-bound mononucleosomes
Sheep cortex tissues were used to produce the mononucleosome-containing S1 and SE chromatin fractions following the procedure described above. To ensure that the antibody binding capacity was saturated, an excess starting material was used for both the S1 and SE coimmunoprecipitations. In paired MeCP2 antibody and normal rabbit IgG pull-down reactions, 350 µg of S1 chromatin was added to 50 mM NaCl, 20 mM Tris pH 7.5 to a final volume of 1.0 ml. In similarly paired pull-down reactions, 250 µg of SE chromatin was added to 100 mM NaCl, 20 mM Tris pH 7.5 and brought to a final concentration of 50 mM NaCl in a total volume of 1.0 ml. An amount of 5.0 µg of salmon sperm DNA (Invitrogen, Carlsbad, CA) was added to block any non-specific interactions. A 5 min pre-clearing of the sample with 15 µl of a 50:50 slurry of protein A/G agarose beads (Pierce/Thermo Scientific, Rockford, IL) at 4°C with tumbling was initially done to reduce any non-specific binding to the beads (34). Following a brief centrifugation to remove the beads (700 g, 3 min, 4°C), 12 µl of each pre-cleared input supernatant was kept aside for later analysis. To the MeCP2 pull-downs, 15 µl of 0.6 mg/ml primary anti-MeCP2 antibody (Sigma, St. Louis, MO) was added to the remaining supernatant, while 9 µl of a 1 mg/ml normal rabbit IgG (Sigma, St. Louis, MO) was added to the non-specific pull-down control. Reactions were tumbled at 4°C overnight. An amount of 100 µl of A/G bead slurry (Pierce/Thermo Scientific, Rockford, IL) were added to all reactions and tumbled at 4°C for 2 h. The suspension was briefly centrifuged as above and the unbound supernatants were kept aside. The pull-down beads were subsequently washed by inversion in 1.0 ml each of 5 mM NaCl, 50 mM NaCl and 100 mM NaCl, in 20 mM Tris (pH 7.5) buffers. The recovered bound beads were then resuspended in 20 µl 2× SDS sample buffer and heated at 100°C for 3 min and analyzed by SDS–PAGE and western blotting.
Immunofluorescence
Wax was removed from slides by heating slides at 60°C, and paraffin removal was achieved by three washes in a xylene bath followed by a reverse ethanol series from 100% ethanol through to water. Slides were then washed with TBS-Tween 20 (0.05%)(TBS-T). For antigen retrieval, a microwave antigen retrieval was performed in 100 mM trisodium citrate with 0.05% Tween 20 pH 6.0. Following antigen retrieval, slides were blocked with 4% BSA in TBS-T for 1 h followed by incubation for 2 h at room temperature with primary antibody [Sigma (St. Louis, MO) M9317] at a dilution of 1:1000. The slides were then washed 3× in TBS-T and then incubated with an Alexa 488-conjugated secondary antibody. After washing, the slide was then mounted in polyvinyl alcohol containing 1 µg/ml DAPI.
Electron microscopy
Male rat brain cortical tissue was fixed overnight in 3% glutaraldehyde/3% formaldehyde in 0.1 M cacodylate buffer and further sliced into small pieces for Epon, processing/embedding. The samples were post-fixed in 1% osmium tetroxide, en bloc stained in 5% uranyl acetate in 50% ethanol, and completely dehydrated through a graded ethanol series and infiltrated with Epon through graded steps with propylene oxide. Individual brain pieces were placed into individual gelatin capsules containing new epon allowed to polymerize at 60°C for 48 h. Ultrathin sections were cut on a Reichert Ultracut E with a diamond knife and placed onto 100 mesh nickel grids containing a carbon-coated formvar film. Sections were treated with rabbit MeCP2 antibody [Sigma (St. Louis, MO), M9317] and probed with 12 nm colloidal Gold-AffinitiPure Goat anti-rabbit IgG (H+L), (Jackson ImmunoResearch Inc, West Grove, PA). The sections were pre-treated with saturated sodium metaperiodate in water for 1 h, blocked for 10 min with PBS-ovalbumin (1%), labeled with primary diluted (1:100 to 1:400) in PBS-ovalbumin for 1 h, washed with PBS-ovalbumin, labeled with secondary diluted 1:100 in PBS-ovalbumin for 1 h, washed in PBS-ovalbumin and distilled water, and stained in 5% uranyl acetate in 50% ethanol and 5% lead citrate. The sections were viewed in a Hitachi H7000 TEM and images captured with an AMT 2 k × 2 k CCD camera.
RESULTS
MeCP2 chromatin distribution does not depend on the extent of global DNA methylation in cultured cells
Using a chromatin fractionation procedure (35,36) (Supplementary Figure S1A), we have previously shown that the majority of MeCP2 cofractionates with the highly nuclease-accessible S1 chromatin fraction from HeLa S3 cells (8). As shown in Supplementary Figure S1B, most of the MeCP2 in this cell-type is of nuclear origin, as only a very small fraction is associated with the cytoplasm. Despite the simplicity of the chromatin fractionation protocol we (26,29), and others (37), have previously shown that the S1 corresponds to euchromatic regions, and to chromatin regions at the periphery of heterochromatin domains that are very accessible to nucleases. The SE consists mainly of facultative heterochromatin, and the pellet (P) fraction consists of constitutive heterochromatin and regions of chromatin associated with large multimeric (transcriptional and DNA-repair) protein complexes (26,35,38). Such distribution is consistent with the histone variants and PTMs observed here (see Supplementary Figure S1A, right panel).
To ensure that the unusual MeCP2 distribution is not an artifact of the buffers used during nuclei isolation and further chromatin digestion, the chromatin preparation and digestion were carried out using three different methods (19,39,40) that utilize different buffers for the micrococcal nuclease digestion (see Supplementary Figure S1C). As shown, a very similar distribution of MeCP2 is observed in all instances.
Early work, and our own in vitro analyses of MeCP2 binding to DNA and nucleosomes, has shown that the protein binds preferentially to methylated DNA templates (8,41). However, based on other in vitro studies (10), it has been argued that MeCP2 may bind to chromatin in a dual way that is dependent and independent of the methylated state of DNA (42). Such conclusions need to be taken cautiously as MeCP2, in addition to containing a methyl DNA-binding domain (MBD), is a very basic protein (pI = 9.9), and under low ionic buffer conditions it may bind to DNA in a non-specific way, whether nucleosomally organized or not. In attempt to address this issue in an in vivo setting, we used different approaches to alter the level of genomic DNA methylation. To this end, HeLa and 3T3 mouse cell cultures were treated with 3-aminobenzamide (3-ABA), and 5-aza-2′-deoxycytidine, which alter the extent of DNA methylation (Figure 1A). Importantly, none of these treatments significantly affected the proportion of chromatin in the S1, SE, and P fractions, which remained at around 11 ± 2% (S1); 30 ± 3% (SE) and 59 ± 5% (P) in HeLa cells treated with +/− 3-ABA and +/− 5-aza-2′-deoxycytidine. In the case of the 3T3 cells the proportions were 11 ± 1% (S1); 34 ± 1% (SE) and 55 ± 1% (P) for the cells treated with +/− 5-aza-2′-deoxycytidine and 10 ± 1% (SI); 36 ± 1% (SE) and 54 ± 1% (P) for the cells treated with +/− 3-ABA.
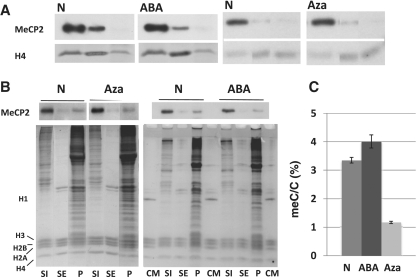
Figure 1.
Distribution of MeCP2in HeLa cells in response to treatment with DNA methylating and de-methylating agents. (A) Western (MeCP2) blot analysis of the S1, SE and P chromatin fractions obtained from the nuclei of HeLa S3 cultures grown in the absence N (native) or in the presence of: 2 mM 3-ABA, methylated (ABA); 1 µM 5-aza-2′-deoxycytidine, de-methylated (Aza). Coomassie blue-stained or antibody detected histone H4 (H4) was used for normalization purposes. (B) Western blot analysis of the S1, SE and P chromatin fractions obtained from the nuclei of mouse 3T3 fibroblast cultures grown in the absence N (native) or in the presence of: 2 mM 3-ABA, methylated (ABA); 1 µM 5-aza-2′-deoxycytidine, de-methylated (Aza). The upper part corresponds to the MeCP2 western and the lower part is a Comassie blue stained of the SDS–PAGE after electroblotting. CM: Chicken erythrocyte histone marker. (C) HPLC-determined relative meC/C percentile of the mouse 3T3 fibroblasts used for the analysis shown in (B).
Global increase in levels of DNA methylation in cultured cells upon exposure to 3-aminobenzamide (3-ABA), an inhibitor of poly(ADP-ribose) polymerases (PARP) (43), has been well documented (44). Although details of the molecular mechanisms involved are not completely understood, PARP inhibition appears to result in the induction of DNMT1, the major maintenance DNA methyltransferase (45). Under our experimental conditions, such treatment resulted in an 11% and a 16% (Figure 1C) increase in DNA methylation in Hela and 3T3 cells respectively, compared to the corresponding DNA from non-treated cultures. As shown in Figure 1A–B, the S1, SE and P fractions of the treated (ABA) and the corresponding untreated (N) cells exhibit an almost indistinguishable MeCP2 distribution and partitioning.
Treatment of the same cell lines with the DNA methylation inhibitor, 5-aza-2′-deoxycytidine, resulted in a 10% decrease in the level of genomic DNA methylation in HeLa S3 and 65 ± 2% decrease in 3T3 (Figure 1C). However, as shown in Figure 1A–B, this did not result in a rearrangement of MeCP2 within chromatin in either one of them. Rather, both the level and distribution of MeCP2 were similar to the untreated and treated cells. Although the partitioning amounts of MeCP2 across the SI/SE/P fractions varied between 3T3 (Figure 1B-Aza) and HeLa cells (Figure 1A-Aza) (simply reflecting different extent of nuclease digestion, see above and next section) the relative amounts of MeCP2 and its distribution were again unaffected by the treatment.
To ensure that none of these treatments affected the amount of MECP2 transcription, real-time RT–PCR was performed (results not shown).
MeCP2 is enriched in the highly nuclease-accessible S1 chromatin fraction of different tissues and cell lines in a way that depends on the extent of nuclease digestion
The next question we wanted to address was whether enrichment of MeCP2 in the S1 fraction is a universal feature of any living cell. In other words, do the cells of different mammalian tissues exhibit a similar distribution of MeCP2 amongst fractionated chromatin that is similar to the one observed in HeLa cells?
To address this question, the chromatin from rat whole brain, and a somatic (liver) and germline (testis) control, were fractionated into the S1, SE and P components following micrococcal nuclease digestion as described above, and the histones and associated DNA of each were analyzed by native- and AUT-PAGE [Supplementary Figure S2 (1) and (2)]. Despite the histone variability (Supplementary Figure S2A-2), and different pattern of nuclease digestion (Supplementary Figure S2A-1), the majority of MeCP2 in all instances was mainly present in the S1 fraction (Supplementary Figure S2-MeCP2). This is in contrast with histone H2A.X, and histone H3K27me3 and H3K9me2, which exhibit a more tissue-dependent distribution (Supplementary Figure S2A-3).
Quite strikingly, and as previously reported, MeCP2 is present in exceedingly large amounts in brain (46,47), when compared to the other tissues investigated (Supplementary Figure S2B-MeCP2). We used a combination of western and electrophoretic approaches (see Supplementary Figure S2C) to quantify the approximate relative amount of MeCP2 in these different tissues, and to estimate the number of MeCP2 molecules per mole of nucleosome in the brain, using histone H4 as a reference. The amount of protein expressed in this tissue (Supplementary Figure S2D-1), determined in this way, is ∼20 - to 30-fold greater than that of any other tissue, and is present at nearly one molecule per every three nucleosomes, in whole brain tissue. Also, the brain had a relative fold difference of 19.5 ± 0.5 (SD) times more MECP2 mRNA than the mixed cDNA comparator. The liver had an MECP2 relative fold difference of 0.33 ± 0.1 (SD), while testis showed a difference of 0.85 ± 0.02 (SD). By this analysis, the brain had more than 22 times the amount of MECP2 mRNA compared to testis, and more than 59 times the amount when compared to the liver.
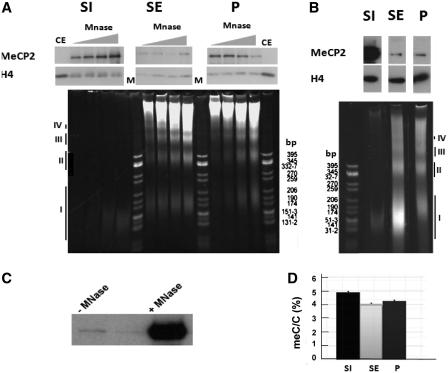
To determine the influence of MNase digestion time in the distribution of MeCP2 amongst brain chromatin, a time-course digestion of isolated nuclei was carried out (Figure 2A) for 2, 4, 8 and 12 min, as described in the figure legend, and fractionated into S1, SE and P. At low MNase concentration (low extent of digestion), the data reveal an interesting redistribution of MeCP2 from fraction P into the mononucleosome containing S1 fraction, as a function of digestion time. Interestingly, the amount of MeCP2 present in the histone H1-containing (Supplementary Figure S2-2), soluble polynucleosome fraction (SE) is relatively low when compared to the other two fractions, at any given digestion time. This suggests that a significant fraction of MeCP2 is loosely bound in the P fraction, and it is readily freed of its DNA interaction upon nuclease digestion. Eventually, most of the MeCP2 bound in the P fraction can be released into S1 (Figure 2B). It is important to note that the majority of nuclear MeCP2 is DNA bound, as very little (2.4%) MeCP2 is released into S1 in the absence of MNase digestion (Figure 2C).
Figure 2.
Preferential fractionation of MeCP2 with the S1 fractionation depends on the extent of micrococcal nuclease digestion. (A) Time-course digestion of rat whole-brain nuclei with MNase at 10 U/mg of DNA for 2, 4, 8 and 12 min followed by fractionation to yield S1, SE and P. The upper panel shows a western blot for MeCP2 that is normalized for H4. Lower panel contains 4% native acrylamide gel of the DNA associated with the S1, SE and P. CE: chicken erythrocyte histones; M: pBR322—CfoI-digested marker. (B) Same as in (A), but using MNase at 30 U/mg of DNA for 15 min. The roman numerals (I–IV) in both (A) and (B) indicate the DNA fragments corresponding to mono-, di-, tri- and tetra-nucleosomes respectively. (C) Western blot analysis of MeCP2 released from brain nuclei without (–) and with (+) micrococcal nuclease digestion treatment reveals that essentially all nuclear MeCP2 is bound to chromatin. (D) HPLC-determined relative meC/C percentile of the fractions shown in (B).
When levels of DNA methylation in S1, SE and P fractions obtained under the digestion conditions used in Figure 2B were determined, we found that it was higher in both S1 and P, in comparison to the SE fraction (20% and 4.6% higher than SE, respectively) (Figure 2D). The level of S1 methylation was about 18% higher than that of chromatin in the nuclei before MNase digestion. Analysis of the levels of DNA methylation in the fraction P of Figure 2A exhibited a decrease in the trend of DNA methylation upon increased digestion time (results not shown).
The higher level of DNA methylation associated with the S1 fraction (20%) suggests that a significant part of the protein is bound to methylated DNA regions of the genome.
A fraction of weakly chromatin-bound MeCP2 is not associated with nucleosomes in brain
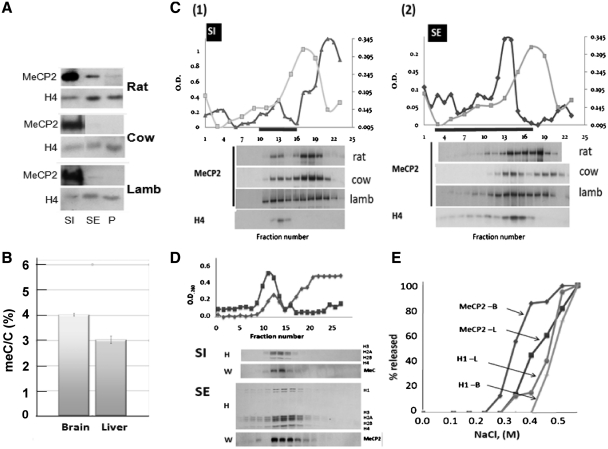
To gain further insight into the differential distribution of MeCP2 amongst chromatin, S1 and SE fractions from the brain of several mammalian species (Figure 3A) were prepared and subjected to sucrose gradient fractionation [Figure 3C(1) and (2)]. As shown by western blot analysis, >50% of MeCP2 in both the S1 and SE fractions partition within the upper part of the gradient, in the region corresponding to free MeCP2.
Figure 3.
A fraction of MeCP2 in the brain is weakly associated with chromatin and is not associated with nucleosomes. (A) Western blot analysis of the MeCP2 partitioning in the S1, SE and P chromatin fractions from the brain of different mammalian species. (B) HPLC-determined relative meC/C percentile in the nuclei isolated from rat brain and liver. (C) Sucrose gradient fractionation of the S1 (1) and SE (2) chromatin fractions from the brain. The upper part depicts the absorbance (260 nm) profile of the fractions collected along the gradient (dark line). The lighter line shows the profile (absorbance at 230 nm) of recombinant MeCP2 run under the same conditions. The lower panel shows the western blot analysis of fractions collected along the gradient using MeCP2 and H4 antibodies. (D) Sucrose gradient fractionation of the S1 and SE chromatin fractions from rat liver. The lower part shows a Coomassie blue-stained SDS–PAGE used for the analysis of the fractions (H) and the corresponding western blot analysis using an MeCP2 antibody (W). (E) Salt extraction of MeCP2 and histone H1 from rat brain and liver.
When using these fractionation procedures, the distribution of brain MeCP2 is quite different from that exhibited by MeCP2 in liver (Figure 3D), or HeLa cells (results not shown). In these two later instances, and in high contrast to what was observed in brain, the majority of MeCP2 in both S1 and SE fractions is found associated with nucleosomes. Importantly, nuclei of whole brain were found to possess ∼30% higher levels of DNA methylation than those of liver (Figure 3B)
The presence of non-nucleosome associated MeCP2 in the sucrose gradient fractionation of brain chromatin suggests the existence of two MeCP2 populations with distinct chromatin-binding profiles, in this tissue. To further confirm this, we performed a salt extraction of rat brain and liver nuclei (Figure 3E) in order to separate sub-populations of MeCP2 exhibiting differential binding, as well as histone H1, in both tissues. Similar to what had been described earlier (41), brain MeCP2 exhibits a bimodal elution that takes place at salt concentrations lower than those required for MeCP2 extraction in liver. For instance, at 0.35 M NaCl, ∼80% of MeCP2 has come free in brain, whereas only 15% has been released in liver. This is in contrast to the behavior of H1, which, in both tissues, exhibits very similar elution trends that begin at salt-concentrations greater than those required to extract MeCP2.
MeCP2 is broadly distributed throughout non-condensed chromatin regions in neuronal nuclei and is also associated to the peripheral regions of heterocromatic domains
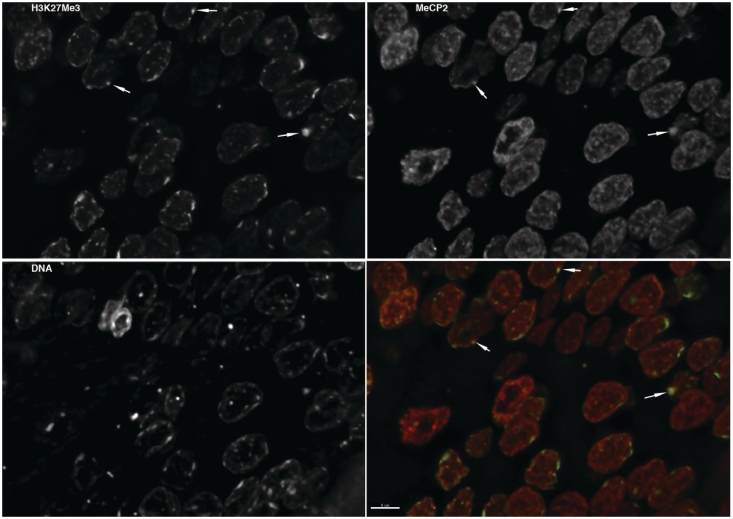
Microscopy techniques were used to investigate the distribution of MeCP2 in neuronal nuclei. Fluorescence microscopy carried out on brain sections stained with an MeCP2 antibody (Figure 4) show that, in contrast to the predominant heterochromatin localization observed in non-neuronal cell lines (5,48), in rat neurons, MeCP2 is broadly distributed throughout both hetero- and euchromatic domains (Figure 4). Only a relatively small fraction of MeCP2 was observed to colocalize with H3K27me3, a well-established heterochromatin marker (Figure 4, green fluorescence).
Figure 4.
MeCP2 is broadly distributed throughout the interphase nucleus of neurons and exhibits a reduced colocalization with H3K27me3. The image shows a thin section of rat brain stained with antibodies recognizing trimethylated lysine 27 of histone H3 and MeCP2. Total DNA was stained with DAPI. A z-series was collected and the image series was processed by deconvolution. A maximum intensity projection image was then generated for each channel. The bottom right panel shows a composite image of the trimethylated lysine 27 of histone H3 (green) and MeCP2 (red). Regions enriched in both appear as yellow in the composite image. The trimethylated lysine 27 was found to be enriched in the most dense regions of chromatin. MeCP2 was broadly distributed throughout the interphase nucleus, showing association with both heterochromatin and euchromatin. Examples where trimethylated lysine 27 and MeCP2 are both enriched are highlighted with arrows.
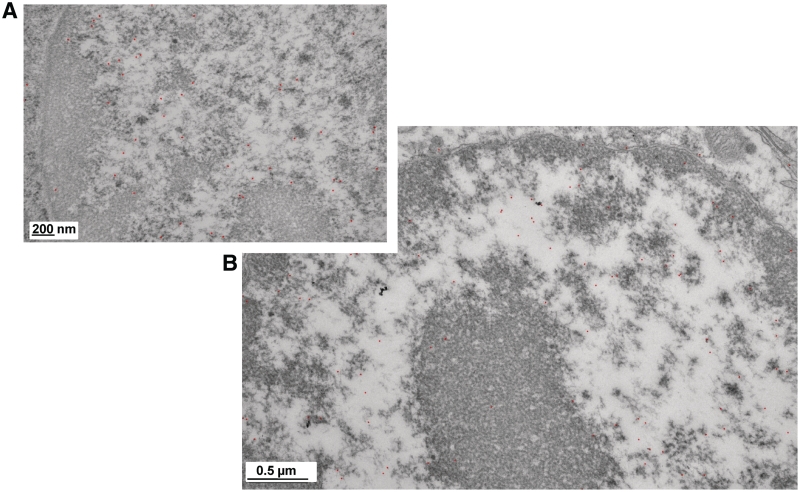
To analyze this result in further detail, we examined the distribution of MeCP2 in neuronal nuclei with immunogold particle localization using TEM. Representative samples of the many images collected using this protocol, are shown in Figure 5. In all the images analyzed, the gold particles were distributed mainly in the regions of the nuclei containing low-density (unfolded) euchromatin, or at the periphery of the highly condensed heterochromatin domains. Very few particles were observed within the tightly packed regions of chromatin, and still fewer in the cytoplasm (not shown).
Figure 5.
A fraction of MeCP2 localizes at the periphery of highly dense chromatin structures. (A and B) Two transmission electron microscopy magnification views of neuron nuclei showing the immunogold (black dots) localization of MeCP2.
In brain, MeCP2 binds to mononucleosomes containing histone H2A.X and specific histone H3 PTMs
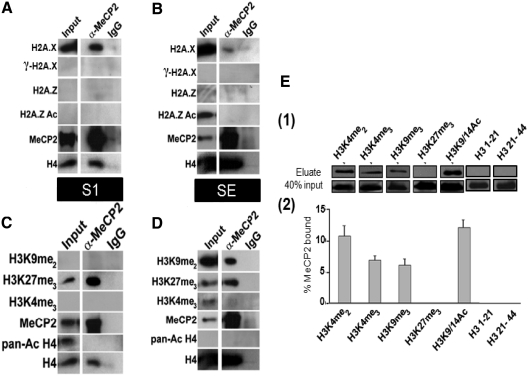
In order to identify epigenetic factors that may influence MeCP2 localization to specific nucleosomes in different chromatin environments, different nucleosome components were investigated (Figure 6). These included histone variants or histone post-translational modifications that may serve as epigenetic signals to facilitate and distinguish MeCP2 binding between heterochromatin and euchromatin. Native coimmunoprecipitations were performed using mononucleosomes obtained from S1 and SE chromatin fractions and an MeCP2 antibody, followed by probing for the respective histone variant or PTM.
Figure 6.
MeCP2 is associated with mononucleosomes containing histone H2A.X and histone H3 PTMs. (A–D) Mononucleosomes from S1 and SE fractions were coimmunoprecipitated under native conditions using a MeCP2 antibody. The immunoprecipitated nucleosomes were then analyzed by western blot using the antibodies indicated. Normal rabbit IgG was used as a non-specific control. The pull-downs for both the MeCP2 antibody and IgG were compared to a normalized initial sample input used in the coimmunoprecipitations. (E) MeCP2 pull-down experiments using H3 N-terminal peptides with several PTMs. (1) MeCP2 western blot analysis after pull-down using biotinylated peptides and streptavidin beads. (2) Quantification of the results in (1). Results are expressed as percent of MeCP2 eluted compared to total input. Error bars represent standard error of the mean of triplicate experiments.
MeCP2 was observed to interact with H2A.X-containing mononucleosomes in both the S1 and SE fractions. This interaction was more enriched in the S1 than in the SE when the precipitated amounts were compared to their respective inputs. (Figure 6A–B). No associations, however, were observed with γ H2A.X, H2A.Z, or acetylated H2A.Z in either S1 or SE (Figure 6A–B). We used coimmunoprecipitated mononucleosomes probed for H4 and MeCP2 by western blotting as a control.
Interactions of MeCP2 with mononucleosomes containing specific H3 PTMs were investigated as well. In the SE, MeCP2 associated with mononucleosomes containing the repressive marks H3K9me2 and in a separate pull-down, H3K27me3. No interactions were observed with H3K4me3 (Figure 6D), a PTM associated with transcriptionally active regions. Similar interactions were also observed in the S1 of MeCP2 pull-downs of mononucleosomes; however, no interactions were observed with H3K9me2 (Figure 6C). Of note, H3K27me3 in the S1 was greatly enriched in the MeCP2-immunoprecipitated nucleosomes compared to the input (Figure 6C).
MeCP2's ability to distinguish nucleosomes containing different H3 PTMs in brain is very interesting. It raises the question as to whether MeCP2 can directly interact with the N-terminal ‘tail’ of H3, or whether this interaction additionally depends on the presence of nucleosomally organized methylated DNA. A recent survey using a SILAC-based proteomics approach (49) has identified a plethora of nucleosome-interacting proteins, including MeCP2. Based on these results, it appears that both DNA and histone methylation are required for the binding of MeCP2. Among the histone H3 PTMs involved with MeCP2 binding within this setting, a strong enrichment for H3K4me3, and a moderate enrichment for H3K9me3 and H3K27me3, was observed. In all these three instances, binding of MeCP2 additionally required the presence of DNA methylation, and no binding could be detected in its absence. To analyze this relationship further, we performed pull-down experiments using streptavidin-immobilized biotinylated peptides corresponding to the N-terminal tail of H3, each containing a unique, or set of PTMs (Figure 6E). While this data confirms the ability of MeCP2 to interact to different extents with histone PTMs, the lack of binding to H3K27me3 strongly suggests that recognition of this signal is strongly dependant on nucleosomally arranged methylated DNA. The ability of MeCP2 to interact with both active (H3K4me3, H3K9/14Ac) and repressive (H3K9me2–3, H3K27me3) PTM markers supports the notion of this protein being a transcriptional regulator (18), rather than a mere transcriptional repressor as it was initially thought (50).
MeCP2 interacts with the N-terminal region of H2A and its in vitro nucleosome binding affinity is decreased in the presence of H2A.X
In an initial attempt to characterize the molecular involvement of histones in the interaction of MeCP2 with the brain S1 fraction nucleosomes, we performed some preliminary cross-linking analysis. We took advantage of a powerful method that uses isotopically (deuterium) labeled amine-reactive crosslinkers which can be cleaved chemically (i.e. D12-EGS) (51), by light (i.e. BiPS) (52), or by collision-induced dissociation (i.e. CBDPS) (53) that was recently developed at the Genome BC Proteomics Centre at UVic. The data thus obtained (Supplementary Figure S3) indicated that a region on the C-terminal transcription repression domain of MeCP2 (KAEADPQAIPK, residues 266–275) is in very close proximity to, and likely interacts with, a region (AKSR, residues 16–19) located at the N-terminal end of H2A.
Also, and because of the association observed between H2A.X and MeCP2 in the previous section, we repeated some of our previous in vitro MeCP2-nucleosome interaction studies (8), but using histone H2A.X-containing nucleosomes. The pattern of MeCP2 binding to H2A.X-containing nucleosomes (Supplementary Figure S4) is very similar to that observed with linker histones (38). Accordingly, the presence of H2A.X/ γH2A.X decreases the ability of histone H1 to bind to the nucleosome (38). Given the similarity with which MeCP2 and linker histones bind to the nucleosome (8,54,55), this is somewhat not surprising.
DISCUSSION
DNA methylation is not the only determinant of MeCP2 binding to chromatin
The observed differential distribution of MeCP2 (Supplementary Figure S2B) may be linked to distinct regulatory roles for MeCP2 in different tissues. A previous study assessed this tissue variability, but using different approaches to normalization (47). However, in contrast to our results, no correlation between the protein and mRNA expression was found (47).
MeCP2 was first identified based on its ability to recognize and bind methylated DNA (41,56), and was proposed to work as a global repressor. In an attempt to discern between this scenario and that of MeCP2 acting as a transcriptional regulator that binds to highly specific methylated regions of the genome we treated HeLa and mouse fibroblast 3T3 cell lines with chemicals that globally change the levels of DNA methylation (Figure 1). The results obtained in this way, show that not only is the chromatin distribution of MeCP2 unaffected by the non-physiological treatments designed to alter the DNA methylation, but they do not seem to have any major impact on the levels of MeCP2 present in the treated cells (compare Figure 1A–B N with Aza and ABA). These results are in agreement with the FRAP experiments performed in the presence of Aza-deoxycytidine (48) which showed that the mobility of MeCP2 in the nucleus was not dependent on the extent of global methylation of DNA. The comparable amounts of MeCP2 observed in 3T3 cells where the levels of DNA methylation have decreased by 65% (Figure 1C) together with the chromatin distribution similarity would support the notion that part of MeCP2 is bound to non-(DNA) methylated regions of the genome (42,57). Alternatively, in these cell types MeCP2 may also bind to nucleosomes containing methylated histones (Figure 6)
However, the above results need to be taken cautiously as they have been carried out in mammalian cell lines where, as in the case of liver (Figure 3D) most of MeCP2 is bound to nucleosomes (results not shown). It is possible that a completely different situation occurs in brain where a large increase in MeCP2 is observed (Supplementary Figure S2D). Not only this, the higher levels of DNA methylation present in this tissue when compared to liver (Figure 3B) or to the above cell lines (Figure 1C) could lead to a different situation where binding to methylated DNA is more prevalent. Indeed, the difference in DNA methylation between brain and liver (Figure 3B) is significant and may account for the greater abundance of MeCP2 observed in this tissue. Statistically, a 1% increase in methylated cytosine can easily provide a binding site every three nucleosome equivalent regions in the brain.
MeCP2 and the organization of brain chromatin
The large amount of MeCP2 in adult brain (Supplementary Figure S2B–D) (47), especially in neurons (46) and our reported MeCP2 chromatin partitioning have implications for MeCP2-mediated changes in chromatin organization. It has been estimated that, in cortical neurons, there is approximately one molecule of MeCP2 per every two nucleosomes (46), a value similar to that observed by us of one molecule per every three nucleosomes in whole unfractionated brain. Also, there is plenty of evidence that the chromatin soluble fraction of cortical neurons upon MNase digestion exhibits a repeat length of ∼160 bp, which is lower than that of 190–200 bp observed in glial cells and other somatic tissues (58,59). Although not the purpose of our current analysis, as no attempt was made to purify cortical neuron nuclei, we consistently observed a shorter DNA repeat with our brain chromatin samples. The median value was centered at ∼170 bp for the SE fraction chromatin containing a full amount of histone H1, when compared to the values obtained from other tissues, or to the P fraction (Supplementary Figure S2A and Figure 2B).
Interestingly, the decrease in nucleosome repeat length observed in mature neurons (59) takes place in a gradual fashion as the brain matures and levels of MeCP2 increase. In quiescent neurons, DNMT3A-mediated changes in DNA methylation during development would presumably be favored at linker regions (60). Taken together, it is tempting to speculate that in such cells, providing the average number of nucleosomes remains relatively constant during maturation (61), a decrease in repeat length would come at expense of exposing longer regions of linker DNA, which may then become bound by MeCP2 (Figure 7). These regions would correspond to the non-nucleosomally associated MeCP2 in fractions S1 and SE (Figure 3B–C). Furthermore, in a situation where one molecule of MeCP2 is present every two nucleosomes, one would predict a more homogeneous distribution throughout the S1, SE and P fractions in these cells. While this is speculative, and requires further attention, one could envisage these regions comprising some of the MeCP2-mediated chromatin looping structures (16) and/or the regions associated with the nuclear matrix (62). The latter could explain the transition observed from the P to S1 fraction observed in Figure 2A.
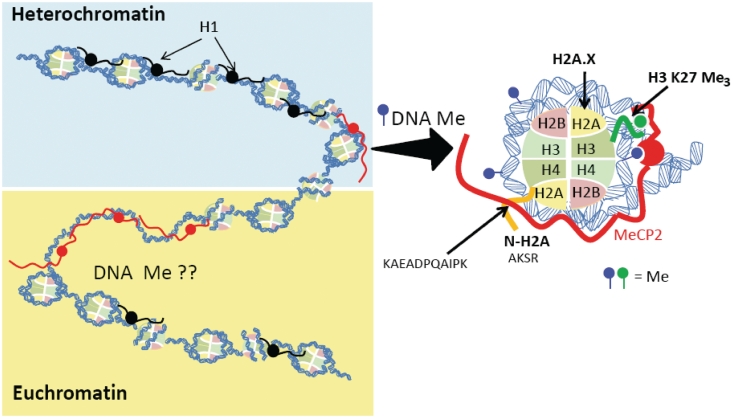
Figure 7.
Schematic representation to illustrate the dual association of MeCP2 with different chromatin domains in the brain. Shown in the right is a magnified image of the interaction of MeCP2 within the nucleosome fraction. Methylation of DNA in the vicinity of the nucleosome (in the proximity of the entry and exit sites of the DNA) as well as histone PTMs (such as H3K27me3) or histone variants may play a role in the interaction. Also shown is a site of interaction of the C-terminal tail of MeCP2 with the N-terminal of histone H2A.
MeCP2 interacts with nucleosomes containing specific histone variants and post-translational modifications
DNA methylation within a sequence specific context has been shown to direct MeCP2 binding (5). However, it is hard to envisage how such a simple mechanism is sufficient to account for the multiple potential roles in regulation of gene expression by MeCP2. Hence, we decided to check for the presence of other nucleosomal components that could allow MeCP2 to distinguish between various binding targets (63). As the H2A C-terminal tail and the H3 N-terminal tails exit the nucleosome (64) close to the site where MeCP2 has been proposed to bind (8,9), candidate variants and PTMs of these two core histones were investigated using coimmunoprecipitations of brain chromatin (Figure 6).
A novel and strong association was observed between MeCP2 and H2A.X-containing mononucleosomes in S1 chromatin (Figure 6A). The association of MeCP2 with H2A.X-containing nucleosomes is interesting. H2A.X has been shown to accumulate during neuron development (65,66) and to affect neuron stem cell proliferation and neuron output (67). MeCP2 interacts with the Brahma subunit of the SWI/SNF remodeling complex (68) and both MeCP2 and SWI/SNF have been shown to cyclically associate with an inducible promoter undergoing transcriptional activation (69). The binding of MeCP2 to H2A.X nucleosomes may be functionally linked through SWI/SNF, rather than to a direct binding preference for nucleosomes containing this variant. Indeed, the results shown in Supplementary Figure S4 indicate that it is highly likely that the association between MeCP2 and H2A.X-containing nucleosomes is of a casual nature.
H3K9me2 had a broad chromatin distribution within whole brain (Supplementary Figure S2A-3), although it was not observed in the cortex S1 immunoprecipitated fraction (Figure 6C), suggesting differential specialization of various brain regions may be regulated by a specific combination of MeCP2, DNA methylation and histone post-translational modifications. MeCP2 interacted with mononucleosomes containing this PTM in the SE fraction, but not in the S1 fraction (Figure 6C). The association of MeCP2 with H3K9 methylation has been described in the IL-6 gene region (70). A direct correlation between the binding of MeCP2 and the presence of H3K9me2 in promoter 1 of the BDNF gene and the IκBα gene has been reported as well (71,72). These coimmunoprecipitation results show, for the first time, that MeCP2 not only directly interacts with nucleosomes containing H3K9me2, but that this interaction is dependent upon the chromatin organization in the cortex. Di- and tri-methylated H3K9 have been shown to be marks for DNA methylation in Arabidopsis (73) and in Neurospora (74).
The presence of divalent PTMs, H3K27me3 and H3K4me3, on the same histone tail has been reported to poise the associated gene regions for either transcriptional activation or silencing (75–77). Interestingly, MeCP2 has been found to associate with methylated nucleosomes containing H3K4me3 (49) (Figure 6E). If activation is triggered, then the H3K27me3 mark is lost while the H3K4me3 PTM is perpetuated. The opposite scenario proceeds in the case of a repressive signal. H3K27me3, a mark of facultative heterochromatin, was present in MeCP2-bound nucleosomes in both S1 and SE (Figure 6C–D). Despite its low presence in S1, a strong enrichment for this signal was observed in the S1 pull-downs of the sheep cortex (Figure 6C). This suggests that a major portion of MeCP2-bound nucleosomes contain H3K27me3 in this particular brain fraction.
Whilst the importance of the association of MeCP2 with nucleosomes consisting of H2A.X, methylated H3K9 and/or H3K27 remains to be determined, our results indicate that in the in vivo setting, DNA methylation is not the sole determinant of MeCP2 binding to chromatin, and that the molecular constraints involved in the binding of MeCP2 to chromatin are more complex than originally envisaged.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary figures S1–S4, Supplementary methods, and Supplementary references [78–82].
FUNDING
Canadian Institute of Health Research (CIHR) (Grant MOP-97878 to J.A.); Natural Sciences and Engineering Research Council (NSERC) CGS doctoral fellowship to A.A.T. Funding for open access charge: CIHR (Grant MOP-97878).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Brian R. Christie from the UVic Island Medical Programme (IMP) for providing the male rats used in the RT–qPCR work and Michael Peterson for the lamb brains. The authors are also very thankful to Bethany McMullen who provided us with experimental assistance during the reviewing process. Thanks also to Dr Nik Veldhoen and Dr Deanna Dryhurst for many helpful qPCR discussions. The authors are also indebted to Dr Susan P. Lees-Miller from the Department of Biochemistry and Molecular Biology at the University of Calgary for providing us with DNA-PK.
REFERENCES
- 1.Crutchley JL, Wang XQ, Ferraiuolo MA, Dostie J. Chromatin conformation signatures: ideal human disease biomarkers? Biomarkers Med. 4:611–629. doi: 10.2217/bmm.10.68. [DOI] [PubMed] [Google Scholar]
- 2.Rett A. [On a unusual brain atrophy syndrome in hyperammonemia in childhood] Wiener medizinische Wochenschrift (1946) 1966;116:723–726. [PubMed] [Google Scholar]
- 3.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 4.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Wakefield RI, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, Bird AP, Barlow PN. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- 7.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 8.Ishibashi T, Thambirajah AA, Ausio J. MeCP2 preferentially binds to methylated linker DNA in the absence of the terminal tail of histone H3 and independently of histone acetylation. FEBS Lett. 2008;582:1157–1162. doi: 10.1016/j.febslet.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 11.Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 12.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 13.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 14.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 15.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 17.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl Acad. Sci. USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, He C, Moore SC, Ausió J. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 2001;276:12764–12768. doi: 10.1074/jbc.M100501200. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Ausió J. Presence of a highly specific histone H1-like protein in the chromatin of the sperm of the bivalve mollusks. Mol. Cell. Biochem. 1992;115:163–172. doi: 10.1007/BF00230327. [DOI] [PubMed] [Google Scholar]
- 22.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 23.Yager TD, van Holde KE. Dynamics and equilibria of nucleosomes at elevated ionic strength. J. Biol. Chem. 1984;259:4212–4222. [PubMed] [Google Scholar]
- 24.Abbott DW, Chadwick BP, Thambirajah AA, Ausio J. Beyond the Xi: macroH2A chromatin distribution and post-translational modification in an avian system. J. Biol. Chem. 2005;280:16437–16445. doi: 10.1074/jbc.M500170200. [DOI] [PubMed] [Google Scholar]
- 25.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 26.Ishibashi T, Dryhurst D, Rose KL, Shabanowitz J, Hunt DF, Ausio J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009;48:5007–5017. doi: 10.1021/bi900196c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davie JR, Saunders CA. Chemical composition of nucleosomes among domains of calf thymus chromatin differing in micrococcal nuclease accessibility and solubility properties. J. Biol. Chem. 1981;256:12574–12580. [PubMed] [Google Scholar]
- 28.Rozhon W, Baubec T, Mayerhofer J, Mittelsten Scheid O, Jonak C. Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 2008;375:354–360. doi: 10.1016/j.ab.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Dryhurst D, Ishibashi T, Rose KL, Eirin-Lopez JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Walzak AA, Veldhoen N, Feng X, Riabowol K, Helbing CC. Expression profiles of mRNA transcript variants encoding the human inhibitor of growth tumor suppressor gene family in normal and neoplastic tissues. Exp. Cell Res. 2008;314:273–285. doi: 10.1016/j.yexcr.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 32.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. BioTechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 33.Kwon MJ, Oh E, Lee S, Roh MR, Kim SE, Lee Y, Choi YL, In YH, Park T, Koh SS, et al. Identification of novel reference genes using multiplatform expression data and their validation for quantitative gene expression analysis. PloS One. 2009;4:e6162. doi: 10.1371/journal.pone.0006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer VA, Sun JM, Li L, Davie JR. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods. 2003;31:67–75. doi: 10.1016/s1046-2023(03)00089-6. [DOI] [PubMed] [Google Scholar]
- 35.Rose SM, Garrard WT. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- 36.Nicolas RH, Wright CA, Cockerill PN, Wyke JA, Goodwin GH. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983;11:753–772. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li A, Yu Y, Lee SC, Ishibashi T, Lees-Miller SP, Ausio J. Phosphorylation of histone H2A.X by DNA-dependent protein kinase is not affected by core histone acetylation, but it alters nucleosome stability and histone H1 binding. J. Biol. Chem. 2010;285:17778–17788. doi: 10.1074/jbc.M110.116426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausió J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J. Mol. Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 40.Kurtz K, Saperas N, Ausio J, Chiva M. Spermiogenic nuclear protein transitions and chromatin condensation. Proposal for an ancestral model of nuclear spermiogenesis. J. Exp. Zool. B. 2009;312B:149–163. doi: 10.1002/jez.b.21271. [DOI] [PubMed] [Google Scholar]
- 41.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen JC, Ghosh RP, Woodcock CL. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010;62:732–738. doi: 10.1002/iub.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satoh MS, Poirier GG, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry. 1994;33:7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- 44.Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- 45.Zardo G, Reale A, De Matteis G, Buontempo S, Caiafa P. A role for poly(ADP-ribosyl)ation in DNA methylation. Biochem. Cell Biol. Biochimie et biologie cellulaire. 2003;81:197–208. doi: 10.1139/o03-050. [DOI] [PubMed] [Google Scholar]
- 46.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum. Mol. Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 48.Kumar A, Kamboj S, Malone BM, Kudo S, Twiss JL, Czymmek KJ, LaSalle JM, Schanen NC. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J. Cell Sci. 2008;121:1128–1137. doi: 10.1242/jcs.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 51.Petrotchenko EV, Serpa JJ, Borchers CH. Use of a combination of isotopically coded cross-linkers and isotopically coded N-terminal modification reagents for selective identification of inter-peptide crosslinks. Anal. Chem. 2010;82:817–823. doi: 10.1021/ac901637v. [DOI] [PubMed] [Google Scholar]
- 52.Petrotchenko EV, Xiao K, Cable J, Chen Y, Dokholyan NV, Borchers CH. BiPS, a photocleavable, isotopically coded, fluorescent cross-linker for structural proteomics. Mol. Cell. Proteomics. 2009;8:273–286. doi: 10.1074/mcp.M800265-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Petrotchenko EV, Serpa JJ, Borchers CH. An isotopically-coded CID-cleavable biotinylated crosslinker for structural proteomics. Mol. Cell. Proteomics. 2010 doi: 10.1074/mcp.M110.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell. Biol. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandler SP, Guschin D, Landsberger N, Wolffe AP. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–7018. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- 56.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 57.Yakabe S, Soejima H, Yatsuki H, Tominaga H, Zhao W, Higashimoto K, Joh K, Kudo S, Miyazaki K, Mukai T. MeCP2 knockdown reveals DNA methylation-independent gene repression of target genes in living cells and a bias in the cellular location of target gene products. Genes Genet. Sys. 2008;83:199–208. doi: 10.1266/ggs.83.199. [DOI] [PubMed] [Google Scholar]
- 58.Thomas JO, Thompson RJ. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977;10:633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- 59.Jaeger AW, Kuenzle CC. The chromatin repeat length of brain cortex and cerebellar neurons changes concomitant with terminal differentiation. EMBO J. 1982;1:811–816. doi: 10.1002/j.1460-2075.1982.tb01252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Felle M, Hoffmeister H, Rothammer J, Fuchs A, Exler JH, Langst G. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 2011;39:6956–6967. doi: 10.1093/nar/gkr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pearson EC, Bates DL, Prospero TD, Thomas JO. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur. J. Biochemi. / FEBS. 1984;144:353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- 62.Stratling WH, Yu F. Origin and roles of nuclear matrix proteins. Specific functions of the MAR-binding protein MeCP2/ARBP. Crit. Rev. Eukaryot. Gene Expr. 1999;9:311–318. doi: 10.1615/critreveukargeneexpr.v9.i3-4.150. [DOI] [PubMed] [Google Scholar]
- 63.Thambirajah AA, Ausio J. A moment's pause: putative nucleosome-based influences on MeCP2 regulation. Biochem. Cell Biol. = Biochimie et biologie cellulaire. 2009;87:791–798. doi: 10.1139/O09-054. [DOI] [PubMed] [Google Scholar]
- 64.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 65.Pina B, Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 1987;123:51–58. doi: 10.1016/0012-1606(87)90426-x. [DOI] [PubMed] [Google Scholar]
- 66.Bosch A, Suau P. Changes in core histone variant composition in differentiating neurons: the roles of differential turnover and synthesis rates. Eur. J. Cell Biol. 1995;68:220–225. [PubMed] [Google Scholar]
- 67.Fernando RN, Eleuteri B, Abdelhady S, Nussenzweig A, Andang M, Ernfors P. Cell cycle restriction by histone H2AX limits proliferation of adult neural stem cells. Proc. Natl Acad. Sci. USA. 2011;108:5837–5842. doi: 10.1073/pnas.1014993108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 69.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 70.Dandrea M, Donadelli M, Costanzo C, Scarpa A, Palmieri M. MeCP2/H3meK9 are involved in IL-6 gene silencing in pancreatic adenocarcinoma cell lines. Nucleic Acids Res. 2009;37:6681–6690. doi: 10.1093/nar/gkp723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian F, Hu XZ, Wu X, Jiang H, Pan H, Marini AM, Lipsky RH. Dynamic chromatin remodeling events in hippocampal neurons are associated with NMDA receptor-mediated activation of Bdnf gene promoter 1. J. Neurochem. 2009;109:1375–1388. doi: 10.1111/j.1471-4159.2009.06058.x. [DOI] [PubMed] [Google Scholar]
- 72.Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Different. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- 73.Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 74.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 75.Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J. Cell. Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- 76.Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cell. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- 77.Soshnikova N, Duboule D. Epigenetic regulation of Hox gene activation: the waltz of methyls. Bioessays. 2008;30:199–202. doi: 10.1002/bies.20724. [DOI] [PubMed] [Google Scholar]
- 78.de Capoa A, Febbo FR, Giovannelli F, Niveleau A, Zardo G, Marenzi S, Caiafa P. Reduced levels of poly(ADP-ribosyl)ation result in chromatin compaction and hypermethylation as shown by cell-by-cell computer-assisted quantitative analysis. FASEB J. 1999;13:89–93. doi: 10.1096/fasebj.13.1.89. [DOI] [PubMed] [Google Scholar]
- 79.Pearson EC, Butler PJ, Thomas JO. Higher-order structure of nucleosome oligomers from short-repeat chromatin. EMBO J. 1983;2:1367–1372. doi: 10.1002/j.1460-2075.1983.tb01593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thompson RJ. Studies on RNA synthesis in two populations of nuclei from the mammalian cerebral cortex. J. Neurochem. 1973;21:19–40. doi: 10.1111/j.1471-4159.1973.tb04222.x. [DOI] [PubMed] [Google Scholar]
- 81.Rose KL, Li A, Zalenskaya I, Zhang Y, Unni E, Hodgson KC, Yu Y, Shabanowitz J, Meistrich ML, Hunt DF, et al. C-terminal phosphorylation of murine testis-specific histone H1t in elongating spermatids. J. Proteome Res. 2008;7:4070–4078. doi: 10.1021/pr8003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Petrotchenko EV, Borchers CH. ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinformatics. 2010;11:64. doi: 10.1186/1471-2105-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.