Abstract
Transfer RNAs (tRNAs) are present in all types of cells as well as in organelles. tRNAs of animal mitochondria show a low level of primary sequence conservation and exhibit ‘bizarre’ secondary structures, lacking complete domains of the common cloverleaf. Such sequences are hard to detect and hence frequently missed in computational analyses and mitochondrial genome annotation. Here, we introduce an automatic annotation procedure for mitochondrial tRNA genes in Metazoa based on sequence and structural information in manually curated covariance models. The method, applied to re-annotate 1876 available metazoan mitochondrial RefSeq genomes, allows to distinguish between remaining functional genes and degrading ‘pseudogenes’, even at early stages of divergence. The subsequent analysis of a comprehensive set of mitochondrial tRNA genes gives new insights into the evolution of structures of mitochondrial tRNA sequences as well as into the mechanisms of genome rearrangements. We find frequent losses of tRNA genes concentrated in basal Metazoa, frequent independent losses of individual parts of tRNA genes, particularly in Arthropoda, and wide-spread conserved overlaps of tRNAs in opposite reading direction. Direct evidence for several recent Tandem Duplication-Random Loss events is gained, demonstrating that this mechanism has an impact on the appearance of new mitochondrial gene orders.
INTRODUCTION
The typical gene complement of metazoan mitochondria is remarkably conserved, comprising genes for 13 proteins, 2 ribosomal RNAs and 22 transfer RNAs (tRNAs), two specific for leucine and serine, respectively, and a single one for each of the other 18 amino acid specificities (1). Some exceptions to this rule have been described for several non-bilaterian animals that feature additional genes (2), and for many bivalve molluscs that exhibit an additional, sex-specific open reading frame of unknown function (3). Most of the deviations, however, involve the loss of tRNAs. In extreme cases, such as Cnidaria (4,5) or Chaetognatha (6), only one or two of the tRNAs are encoded in the mitochondrial genome (mitogenome), the missing ones being functionally replaced by nuclear tRNAs (7,8). In addition, the tRNA genes of metazoan mitogenomes often appear degenerated. In many cases, they still show the famous cloverleaf structure, but lack the otherwise highly conserved D-loops and/or T-loops (9). Some tRNAs lost complete arms (10,11). This is the case for all tRNAs in several mitogenomes from Nematoda (12). Losses of complete D- and T-domains were also reported in Chelicerata (13–16). Due to the lack of the systematic investigation of mitochondrial tRNAs (mt-tRNAs), however, no overview of these features throughout the metazoan Tree of Life has become available so far.
The order and reading direction of the genes on (typically) circular mitogenomes varies throughout Metazoa and hence constitutes a valuable source of information for phylogenetic reconstructions (17–19). The mechanisms causing genome rearrangements, however, are poorly understood. Most computational approaches assume that either inversions or transpositions are the elementary operations taking place. Inversions can be explained by inter-mitochondrial recombination (20,21). Tandem duplications of parts of the genome with subsequent random loss of duplicates, on the other hand, were suggested in an analysis of lizard mitogenomes (22). Investigation into the mechanisms of mitogenome rearrangements require examples of very recent rearrangement events since in such cases it is likely that the genomic sequence will have maintained information that can be used to distinguish between different hypotheses. Since the genomic positions of mt-tRNAs are rearranged much more frequently than the larger protein-coding genes and rRNA genes [as shown e.g. by the data compiled in (23)], a correct and complete annotation of mt-tRNAs is an important prerequisite for a systematic investigation into rearrangement mechanisms.
Typically, non-mt-tRNAs are among most highly conserved genes (24). Despite their short size and their divergence predating the last universal common ancestor, their homology is still clearly recognizable (25). The preservation of a common structural layout, and the extreme sequence conservation makes it possible to use a single tool, tRNAscan–SE, to identify tRNAs with nearly perfect accuracy in the nuclear DNA of eukaryotes and in the genomes of prokaryotes alike (26). Mt-tRNAs, however, are often structurally diverged (27,28). This makes their detection and annotation a challenging computational problem (29) and has lead to the development of specialized tools such as ARWEN (30) for this purpose. In contrast to tRNAscan–SE that searches for a complete cloverleaf structure, ARWEN (30) first identifies only the most conserved domain, the anticodon stem. The subsequent evaluation of possible D-stem and T-stem structures and the search for an acceptor stem then provides specificity. Nevertheless, ARWEN buys its increased sensitivity at the expense of a substantial false discovery rate. In its normal mode of operation, tRNAscan–SE uses covariance models (CMs) (specific to the three domains of life) to investigate the initial candidates. Instead, the mitogenome can be searched directly with the CMs, leading to an increase in sensitivity. State of the art annotation pipelines thus use results of both programs followed by inspection by eye and manual curation of the results (31). This is in particular the case for the 1876 metazoan mitochondrial RefSeq genomes (32) used in the present study. We restricted ourselves to RefSeq genomes because this database is the best source for a test set of non-redundant metazoan mitogenomes. All these genomes are curated by NCBI staff, feature a consistent format, and fulfill minimum quality standards. We may expect therefore, that annotation errors in this data set are rare enough to allow a meaningful statistical comparison of annotation tools.
Both ARWEN and tRNAscan–SE use common models for all tRNAs hence employ a consensus of the features specific to individual tRNA families. Given the moderate size of metazoan mitogenomes of usually <20 kb it is well within reach to use a covariance model customized to each of the 22 tRNA families. With the recent improvements of the Infernal software (33), the required computational resources have been reduced to a level that poses no restrictions in the context of mitogenomes any more. The strategy followed here is therefore to use Infernal as search engine for specialized covariance models for each of the 22 mt-tRNAs and for some of the aberrant tRNA structures. We implemented a script called MiTFi (mitochondrial tRNA finder) that invokes Infernal-1.0.2 using all covariance models. It predicts anticodons for all candidates and then selects plausible hits that are most likely true mt-tRNAs. This pipeline is intended to be used automatically for all metazoan mitogenomes without specific adjustments for individual taxonomical families. More precisely, no prior knowledge about expected tRNA sequences or structures is required since we use a single set of generic CMs to annotate all metazoan genomes. An alternative strategy would be to use specific CM models for particular clades, such as the nematode-specific model of tRNAscan–SE, or to modify the thresholds and parameters of the other search tools in a clade-specific way. However, this would implicitly make additional assumptions and also reduce the specificity of the search tools on other clades. Hence, in order to build a generally applicable pipeline, we opt for generic CMs that are phylogenetically agnostic.
MATERIALS AND METHODS
Alignments and covariance models
For the construction of the covariance models we started from an initial set of tRNAs obtained by scanning all available metazoan mitogenomes of the NCBI RefSeq version 39 (32) with both tRNAscan-SE-1.23 and ARWEN-1.2.3 tRNAscan–SE annotations were computed invoking the options –O and –X 5 to ensure that the program searches only with the built-in CM and that the number of false negatives is reduced to a minimum. After removing duplicates we sorted the sequences according to their corresponding amino acid as defined by the anticodon. For both serine and leucine there are two groups of tRNAs recognizing two distinct anticodons classes. In the case of serine the two groups are very different and can be easily distinguished by the codons they recognize (UCN versus AGY). For the leucine tRNAs, however, multiple duplication/deletion events occurred throughout metazoan evolution, in which remolded Leu-UUR tRNA genes have taken over the role of isoaccepting Leu-CUN tRNAs (34,35). Since this makes it impossible to determine orthology by the anticodon alone we initially treated the leucine tRNAs as a single set.
We constructed 21 initial alignments corresponding to the 21 tRNA classes using ClustalW2 (36). The NCBI taxonomy (www.ncbi.nlm.nih.gov/sites/entrez?db=taxonomy) was used as guide tree since we observed that this leads to an improvement of the alignment compared to ClustalW2's estimate of the guide tree. Nevertheless, extensive manual editing was required to rearrange poorly aligned sequences and to exclude likely false positives. These alignments were used to build a first set of CMs using Infernal. For the leucine tRNA we used the integrated function calling the ––ctarget option of Infernal to build two separate CMs. These correspond to the two major tRNA-Leu classes, namely the ancestral Leu-CUN group and the Leu-UUR together with all their secondarily remodelled descendants.
The complete collection of metazoan mitogenomes was then scanned again with these 22 CMs. The resulting new set of predictions was aligned with cmalign to the covariance models of the corresponding tRNA family. Manual editing again lead to a noticeable improvement of the structural alignments. Although Infernal already implements strategies to compensate for biased sampling, we excluded nearly identical sequences and kept only a subset with approximately uniform phylogenetic distribution in the final seed alignments, which, depending on primary sequence conservation of the tRNA family, consist of 33–69 sequences. The 22 final CMs were calibrated to enable Infernal to compute P-values and E-values of matches.
Mitochondrial tRNA finder MiTFi
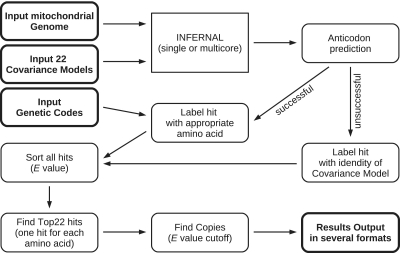
Since mt-tRNAs of the different families are distant homologues of each other, a search with one CM typically not only recognizes members of the tRNA family on which it was trained but also reports several other tRNA genes. The mitochondrial tRNA finder (MiTFi) is a script that invokes Infernal to search the target mitogenome with all 22 CMs and then employs a step-wise procedure (Figure 1) to evaluate and summarize the search results. Its output is a comprehensive annotation of tRNA genes.
Figure 1.
The MiTFi annotation pipeline for complete metazoan mitogenomes. Starting from all Infernal-hits, overlapping (i.e. conflicting) predictions are reconciled in a step-wise procedure.
For all Infernal-hits, MiTFi attempts to predict an anticodon. To this end, the number of interior stems and the length of the loops is evaluated. If only two interior stem loops are predicted, i.e. in the case of tRNAs which lost a secondary domain (e.g. the D-domain or the T-domain), first the loops are scanned for unpaired regions of 7 nt. If only one loop has this expected size, it is interpreted as the anticodon loop. If both loops have 7 unpaired nt, the loop closest to the mean of the sequence is regarded as the anticodon loop. If no loop containing exactly 7 nt is found, also a loop size of 9 is considered. If no candidate for an anticodon loop can be found according to these rules, the corresponding data fields for anticodon are left empty and the hit is tagged with the amino acid of the CM that found this hit.
Typically the CMs for specific tRNAs also recognize several other tRNAs, although in most cases with much larger E-values. For each locus, the MiTFi pipeline accepts only the hit of the CM matching with the smallest E-value. In practice, this simple rule is sufficient to disambiguate overlapping CM hits. Note that no score cutoffs are used for the 22 top hits at this point. In order to accommodate overlaps of tRNA genes, several cases of which are well documented in mitogenomes (9,37,38), MiTFi by default regards predictions that overlap not more than 10 nt as distinct loci. After this first iteration, in which best hits are accepted according to their identity, MiTFi tries to annotate copies of tRNA genes in remaining genomic locations. Hits without a specified anticodon are also annotated during this second step.
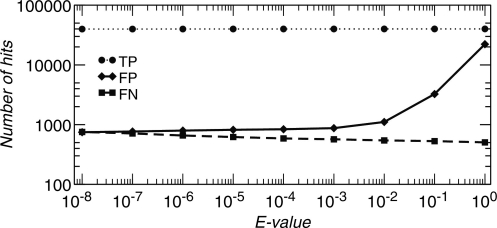
Almost all tRNA families exhibit a large diversity and in particular include aberrant sequences that lack complete structural domains. As a consequence there is no natural cutoff value for the Infernal bit-score that would be analogous to the COVE score threshold used in tRNAscan–SE. In order to determine an appropriate cutoff for the Infernal predictions, we therefore compared the predictions of the 22 CMs to the existing RefSeq annotations. Figure 2 shows that true positives are nearly unaffected at E = 0.001, while the false positives drop to a nearly constant value at this level. For this reason we used this E-value as a cutoff to predict remaining tRNA genes in the second step. We note that, in contrast to the bitscore, the E-value is computed using a model-specific calibration.
Figure 2.
Comparison of E-values of MiTFi hits as compared to RefSeq annotation: true positive hits (TP, circles), false positive hits (FP, diamonds), and false negative hits (FN, squares). By default, MiTFi uses an E-value cutoff of E ≤ 0.001 for finding copies of tRNA genes as there is no significant change for false positive hits below this limit.
Due to the variability of mitochondrial genetic codes (28) the correspondence of anticodon and isoacceptor class is ambiguous. Thus MiTFi allows the user to specify a code from the NCBI genetic code page (http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi) or to supply modified codes. Finally, MiTFi offers a variety of output options to facilitate the manual inspection of the results. It is also possible to distinguish between genes and degrading pseudogenes as calculated E-values allow comparisons of all hits. The re-annotation of the mitogenomes with MiTFi was performed at the High Performance Cluster of the TU Dresden (http://tu-dresden.de/die_tu_dresden/zentrale_einrichtungen/zih/hpc). MiTFi is available for download at our website (http://www.bioinf.uni-leipzig.de/software.html) including all required CMs.
Data evaluation
The complete dataset was stored in a MySQL (http://www.mysql.com/) database server based on the tRNAdb system (39,40) allowing further investigations. The complete data analysis was performed with the help of internal functions of the database server. In addition, we used Infernal and the RALEE Emacs mode (41) for detailed alignment studies, e.g. to distinguish false and true positive hits. Plots of secondary structures within this study were performed using the RNAplot program (42).
RESULTS AND DISCUSSION
Re-annotation of mt-tRNA genes
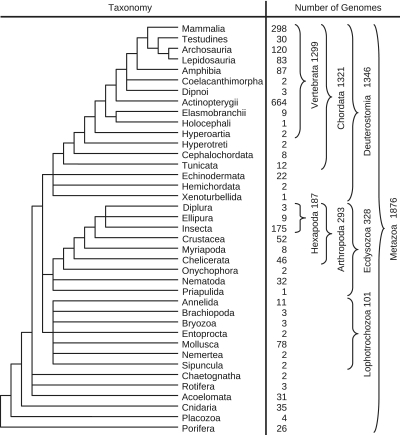
The complete set of 1876 metazoan mitogenomes (Figure 3) was annotated independently with tRNAscan–SE, ARWEN and MiTFi and then compared to the RefSeq annotation. An annotation item computed by one of the three methods was counted as true positive if it overlapped a RefSeq entry with the same identity. We disregarded strand information and the distinction between the two serine and leucine tRNAs since the RefSeq annotation shows a high level of misannotations of this type, see e.g. (43). All hits without an overlap with RefSeq were counted as false positives. Table 1 summarizes the results, showing that the use of family-specific CMs increases sensitivity above the level of ARWEN while at the same time reaching the same precision rates as those of tRNAscan–SE. We note that our estimates of the precision rate of ARWEN (86.9%) is more favorable than the 80.2% reported by its authors (30).
Figure 3.
Taxonomic distribution of metazoan mitogenomes investigated in this study.
Table 1.
Comparisons of mt-tRNA predictions and RefSeq annotation
| Method | RefSeq (40 521) |
||||
|---|---|---|---|---|---|
| TP | FP | FN | Sens. | Prec. | |
| tRNAscan–SE | 36 374 | 688 | 4147 | 0.898 | 0.981 |
| ARWEN | 39 569 | 5957 | 952 | 0.977 | 0.869 |
| MiTFi | 39 953 | 873 | 568 | 0.986 | 0.979 |
The data covers 40 521 mt-tRNA gene annotations of 1876 RefSeq genomes. Numbers of true positives (TP), false positives (FP), false negatives (FN), sensitivity (Sens.) and precision rate (Prec.) are counted relative to the RefSeq annotation.
The NCBI RefSeq is currently the most comprehensive data source for mitogenomes and their annotation. It is not a perfect gold standard, however. A detailed analysis of mitogenomes, for instance, revealed more than a dozen annotation errors including missing tRNAs, inaccurate positions, wrong reading directions and incorrect anticodons and isoacceptor families affecting 7 of the 16 echinodermate mitogenomes (43). In order to obtain more realistic performance estimates, we thus manually inspected about 3250 false positive hits. These consist of the best hits for individual tRNAs from the first step of MiTFi and other tRNAs with E < 0.1. We first created alignments for each isoacceptor family using Infernal. Within each of these alignments, MiTFi hits were sorted taxonomically such that known tRNAs and putative false positives from the most closely related species are located in adjacent rows to facilitate the manual inspection. We found 272 tRNA candidates in 170 organisms that closely match a homologous known tRNA gene in both its conserved primary and secondary structures, 145 of which are in addition supported by CM E-values <10−6. About 30 sequences from Metatheria were originally tagged as false positives due to an incorrect anticodon assignment, the other 242 hits were newly identified. Examples of corrected and newly found tRNAs are given in Supplementary Figure S1. All alignments containing newly identified mt-tRNAs and their homologs in related organisms are compiled in Supplementary Dataset S1. We reclassified these cases as true mt-tRNAs.
Many of the remaining false positive hits are introduced because MiTFi includes at least one hit for each of the 22 canonical tRNAs. Some clades, however, have lost most of their mitochondrially encoded tRNAs. Loss of tRNAs in Cnidaria, for instance, accounts for about 283 of the false predictions. Other false positive hits occur in Arthropoda (71 hits), Nematoda (31 hits) and other basal metazoans (except Cnidaria, 59 hits). A further group of 264 false positives is easily recognizable by large overlaps with mitochondrial gene annotations and a lack of conserved secondary structures. Several additional false positives are the result of an unusual genetic code or of RNA editing of the anticodon (44), since this leads to an assignment of the tRNA candidate to an incorrect amino acid specificity.
All tRNA genes annotated in RefSeq that were not recovered by MiTFi were also inspected manually on the basis of structure-annotated multiple alignments. We eliminated 146 annotations that showed neither recognizable sequence similarity nor a plausible structural conservation. Most of the false negatives that were not detected or only found with E-values larger than the cutoff lack one arm of the cloverleaf structure. These cases are concentrated in a few taxonomic groups: arthropods (127 hits), nematodes (102 hits), molluscs (14 hits) and basal metazoans, in particular poriferans (22 hits).
Some of the most unusual mt-tRNAs are found in Arachnida (14). Therefore, we evaluated all three programs in more detail on these genomes. ARWEN was able to detect 82.8% and tRNAscan recovered only 50.4% of the 9191 annotated mt-tRNAs of Arachnida in RefSeq while MiTFi performed best with 89.7%. A similar situation was reported for tRNA sequences in Cecidomyiidae (45), where tRNAs lack the 3′-end. In the two available genomes, MiTFi retrieved the majority (24 hits) while ARWEN reported 21 and tRNAscan–SE recovered only 7 of the RefSeq tRNA annotations. For both families together, MiTFi produced 62 false positive hits, ARWEN 233 and tRNAscan–SE 22. As MiTFi always reported most true positive and fewer false positive hits compared to ARWEN, its results are the best starting point for annotating genomes featuring completely truncated tRNA sequences. These results also show that tRNAscan–SE is not suitable to deal with such highly divergent sequences.
Loss of mt-tRNA genes
Some animals do not encode the full set of 22 mt-tRNA genes. Instead, they import the missing tRNAs from the cytosol. Cnidarians (46) and some Ceractinomorpha (sponges, belonging to Porifera) (47) lost up to 21 tRNA genes and only encode tRNAMet. Some members of these clades encode tRNATrp or copies of tRNAMet and import the remaining tRNAs. Another well-known case is the loss of a single mt-tRNALys gene in marsupials (48). Our data are entirely consistent with these findings: we did not predict any previously unknown tRNA genes within these three taxonomic groups, although MiTFi recovered some of the reported pseudogenes of the highly variable tRNALys-like sequences in marsupials, although only with a larger E-value cutoff (E > 0.1). Similarly, most of the putative candidates in Cnidaria and sponges detected in MiTFi's first search step are most likely false positives.
Within the Sciaroidea, a subfamily of the Insecta, where dramatically truncated sequences are described (45), we found only a subset of tRNA genes, many with very poor E-values. These tRNA sequences completely lack the 3′-end, including the full T-stem region. This severe degradation suggests that these organisms feature an unknown mechanism for repairing these tRNAs and/or for attaching amino acids to them. At present, it is unknown whether these small fragments still encode functional tRNAs, or whether the degraded mt-tRNAs are functionally replaced by tRNAs imported from the nucleus (7). A similar situation is observed in Onychophora, where only incomplete sets of truncated mt-tRNA genes were found (49). Here, extensive tRNA editing is capable of repairing large fragments of truncated tRNA molecules (50). Our data also reflect previous reports on the loss of tRNA genes in other taxonomic families, including Chaetognatha (51) and Rotifera (52). For these clades, we did not find complete sets of 22 tRNA genes and some of the predicted tRNAs have extremely poor E-values. Since we found no other tRNA genes in corresponding genomes, one can conclude that, once a nuclear tRNA has replaced a mt-tRNA, the lost tRNA genes are not restored in the mitogenome. This implies that the absence of mt-tRNA genes are phylogenetically informative markers that could help to clarify ambiguities. In basal metazoans, for instance, some clades lost the gene for tRNATrp, while others still encode it.
Overlapping mt-tRNA genes
Overlapping mt-tRNA genes have long been known throughout metazoans (9,37,38). In order to investigate how wide-spread such overlaps are, we considered overlaps of up to 10 nt as distinct tRNA loci. From candidates with a pairwise overlap of >10 nt MiTFi selects only the one with the largest E-value. The MiTFi script allows the user to change this default value of 10 and to consider even larger levels of overlap.
Our systematic analysis revealed more than 3700 cases of overlaps between tRNA genes in all 1876 metazoan mitogenomes. A summary of the taxonomically most conserved overlapping tRNA genes is given in Table 2. Single nucleotide overlaps are most common. Taxonomically conserved overlaps are mostly found for tRNAs encoded on different strands. This may be correlated to the fact that no alternative cleaving of the primary transcript is needed in this situation. For example, this is the case for the highly conserved tRNAIle and tRNAGln overlaps on different strands for up to 3 nt in arthropods and vertebrates. Hyperoartia seems to be an exception as it is the only group within the vertebrates where no overlaps could be detected.
Table 2.
Conserved overlaps of mt-tRNA genes that have been observed more than 50 times in the dataset
| No. | tRNA Genes | Overlap | Taxonomy |
| 1189 | tRNAIle+-tRNAGln− | 1,3,2 | Vertebrata (1056) |
| 3 | Arthropoda (131) | ||
| 3 | Priapulida (1) | ||
| 1 | Xenoturbellida (1) | ||
| 902 | tRNAGln−-tRNAMet+ | 1 | Vertebrata (825) |
| 1 | Arthropoda (76) | ||
| 3 | Onychophora (1) | ||
| 639 | tRNAThr+-tRNAPro− | 1 | Vertebrata (607) |
| 1,2 | Arthropoda (24) | ||
| 1 | Cephalochor. (8) | ||
| 230 | tRNASer1+-tRNALeu1+ | 1 | Vertebrata (230) |
| 188 | tRNATrp+-tRNACys− | 8,1 | Arthropoda (187) |
| 1 | Priapulida (1) | ||
| 119 | tRNAGlu+-tRNAPhe− | 2,1 | Arthropoda (119) |
| 53 | tRNAArg+-tRNAAsn+ | 1,3 | Arthropoda (53) |
| 51 | tRNAAsn+-tRNASer1+ | 1,3 | Arthropoda (51) |
The size of the overlaps is given as number of overlapping nucleotides. Where multiple values are given, they are sorted by the frequency with which they appear. Overlaps that appear in <10% of the mitogenomes in a listed clade are omitted from the table. The orientation of genes are indicated by ‘+’ (plus strand) and ‘−’ (minus strand).
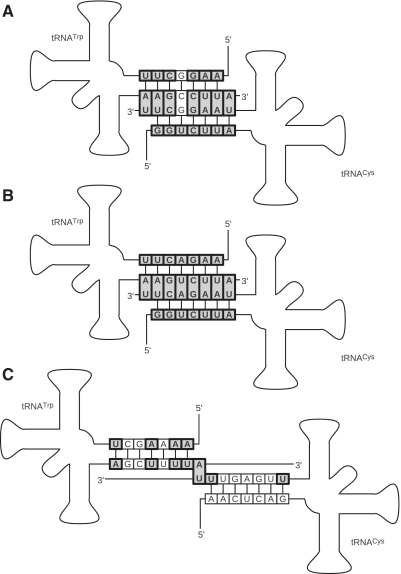
The most remarkable example of overlapping tRNA genes are tRNATrp and tRNACys in Arthropoda (53) (Figure 4). We investigated this link of two tRNA genes systematically and confirmed examples in every subphylum of Arthropoda. In contrast to this general picture, there are many species that independently lost the overlap. The two genes are located on different strands and overlap by up to 8 nt. They show a very high level of sequence conservation of the acceptor stem. Mutations in this short region would simultaneously affect a stem region in each of the tRNAs. Arthropoda genomes that lost this correlation do not show this strong sequence conservation any more. The difference of overlapping and non-overlapping acceptor stems are illustrated in Figure 4. While acceptor stems of Drosophila melanogaster (Hexapoda) in comparison to Eremobates palpisetulosus (54) (Chelicerata), that overlap by 8nt, are nearly perfectly conserved, the same region is much more variable, e.g. in other Hexapoda like Damon diadema (55), where the overlap is reduced to a single nucleotide.
Figure 4.
Overlapping tRNATrp and tRNACys genes in Arthropoda. Drosophila melanogaster [Hexapoda, (A)] and Eremobates palpisetulosus [Chelicerata, (B)] feature overlapping genes while Damon diadema [Chelicerata, (C)] encodes both genes with an overlap of only 1 nt. As a result the conservation of the stem region between the two Chelicerata species is much less pronounced than between the two organisms featuring overlapping genes of 8 nt at their 3′-ends even though they are members of completely different subphyla. Conserved nucleotides are highlighted in bold.
Our data demonstrate that overlapping tRNAs have a profound effect on primary sequence conservation, which needs to be taken in account e.g. in the context of phylogenetic studies based on (single) tRNA genes such as recently reported (56). Also when concatenated tRNAs are used (57), overlaps cannot be neglected. Like loss events, overlaps can also be used as a phylogenetic marker as once the overlapping link between two genes is broken [e.g. by a tandem duplication-random loss (TDRL) event], the two genes rapidly diverge making it unlikely to regain an overlapping configuration.
A dramatic type of overlap, suggesting that functional tRNAs could also be expressed from the reverse strand of known tRNA genes, was postulated (58). We searched the complete Infernal output, i.e. all candidate predictions used by MiTFi, for predictions that nearly perfectly overlap with opposite reading direction, although without success.
Exceptional structures of mt-tRNAs
More than 90% of mt-tRNAs share the common global cloverleaf secondary structure of nuclear-encoded tRNA sequences, i.e. a structure with four stems and three loops. A large number of exceptional mt-tRNAs have been described previously that lack either the D-domain or the T-domain. The CM-based approach greatly facilitates a comprehensive detection and analysis, since it provides efficient and accurate structural alignments of individual tRNAs to the family-specific norm. Using the NCBI taxonomic tree as an approximation of the phylogeny, we mapped all tRNA sequences and their characteristics to generate an overview of the distribution of exceptional structures and manually checked spots of structural divergences. As summarized in Table 3, hotspots of diversity in presence or absence of D- and T-domains are found throughout the two major groups of protostomes (Ecdysozoa and Lophotrochozoa). In contrast, both Deuterostomia and diploblasts (Placozoa, Porifera and Cnidaria) show classical cloverleaf structures with only a few exceptions.
Table 3.
Exceptional structures of mt-tRNA genes and loss of tRNA genes
| Taxonomy | Ser1 | Ser2 | Cys | others | missing |
|---|---|---|---|---|---|
| Deuterostomia | |||||
| Mammalia | #D | – | ○D | – | ○ |
| Testudines | #D | – | – | – | – |
| Archosauria | #D | – | – | – | – |
| Lepidosauria | #D | – | ○D | ○D | – |
| Amphibia | #D | – | ○D | – | – |
| Coelacanthimorpha | #D | – | – | – | – |
| Dipnoi | #D | – | – | – | – |
| Actinopterygii | ○Cl | – | – | – | – |
| Elasmobranchii | #D | – | – | – | – |
| Holocephali | #D | – | – | – | – |
| Hyperoartia | #D | – | – | – | – |
| Hyperotreti | #D | – | – | – | – |
| Cephalochordata | #D | – | #D | – | – |
| Tunicata | ○Cl | – | ○D | ○T | – |
| Echinodermata | #D | – | – | – | – |
| Hemichordata | #D | – | – | – | – |
| Xenoturbellida | #D | – | – | – | – |
| Ecdysozoa | |||||
| Diplura | #D | +D | ○D | ○D | – |
| Ellipura | #D | – | ○D | – | – |
| Insecta | ○Cl | ○D/T | – | ○D/T | ○ |
| Crustacea | #D | ○D/T | ○D/T | ○D/T | – |
| Myriapoda | #D | ○D | ○D/T | ○D/T | – |
| Chelicerata | #D | ○D | +D/T | ○D/T | – |
| Onychophora | #D | – | – | – | + |
| Nematoda | #D | #D | #D/T | +D/T | – |
| Priapulida | #D | – | – | – | – |
| Lophotrochozoa | |||||
| Annelida | #D | ○D | – | ○D | – |
| Brachiopoda | #D | +D | – | ○D/T | – |
| Bryozoa | #D | #D | ○D | ○D/T | – |
| Entoprocta | #D | – | – | – | – |
| Rotifera | #D | – | – | ○T | + |
| Mollusca | ○Cl | ○D | – | ○D/T | – |
| Nemertea | #D | – | – | – | – |
| Sipuncula | ○Cl | – | – | – | – |
| Platyhelminthes | #D | +D | +D | ○D/T | – |
| Basal Metazoa | |||||
| Chaetognatha | – | – | – | – | + |
| Cnidaria | – | – | – | – | + |
| Placozoa | +Cl | – | – | – | – |
| Porifera | +Cl | – | – | ○D | + |
‘○’ indicates occasional events, ‘+’ frequent (>50%) events and ‘#’ highlights taxa that all share the same abnormality. The ‘Ser1’ column summarizes tRNASer1 genes exceptionally featuring the classical cloverleaf (‘Cl’) or commonly lost the D-domain (‘D’). Columns ‘Ser2’, ‘Cys’ and ‘others’ indicate tRNA genes that lost the D-domain (‘D’), the T-domain (‘T’) or one of both domains (‘D/T’). The ‘missing’ column summarizes where it was not possible to find a complete set of 22 tRNA genes within the genomes.
We detected the well known lack of a D-domain (and innovation of a D-arm replacement domain) in mt-tRNASer1 (9) in nearly all Metazoa. In a few exceptions, a classical cloverleaf was retrieved. Frequent exceptions were found in basal metazoan lineages. Mt-tRNASer2 lacks also a D-domain, but however, only in Lophotrochozoa and Ecdysozoa, with highest penetrance in Bryozoa and Nematoda (in Deuterostomia this tRNA is always of complete 4-arm type). Accordingly, these loss events appear to be independent. Our data also revealed independent losses of the D-domain in tRNACys in Amphibia, Tunicata, Bryozoa, Platyhelminthes and Arthropoda in addition to those previously reported in Lepidosauria (59,60) and Mammalia (61). Further, while the compensation of the loss of the D-domain by a D-arm replacement loop seems to be a very common event in all Cephalochordata tRNACys, the absence of either the D-domain or the T-domain is the rule for Nematoda tRNACys. A particularly nice case of variability in domain loss concerns Campodea lubbocki that lacks the D-domain of tRNACys while a normal cloverleaf structure is present in the closely related Campodea fragilis (62). The high frequency of these events suggests that the abnormal tRNACys should be still functional. The widespread loss of either the D- or the T-domain leads to the well-known large diversity in structures for Arthropod mt-tRNAs (14,15,63). Our taxonomic overview now identified that this variability is focused on only three hotspots. Chelicerata, Crustacea and Myriapoda mt-tRNAs numerously lost arms of the cloverleaf structure, with different patterns even within each group, indicating a large number of independent events. Figure 5 illustrates these parallel events for all three hotspots. In contrast, other Arthropoda groups such as Insecta show only very occasional deviations from the classical cloverleaf structure.
Figure 5.
Examples of tRNACys secondary structures derived genes in (A) Crustacea (left: Tigriopus japonicus, middle: Daphnia pulex, right: Lepeophtheirus salmonis), (B) Myriapoda (Scutigerella causeyae, Narceus annularus, Antrokoreana gracilipes), and (C) Chelicerata (Buthus occitanus, Damon diadema, Haemaphysalis flava). In each family, some organisms present four-arm cloverleafs (middle column), others present tRNAs missing the T-domain (left) or the D-domain (right). Anticodons are highlighted in bold.
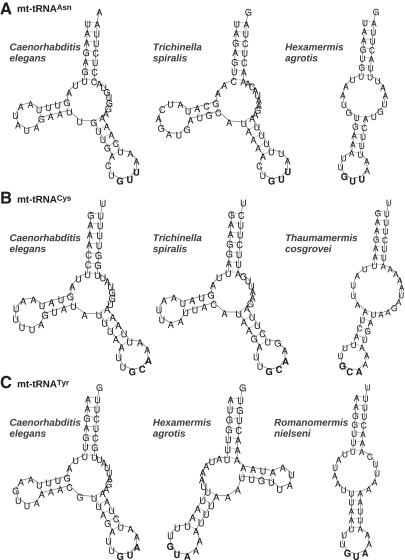
In addition to structures missing either the D- or the T-domains, we retrieved structures with truncated acceptor stems. This unusual situation discovered in Lithobius forficatus and calling for specific editing mechanisms to gain mature tRNAs (64), could now be confirmed also for other related organisms in Myriapoda. The case of Nematoda mt-tRNAs was also analyzed in details. Rather than being the exception (65), bizarre tRNAs appear to be the rule. Interestingly, even with the reduced sensitivity of MiTFi for shorter sequences in general (due to their reduced information content) and for tRNAs lacking individual arms in particular (as the deletion incurs a score penalty), we found tRNAs without T-domains throughout the whole taxonomic group. This led us to the subfamily Enoplea where we obtained a very low sensitivity and, in addition, hits with minimal tRNA structures featuring both D- and T-stem replacement loops. Therefore, we constructed group-specific new covariance models built only from nematode sequences and searched for missing genes. This lead us to predict extremely truncated sequences of only acceptor- and anticodon stems. We were not able to find other candidates featuring D- or T-stems in the same genomes. These truncated structures could be predicted for several tRNA families, including tRNAAsn, tRNACys and tRNATyr (Figure 6). Some hits overlap with previous tRNA annotations in RefSeq, others were newly found. Interestingly, since gene overlaps could be reduced to a minimum, some of our hits fit much better with the annotations of the adjacent genes than in prior tRNA annotation. The newly detected structures present rather conserved stems as compared within Enoplea or to Caenorhabditis elegans. As acceptor stems define the 3′/5′-ends of tRNA genes, their high conservation strongly suggests that we found a correct annotation. These nematode specific results are not included in the statistical evaluation of the previous section since it required significant manual post-processing. As more information becomes available it may be worth while, however, to append a search with specific CMs for aberrant structures as a further step in the MiTFi pipeline.
Figure 6.
Examples of tRNAs without D- and T-domains in several Enoplea in comparison to known mitochondrial tRNAs in C. elegans. Sequences were found with refined nematode-specific covariance models. Anticodons are highlighted in bold.
The results of this systematic analysis of exceptional structures illustrates major features of the evolution of metazoan mt-tRNAs. All basal metazoan mt-tRNAs fold into the common cloverleaf, supporting a secondary structure from which all metazoan mt-tRNAs originate from. Import mechanisms appeared also very early in evolution as Chaetognatha and Cnidaria already lost most of their mitochondrial encoded tRNAs and need to import them from the cytosol. Mechanisms to compensate/adapt to tRNAs with lost D-domains emerged shortly afterwards as nearly all Bilateria (Ecdysozoa, Lophotrochozoa and Deuterostomia) encode at least for one tRNA missing a D-domain. Equivalent mechanisms for mt-tRNAs lacking the T-domain appeared only in Ecdysozoa and Lophotrochozoa, finally leading to mitochondrial translation machineries in Enoplea tolerating minimal tRNAs lacking both domains. These further developments seem to have arisen after the split from the Deuterostomia (showing only sequences lacking the D-domain). Only Tunicata exceptionally encode mt-tRNAs lacking the T-domain that suggests an independent evolutionary event.
TDRL events in mitogenomes
The increased sensitivity of the CM-based approach frequently reveals additional hits of duplicated mt-tRNAs. In most cases these additional candidates appear to be degrading and most likely constituting pseudogenes as they show larger E-values than the best scoring copy of the homologous gene (Figure 7). Such cases provide direct evidence for the mechanisms of mitogenome rearrangements (66). The systematic survey reported here, therefore, provides direct evidence for the profound impact of TDRL events on the appearance of new gene orders in several sub-phyla. According to the orders of tRNA genes, we identified 77 genomes showing patterns of tandem duplications. We recovered, in addition to the well-studied examples, such as those in Heteronotia binoei and other Lepidosauria (67) also unknown events. To our knowledge this is the first systematic survey for TDRL events throughout the Metazoa.
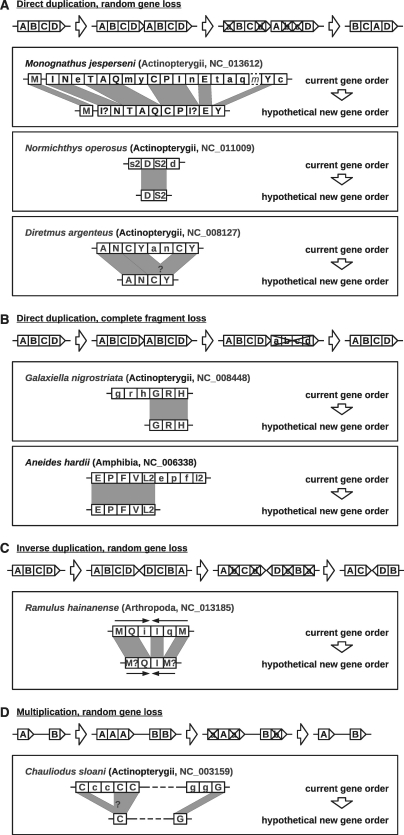
Figure 7.
TDRL events in metazoan mitogenomes. Only duplicated tRNA genes are shown, lower case letters indicate degrading genes (with larger E-values than the best scoring copy of the homologous gene copy). The one-letter code is used for abbreviating amino acids. Boxes with dashed outlines show pseudogenes that were not detected by MiTFi but by manual inspection. Dashed lines illustrate large genome segments containing other genes. Unknown hypothetical new gene orders are visualized by ‘?’ as the duplicated tRNA genes do not have acquired mutations yet.
Most tandem duplications seem to occur directly on the same strand (Figure 7A). Several examples could be retrieved in mitogenomes of Actinopterygii. The mitogenome of the deep sea eel-like fish Monognathus jesperseni (68) shows a large tandem duplication including at least nine tRNA genes which were, so far, incorrectly annotated as a control region. In fact, this large duplication is comparable in terms of the number of duplicated genes to previously reported events in Plethodon (69). The duplicated parts of the genome still show the same gene order. One copy of mt-tRNAMet is missing in our predictions. Its remnant, which can be identified by direct sequence alignment, lacks parts of both the D-domain and the anticodon region. For the other mt-tRNAs we observe large differences in the E-values of the two copies, clearly distinguishing the intact tRNAs from their error-ridden copies which most likely are not functional any more. As a result of these events, the gene order of the remaining 22 best-scoring tRNA genes has been completely rearranged. A similar situation has been reported for Normichthys operosus, another bony fish (70). Again we can clearly distinguish functional and degrading copies in the small cluster of tRNASer and tRNAAsp resulting in an inverted gene order compared to the ancestral observed for many other Actinopterygii (71). In Diretmus argenteus, for instance, already half of the duplicated fragment is degenerated. It is part of the WANCY region, which has been identified as a hotspot for tandem duplications in vertebrate genomes (72). The eventual outcome does not appear to be decided yet as at least half of the duplicated tRNA genes do not have acquired mutations that distinguish the copies.
Some mitogenomes containing tandem duplications seem to be losing a complete fragment with all duplicated tRNA genes (Figure 7B). We found this case in mitogenomes of the black-stripe minnow Galaxiella nigrostriata and the Sacramento mountain salamander Aneides hardii (73). A reason for the disappearance of these large fragments but not of randomly selected genes is may be due to different transcription rates of parts of the mitogenome as it is known in human (74).
We found the first convincing case of an inverse TDRL in the walking stick Ramulus hainanense (Figure 7C). It is an inverse TDRL in progress whose comparison of the E-values suggests that at least one tRNA will survive in each copy of the cluster, while the two copies mt-tRNAMet do not yet show any differences.
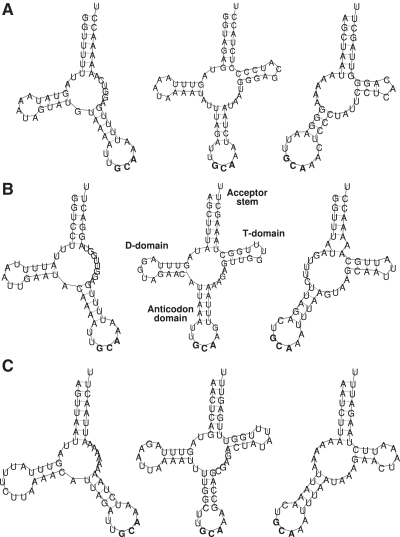
An extension of TDRLs is the occurrence of ‘multiplications’, i.e. the inclusion of multiple copies followed by random loss of duplicates. The mitogenome of Chauliodus sloani has two loci with up to 5 copies of the same tRNA. In this case there is no effect on the gene order.
Results of TDRL events can be studied in closely related mitogenomes that still have duplicated tRNA genes (Figure 8). Nice examples are the salamander species Plathodon cinereus, P. elongatus and P. petraeus, which exhibit numerous duplications (69) of the region containing tRNAsGlu, tRNAsThr, tRNAsPro and others. A comparison of E-values again clearly shows an ongoing change of the gene order in P. elongatus. Even though the two copies of tRNAsThr, either TEP or EPT, will be different from the ancestral state ETP as found also in other vertebrates (1).
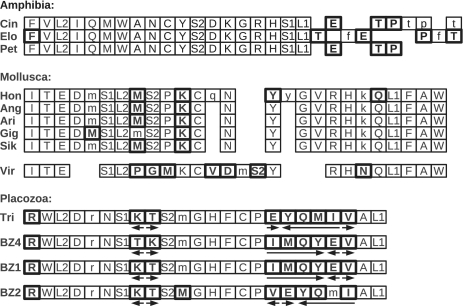
Figure 8.
Results of TDRL events in metazoan mitogenomes of closely related organisms in Amphibia (Elo: P. elongatus, Cin: P. cinereus, Pet: P. petraeus), Mollusca (Hon: Crassostrea hongkongensis, Ang: C. angulata, Ari: C. ariakensis, Gig: C. gigas, Sik: C. sikamea, Vir: C. virginica) and Placozoa (Tri: Trichoplax adhaerens, BZ4: Placozoan sp. BZ49, BZ1: Placozoan sp. BZ10101, BZ2: Placozoan sp. BZ2423). Different gene orders and non-degrading candidates of duplicated genes are shown in bold. Lower case letters indicate degrading genes (lower E-values than the best scoring copy of the homologous gene copy). Arrows indicate inverted duplicated genome fragments that are only present in Placozoa.
Similar events occurred in Mollusca where gene orders of six genomes show partial differences. Crassostrea hongkongensis, C. angulata, C. ariakensis, C. gigas and C. sikamea, show nearly the same gene orders, only in C. gigas another copy of tRNAMet seems to degrade. In contrast, C. virginica shows a gene order pattern different from related organisms probably because it forms the most basal branch of the group. At least one large stretch of duplicated DNA is shared by all six Crassostrea mitogenomes. The fact that all homologous gene copies are encoded on the same strands, further supports the hypothesis that they arose through a common TDRL event.
Another example of fast evolving genome organizations are Placozoa. Here, duplications and inversions of whole tRNA clusters can be observed. The single duplicated tRNA genes in Trichoplax adhaerens, Placozoan sp. BZ49 and Placozoan sp. BZ10101 show similar patterns, only Placozoan sp. BZ2423 differs from them as another tRNAMet gene is slightly more degenerated. In addition, an inversion of the tRNALys–tRNAThr cluster is present in Placozoan sp. BZ49. Most interesting in terms of TDRL events is the EYQMIV region of T. adhaerens. The most likely explanation of the different positions of tRNAVal in the trichoplax strains is a single inverse duplication followed by random loss events (iTDRL hypothesis). The most plausible alternative explanation requires two independent inversions of different parts of this regions without destroying any of the tRNA genes in the process. The EMBOSS tool equicktandem (75) identifies 12 repeated sequences with a length up to 25 nt within the EYQMIV region of the T. adhaerens genome. Together with the degrading copies of tRNAMet and tRNAArg this constitutes compelling evidence for the iTDRL hypothesis.
Over all, duplication events occur more often than previously expected: MiTFi annotated 329 potential isoacceptor tRNA genes in 210 mitogenomes. This number includes only copies with plausible E-values (E < 0.001). We expect that there are many additional tRNA copies that are already degraded beyond this cutoff. Our analysis thus most probably underestimates the number of TDRL events. This emphasizes the impact of TDRL events to the evolution of mt-tRNA genes as every duplication event is a potential starting point for changing the gene order of these mitogenomes. While the standard model, i.e. a tandem duplication followed by a complete loss of one of the redundant copies, is well understood from a formal/bioinformatic point of view (76,77), our results motivate also for further studies of the TDRL model. In particular, this includes multiplications, cases with inverse duplications and especially the possibility of partial loss. It is generally believed that different kinds of rearrangement operations have modified the gene order of metazoan mitogenomes throughout evolution, including inversions, transpositions, inverse transposition and TDRL (1). These operations have different mechanistic explanations. We suggest that a rearrangement model consisting of tandem duplication or inverse tandem duplication followed by random loss is more parsimonious in the number of necessary explanations for the observed rearrangements.
CONCLUSION
The use of specific covariance models for the 22 types of tRNAs occurring in the mitogenomes of Metazoa leads to a significant improvement of tRNA predictions, in particular regarding tRNAs with missing domains and/or other structural aberrations. Implemented in the MiTFi pipeline the approach sets the stage for a consistent re-annotation of mt-tRNAs in animals. In addition to recovering nearly all known mt-tRNAs, MiTFi discovered 242 previously unannotated tRNAs. Overall, MiTFi provides a substantial improvement in both sensitivity and precision rate for tRNA annotation in animal mitogenomes. The pipeline can also be used as an efficient way to check existing tRNA annotation. We do not employ clade-specific covariance models for truncated tRNAs because this would imply a prior knowledge of the expected structural variations. Furthermore, the use of specific CMs in other taxonomic families would lead to incorrect predictions as these unrelated CMs would only find truncated tRNAs. Such a procedure would require extensive manual post-processing. It appears more efficient, thus, to restrict the pipeline to generic, phylogenetically agnostic models.
The comparative analysis of mt-tRNAs across Metazoa reveals systematic patterns of tRNA loss, aberrant tRNA structures, and overlapping tRNA genes. While loss of tRNAs is particularly prevalent in basal metazoan clades, we observe that both tRNA overlap and deviant tRNA secondary structures are particularly frequent in Arthropoda. We found a surprising number of independent loss events for secondary structure elements and for overlapping patterns. In particular, there is compelling evidence for several functional tRNAs that lack both the T-domain and the D-domain in Enoplea.
The sensitivity of the CM-based approach made it possible to detect hundreds of tRNA pseudogenes. Our data imply that tandem duplications of stretches of mitogenomic DNA are a frequent phenomenon. Consequently, TDRLs are common mechanism leading to major reorganizations of mitochondrial gene orders. In addition to conventional TDRLs, we also found evidence for inverse tandem duplications with subsequent random loss of duplicate gene copies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1, Supplementary Dataset 1.
FUNDING
The Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg, Association Française contre les Myopathies (MNM1 2009), ANR MITOMOT (ANR-09-BLAN-0091-01); Deutsche Forschungsgemeinschaft [SPP-1174 (‘Deep Metazoan Phylogeny’) project STA 850/3-2 and STA 850/2]; French-German PROCOPE program (DAAD D/0628236, EGIDE PHC 14770PJ); French-German University (DFH-UFA, Cotutelle de thèse CT-08-10); doctoral fellowship of the German Academic Exchange Service (DAAD D/10/43622); bridge scholarship of the Collège Doctoral Européen (CDE), Université de Strasbourg. Funding for open access charge: Centre National de la Recherche Scientifique (CNRS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Stimulating discussions with Richard Giegé and Hagen Schwenzer are gratefully acknowledged. We furthermore thank the HPC at the ZIH of the TU Dresden for providing computational facilities.
REFERENCES
- 1.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavrov DV. Key transitions in animal evolution: a mitochondrial DNA perspective. Integr. Comp. Biol. 2007;47:734–743. doi: 10.1093/icb/icm045. [DOI] [PubMed] [Google Scholar]
- 3.Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol. Biol. Evol. 2011;28:1645–1659. doi: 10.1093/molbev/msq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beagley CT, Macfarlane JL, Pont-Kingdon GA, Okimoto R, Okada N, Wolstenholme DR. Mitochondrial genomes of Anthozoa (Cnidaria) Prog. Cell Res. 1995;5:149–153. [Google Scholar]
- 5.Kayal E, Lavrov DV. The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene. 2008;410:177–186. doi: 10.1016/j.gene.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Helfenbein KG, Fourcade HM, Vanjani RG, Boore JL. The mitochondrial genome of Paraspadella gotoi is highly reduced and reveals that chaetognaths are a sister group to protostomes. Proc. Natl Acad. Sci. USA. 2004;101:10639–10643. doi: 10.1073/pnas.0400941101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfonzo JD, Söll D. Mitochondrial tRNA import—the challenge to understand has just begun. Biol. Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr. Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Arcari P, Brownlee GG. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980;8:5207–5212. doi: 10.1093/nar/8.22.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruijn MH, Schreier PH, Eperon IC, Barrell BG, Chen EY, Armstrong PW, Wong JF, Roe BA. A mammalian mitochondrial serine transfer RNA lacking the ‘dihydrouridine’ loop and stem. Nucleic Acids Res. 1980;8:5213–5222. doi: 10.1093/nar/8.22.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolstenholme DR, Okimoto R, Macfarlane JL. Nucleotide correlations that suggest tertiary interactions in the TV-replacement loop-containing mitochondrial tRNAs of the nematodes Caenorhabditis elegans and Ascaris suum. Nucleic Acids Res. 1994;22:4300–4306. doi: 10.1093/nar/22.20.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masta SE. Mitochondrial sequence evolution in spiders: intraspecific variation in tRNAs lacking the TΨC arm. Mol. Biol. Evol. 2000;17:1091–1100. doi: 10.1093/oxfordjournals.molbev.a026390. [DOI] [PubMed] [Google Scholar]
- 14.Masta SE, Boore JL. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol. Biol. Evol. 2004;21:893–902. doi: 10.1093/molbev/msh096. [DOI] [PubMed] [Google Scholar]
- 15.Qiu Y, Song D, Zhou K, Sun H. The mitochondrial sequences of Heptathela hangzhouensis and Ornithoctonus huwena reveal unique gene arrangements and atypical tRNAs. J. Mol. Evol. 2005;60:57–71. doi: 10.1007/s00239-004-0010-2. [DOI] [PubMed] [Google Scholar]
- 16.Klimov PB, OConnor BM. Improved tRNA prediction in the American house dust mite reveals widespread occurrence of extremely short minimal tRNAs in acariform mites. BMC Genomics. 2009;10:598. doi: 10.1186/1471-2164-10-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watterson GA, Ewens WJ, Hall TE, Morgan A. The chromosome inversion problem. J. Theor. Biol. 1982;99:1–7. [Google Scholar]
- 18.Sankoff D, Leduc G, Antoine N, Paquin B, Lang BF, Cedergren R. Gene order comparisons for phylogenetic inference: evolution of the mitochondrial genome. Proc. Natl Acad. Sci. USA. 1992;89:6575–6579. doi: 10.1073/pnas.89.14.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- 20.Shao R, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- 21.Miller AD, Nguyen TTT, Burridge CP, Austin CM. Complete mitochondrial DNA sequence of the Australian freshwater crayfish Cherax destructor (Crustacea: Decapoda: Parastacidae): a novel gene order revealed. Gene. 2004;331:65–72. doi: 10.1016/j.gene.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Moritz C, Brown WM. Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc. Natl Acad. Sci. USA. 1987;84:7183–7187. doi: 10.1073/pnas.84.20.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernt M, Middendorf M. A method for computing an inventory of metazoan mitochondrial gene order rearrangements. BMC Bioinformatics. 2011;12(Suppl. 9):S6. doi: 10.1186/1471-2105-12-S9-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giegé R. Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- 25.Eigen M, Lindemann BF, Tietze M, Winkler-Oswatitsch R, Dress A, von Haeseler A. How old is the genetic code? Statistical geometry of tRNA provides an answer. Science. 1989;244:673–679. doi: 10.1126/science.2497522. [DOI] [PubMed] [Google Scholar]
- 26.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helm M, Brulé H, Friede D, Giegé R, Pütz J, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K. Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:11–39. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyman SK, Boore JL. Technical Report. LBNL-53615: Lawrence Berkeley National Laboratory; 2003. Annotating animal mitochondrial tRNAs: A new scoring scheme and an empirical evaluation of four methods. [Google Scholar]
- 30.Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 31.Sheffield NC, Hiatt KD, Valentine MC, Song H, Whiting MF. Mitochondrial genomics in Orthoptera using MOSAS. Mitochondrial DNA. 2010;21:87–104. doi: 10.3109/19401736.2010.500812. [DOI] [PubMed] [Google Scholar]
- 32.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA Alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgs PG, Jameson D, Jow H, Rattray M. The evolution of tRNA-Leu genes in animal mitochondrial genomes. J. Mol. Evol. 2003;57:435–445. doi: 10.1007/s00239-003-2494-6. [DOI] [PubMed] [Google Scholar]
- 35.Rawlings TA, Collins TM, Bieler R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl Acad. Sci. USA. 2003;100:15700–15705. doi: 10.1073/pnas.2535036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Flook PK, Rowell CH, Gellissen G. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J. Mol. Evol. 1995;41:928–941. doi: 10.1007/BF00173173. [DOI] [PubMed] [Google Scholar]
- 38.Boore JL, Brown WM. Complete DNA sequence of the mitochondrial genome of the black chiton Katharina tunicata. Genetics. 1994;138:423–443. doi: 10.1093/genetics/138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pütz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA. 2007;13:1184–1190. doi: 10.1261/rna.588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S. RALEE–RNA ALignment editor in Emacs. Bioinformatics. 2005;21:257–259. doi: 10.1093/bioinformatics/bth489. [DOI] [PubMed] [Google Scholar]
- 42.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer SL, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 43.Perseke M, Fritzsch G, Ramsch K, Bernt M, Merkle D, Middendorf M, Bernhard D, Stadler PF, Schlegel M. Evolution of mitochondrial gene orders in echinoderms. Mol. Phylog. Evol. 2008;47:855–864. doi: 10.1016/j.ympev.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 44.Börner GV, Mörl M, Janke A, Pääbo S. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 1996;15:5949–5957. [PMC free article] [PubMed] [Google Scholar]
- 45.Beckenbach AT, Joy JB. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): rearrangement and severe truncation of tRNA genes. Genome Biol. Evol. 2009;1:278–287. doi: 10.1093/gbe/evp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haen KM, Pett W, Lavrov DV. Parallel loss of nuclear-encoded mitochondrial aminoacyl-tRNA synthetases and mtDNA-encoded tRNAs in Cnidaria. Mol. Biol. Evol. 2010;27:2216–2219. doi: 10.1093/molbev/msq112. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Lavrov DV. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae. PLoS ONE. 2008;3:e2723. doi: 10.1371/journal.pone.0002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dörner M, Altmann M, Pääbo S, Mörl M. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell. 2001;12:2688–2698. doi: 10.1091/mbc.12.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braband A, Cameron SL, Podsiadlowski L, Daniels SR, Mayer G. The mitochondrial genome of the onychophoran Opisthopatus cinctipes (Peripatopsidae) reflects the ancestral mitochondrial gene arrangement of Panarthropoda and Ecdysozoa. Mol. Phylogenet. Evol. 2010;57:285–292. doi: 10.1016/j.ympev.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 50.Segovia R, Pett W, Trewick S, Lavrov DV. Extensive and evolutionarily persistent mitochondrial tRNA editing in velvet worms (phylum Onychophora) Mol. Biol. Evol. 2011;28:2873–2881. doi: 10.1093/molbev/msr113. [DOI] [PubMed] [Google Scholar]
- 51.Miyamoto H, Machida RJ, Nishida S. Complete mitochondrial genome sequences of the three pelagic chaetognaths Sagitta nagae, Sagitta decipiens and Sagitta enflata. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2010;5:65–72. doi: 10.1016/j.cbd.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Suga K, Welch DBM, Tanaka Y, Sakakura Y, Hagiwara A. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol. Biol. Evol. 2008;25:1129–1137. doi: 10.1093/molbev/msn058. [DOI] [PubMed] [Google Scholar]
- 53.Satta Y, Ishiwa H, Chigusa SI. Analysis of nucleotide substitutions of mitochondrial DNAs in Drosophila melanogaster and its sibling species. Mol. Biol. Evol. 1987;4:638–650. doi: 10.1093/oxfordjournals.molbev.a040464. [DOI] [PubMed] [Google Scholar]
- 54.Masta SE, Boore JL. Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol. Biol. Evol. 2008;25:949–959. doi: 10.1093/molbev/msn051. [DOI] [PubMed] [Google Scholar]
- 55.Fahrein K, Masta SE, Podsiadlowski L. The first complete mitochondrial genome sequences of Amblypygi (Chelicerata: Arachnida) reveal conservation of the ancestral arthropod gene order. Genome. 2009;52:456–466. doi: 10.1139/g09-023. [DOI] [PubMed] [Google Scholar]
- 56.Widmann J, Harris JK, Lozupone C, Wolfson A, Knight R. Stable tRNA-based phylogenies using only 76 nucleotides. RNA. 2010;16:1469–1477. doi: 10.1261/rna.726010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jow H, Hudelot C, Rattray M, Higgs PG. Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Mol. Biol. Evol. 2002;19:1591–1601. doi: 10.1093/oxfordjournals.molbev.a004221. [DOI] [PubMed] [Google Scholar]
- 58.Seligmann H. Undetected antisense tRNAs in mitochondrial genomes? Biol. Direct. 2010;5:39. doi: 10.1186/1745-6150-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seutin G, Lang BF, Mindell DP, Morais R. Evolution of the WANCY region in amniote mitochondrial DNA. Mol. Biol. Evol. 1994;11:329–340. doi: 10.1093/oxfordjournals.molbev.a040116. [DOI] [PubMed] [Google Scholar]
- 60.Macey JR, Larson A, Ananjeva NB, Papenfuss TJ. Replication slippage may cause parallel evolution in the secondary structures of mitochondrial transfer RNAs. Mol. Biol. Evol. 1997;14:30–39. doi: 10.1093/oxfordjournals.molbev.a025699. [DOI] [PubMed] [Google Scholar]
- 61.Arnason U, Gullberg A, Janke A. Phylogenetic analyses of mitochondrial DNA suggest a sister group relationship between Xenarthra (Edentata) and Ferungulates. Mol. Biol. Evol. 1997;14:762–768. doi: 10.1093/oxfordjournals.molbev.a025816. [DOI] [PubMed] [Google Scholar]
- 62.Podsiadlowski L, Carapelli A, Nardi F, Dallai R, Koch M, Boore JL, Frati F. The mitochondrial genomes of Campodea fragilis and Campodea lubbocki (Hexapoda: Diplura): High genetic divergence in a morphologically uniform taxon. Gene. 2006;381:49–61. doi: 10.1016/j.gene.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Machida RJ, Miya MU, Nishida M, Nishida S. Complete mitochondrial DNA sequence of Tigriopus japonicus (Crustacea: Copepoda) Mar. Biotechnol. 2002;4:406–417. doi: 10.1007/s10126-002-0033-x. [DOI] [PubMed] [Google Scholar]
- 64.Lavrov DV, Brown WM, Boore JL. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl Acad. Sci. USA. 2000;97:13738–13742. doi: 10.1073/pnas.250402997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolstenholme DR, Macfarlane JL, Okimoto R, Clary DO, Wahleithner JA. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl Acad. Sci. USA. 1987;84:1324–1328. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boore JL. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In: Sankoff D, Nadeau JH, editors. Dordrecht, NL: Computational Biology Series vol 1, Kluwer Academic Publishers; 2000. pp. 133–147. [Google Scholar]
- 67.Fujita MK, Boore JL, Moritz C. Multiple origins and rapid evolution of duplicated mitochondrial genes in parthenogenetic geckos (Heteronotia binoei; Squamata, Gekkonidae) Mol. Biol. Evol. 2007;24:2775–2786. doi: 10.1093/molbev/msm212. [DOI] [PubMed] [Google Scholar]
- 68.Inoue JG, Miya M, Miller MJ, Sado T, Hanel R, Hatooka K, Aoyama J, Minegishi Y, Nishida M, Tsukamoto K. Deep-ocean origin of the freshwater eels. Biol. Lett. 2010;6:363–366. doi: 10.1098/rsbl.2009.0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mueller RL, Boore JL. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol. Biol. Evol. 2005;22:2104–2112. doi: 10.1093/molbev/msi204. [DOI] [PubMed] [Google Scholar]
- 70.Lavoué S, Miya M, Poulsen JY, Møller PR, Nishida M. Monophyly, phylogenetic position and inter-familial relationships of the Alepocephaliformes (Teleostei) based on whole mitogenome sequences. Mol. Phylogenet. Evol. 2008;47:1111–1121. doi: 10.1016/j.ympev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2003;26:121–138. doi: 10.1016/s1055-7903(02)00332-9. [DOI] [PubMed] [Google Scholar]
- 72.San Mauro D, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- 73.Mueller RL, Macey JR, Jaekel M, Wake DB, Boore JL. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc. Natl Acad. Sci. USA. 2004;101:13820–13825. doi: 10.1073/pnas.0405785101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 75.Olson SA. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief. Bioinformatics. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 76.Chaudhuri K, Chen K, Mihaescu R, Rao S. In: Proceedings of the Seventeenth Annual ACM-SIAM Symposium on Discrete Algorithms, SODA 2006. ACM New York, USA: 2006. On the tandem duplication-random loss model of genome rearrangement; pp. 564–570. [Google Scholar]
- 77.Bernt M, Chen K-Y, Chen M-C, Chu A-C, Merkle D, Wang H-L, Chao K-M, Middendorf M. Finding all sorting tandem duplication random loss operations. J. Discr. Alg. 2011;9:32–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.