Abstract
Chloroplast RNA metabolism is controlled and excecuted by hundreds of nuclear-encoded, chloroplast-localized RNA binding proteins. Contrary to the nucleo-cytosolic compartment or bacteria, there is little evidence for non-coding RNAs that play a role as riboregulators of chloroplasts. We mined deep-sequencing datasets to identify short (16–28 nt) RNAs in the chloroplast genome and found 50 abundant small RNAs (sRNAs) represented by multiple, in some cases, thousands of sequencing reads, whereas reads are in general absent from the surrounding sequence space. Other than sRNAs representing the most highly abundant mRNAs, tRNAs and rRNAs, most sRNAs are located in non-coding regions and many are found a short distance upstream of start codons. By transcript end mapping we show that the 5′ and 3′ termini of chloroplast RNAs coincide with the ends of sRNAs. Sequences of sRNAs identified in Arabidopsis are conserved between different angiosperm species and in several cases, we identified putative orthologs in rice deep sequencing datasets. Recently, it was suggested that small chloroplast RNA fragments could result from the protective action of pentatricopeptide repeat (PPR) proteins against exonucleases, i.e. footprints of RNA binding proteins. Our data support this scenario on a transcriptome-wide level and suggest that a large number of sRNAs are in fact remnants of PPR protein targets.
INTRODUCTION
In the last decade, small non-coding RNAs (ncRNA) have been shown to act as important regulators of gene expression in a broad variety of organisms. Few studies have addressed the presence of ncRNAs specifically in chloroplasts. A first ncRNA candidate called sprA was discovered in tobacco (1). In the same species, a bulk cloning approach of short RNAs led to the identification of 18 chloroplast ncRNAs (2). Recently, deep sequencing of the Chinese cabbage (Brassica rapa ssp. Chinensis) identified hundreds of chloroplast small RNAs (sRNAs) that mostly corresponded to 5′-ends of chloroplast tRNAs and 3′-ends of chloroplast ribosomal RNAs (3). It is at present unclear, whether any of these chloroplast ncRNAs serve a specific function. Recently, a mode for the biogenesis of chloroplast sRNAs was suggested that implies protection from RNA degradation as a major driving force behind the accumulation of specific RNA fragments (4). This proposal was based on current knowledge of a specific class of RNA binding proteins, called pentatricopeptide repeat (PPR) proteins. Hundreds of genes for PPR proteins are found in the nuclear genome of higher plants (5). The proteins are almost exclusively targeted to either mitochondria or chloroplasts (5). The family has no known counterpart in bacteria, and various members have been shown to be essential for specific RNA processing steps, e.g. RNA editing or RNA splicing (6). Where investigated, PPR proteins display exquisitely specific binding to one or few organellar RNAs (6). For example, PPR10 was shown to contact ∼20 nt long RNA elements in two independent inter-cistronic spacers (psaJ–rpl33; atpI–atpH) and acts as a road block against exonucleolytic decay both in 5′-to-3′, as well as in 3′-to-5′ direction (4). The exonucleolytic decay is initiated distal to the PPR10 binding site, probably by an endonucleolytic event, e.g. by chloroplast RNaseE. Eventually, RNA degradation leads to complete elimination of the psaJ–rpl33 and atpI–atpH mRNAs with the exception of the small ∼25 nt stretch protected by PPR10. Such potential footprints have been found in sRNA databases in cereals, among them those attributed to the PPR proteins CRR2 and PPR10 (4,7). Moreover, sRNAs cloned in tobacco (2) and in rice (7) correspond to a site in the psbH–petB intergenic spacers suspected to bind the PPR protein HCF152 (4). We set out to comprehensively identify sRNAs in the chloroplast transcriptome of Arabidopsis and rice and evaluate, whether they could indeed be footprints of RNA binding proteins.
MATERIALS AND METHODS
Alignment of sRNAs to the chloroplast genome
Arabidopsis thaliana sRNA data from Rajagopalan et al. (8), (available as Platform GPL3968 at http://www.ncbi.nlm.nih.gov/geo) including reads from four different tissues/developmental stages (seedlings, rosette leaves, flower buds and siliques) were assembled to the chloroplast genome (NC_000932) using the Geneious Pro Software (Version 5.3.6) allowing only perfect matches. The Inverted Repeat B was excluded from the reference sequence. Small chloroplast accumulating RNAs (sRNAs) were extracted by hand. The sRNAs had to be covered by at least three independent sequences, either from different libraries or with slightly different sequences with a minimum of ten reads in total.
The core sRNA was defined as the sequence element present in at least 50% of the sequences.
Images were extracted from the alignment files as eps files and modified using Adobe Illustrator.
Rice sRNA Data were obtained from the Cereal Small RNA Database (CSRDB), (9). Data included in this analysis are run1 and run2, as well as the sRNA derived from three runs for leaf and inflorescence tissues. The sRNA reads were assembled using the rice chloroplast genome (NC_001320) as reference sequence, the same way as for the Arabidopsis reads.
5′-rapid amplification of cDNA ends
Total RNA was isolated using TRIzol Reagent (Invitrogen) from 3-week-old Arabidopsis plants (Col-0 ecotype). A quantity of 2 µg RNA was ligated to 4 pmol RNA Oligo 5′-GUGAUCCAACCGACGCGACAAGCUAAUGCAAGANNN-3′ using T4 RNA Ligase I (NEB) at 37°C for 1 h. Reaction was Heat inactivated at 65°C for 15 min and RNA purified by standard phenol/chloroform extraction. The RNA was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) using random Primers (Hexa/Nona-Mix). PCR analysis was performed using Primers Rumsh1 5′-TGATCCAACCGACGCGAC-3′ and gene-specific primers. PCR Products were gel-eluted if necessary, using the JETSORB Gel Extraction Kit (Genomed) and cloned in the pDrive Vector using the PCR Cloning Kit (Qiagen). Clones were sequenced using Sanger Sequencing (SMB Berlin).
Primers used:
| rps15 5′ | CCAAATGTGAAGTAAGTCTTCG |
| ndhB 5′ | TATCCAGATAATAGGTAGGAGC |
| psbC.T7 | GTAATCGACTCACTATAGGGCCCCCAAAGGGA GATTTTAG |
| rps12 5′ | TTTCGTGACGTTTCGGATTGG |
| petA 5′ | ATCAGGAAGTACCGTTGTGG |
3′-rapid amplification of cDNA ends
Total RNA was isolated using TRIzol Reagent (Invitrogen) from 3-week-old Col-0 plants. A quantity of 2 µg RNA was ligated to 0.6 µl SRA 3′-Adapter (Illumina) using T4 RNA Ligase I (NEB) at 37°C for 1 h. The reaction was heat inactivated at 65°C for 15 min and RNA purified by standard phenol/chloroform extraction. The RNA was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) using the Primer Adapter RT Primer 5′-CAAGCAGAAGACGGCATA-3′. PCR Analysis was performed using Adapter PCR Primer 5′-CAAGCAGAAGACGGCATACG-3′ and gene-specific primers. PCR Products were gel-eluted if necessary, using the JETSORB Gel Extraction Kit (Genomed) and cloned in the pDrive Vector using the PCR Cloning Kit (Qiagen). Clones were sequenced using Sanger Sequencing (SMB Berlin).
| ycf1 3′ | AGCTTGTATGAATCGCTATTGG |
| rps7 3′ | CGATGCCATACGCAAAAAGG |
| psaI 3′ | TTTTTTAGATCGGCTGAGACC |
| clpP 3′ | TGTACAAAGAACGGGCAAACC |
| cemA 3′ | TTAAATCGTGTATCTCCGTCAC |
| rps18 3′ | TTGAAAGAAGTGAGTCGACTCC |
| atpH 3′ | CTTAGTTTGGCTTTTATGGAAGC. |
RNase protection
Seeds of mutant lines hcf7-2 and hcf152-1 were grown on MS-medium supplemented with 3% sucrose for 2 weeks and screened for high chlorophyll fluorescence. RNA was extracted from mutant plants with TRIzol (Invitrogen).
As a template for radioactive in vitro transcription, two DNA Oligos (hcf152 footprint: TCCTTTTTTTCTGCACCTGTCTC and T7 overlap: TAATACGACTCACTATAGGGAGACAGG) were annealed and double stranded template was created by a ‘fill-in’ reaction using Klenow Exo- (Fermentas). Antisense RNA was synthesized using T7 Polymerase (Fermentas) with radioactive α-32P-UTP (Hartmann Analytic). The reaction was digested with DNase and gel-purified.
The RNase Protection Assay was performed according to the instructions of the mirVana™ miRNA Detection Kit (Ambion) using 5 -µg RNA.
Phylogeny of sRNAs
Intergenic spacers for which sRNAs in Arabidopsis were found were aligned using ClustalW2 algorithm (10) using default settings and visualized using Jalview Software (11). Organellar genome sequences were obtained from NCBI under following accession numbers A. thaliana (NC_000932), Adiantum capillus-veneris (NC_004766), Oryza sativa (NC_001320), Hordeum vulgare (NC_008590), Physcomitrella patens (NC_005087), Nicotiana tabacum (NC_001879), Pinus thunbergii (NC_001631) and Zea mays (NC_001666).
For comparison of sRNAs between Arabidopsis and rice or of all Arabidopsis sRNAs sequences were aligned using ClustalW2 algorithm and a neighbor joining tree by percentage identity was created using the Jalview Software Package (11). Pairwise alignments of two sequences were perfomed with the same software. Image files were exported as eps files and modified using Adobe Illustrator.
RESULTS
Identification of sRNAs within the chloroplast transcriptome by mining Arabidopsis deep-sequencing datasets
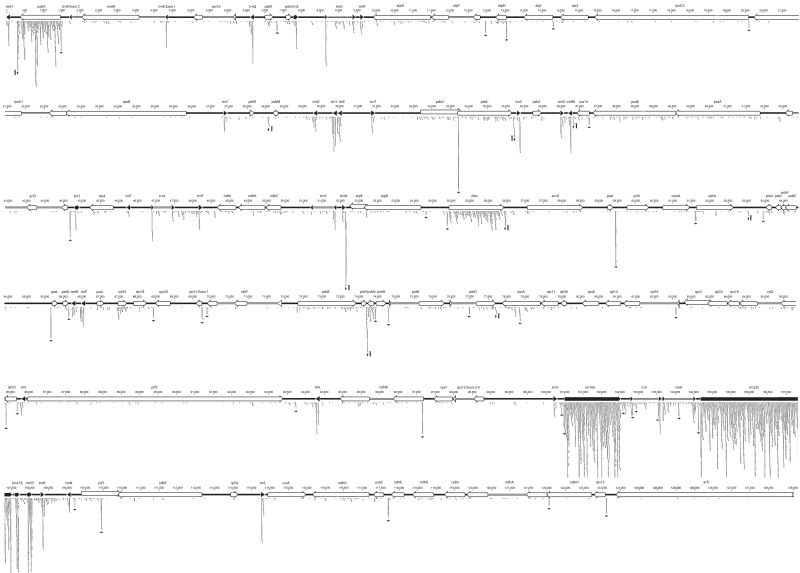
Small regulatory RNAs like miRNAs can be detected by RNA-Seq analysis of gel-fractionated total RNA. Such analyses have also been carried out in Arabidopsis in order to identify nucleo-cytosolic miRNAs. Little attention has so far been paid to sRNAs from mitochondria or chloroplasts that are part of these datasets. We have screened a miRNA deep-sequencing dataset from Arabidopsis generated by the Bartel group (8) for small chloroplast RNAs: The four RNA seq libraries were assembled from RNA samples taken from whole seedlings, rosette leaves, flowers and siliques, harvested at 6 days, 4 weeks, 6 weeks and 2 months after planting, respectively (8). The RNAs were eluted from PAGE gel fragments that represent pools of RNA fragments between 16 and 28 nt in length (8) (Supplementary Figure S1). We identified sequences that match the chloroplast genome with 100% identity using the Geneious software package (http://www.geneious.com). A sequence is defined as a unique set of identical reads from only a single library. In extreme cases, a sequence can represent hundreds of individual reads, i.e. hundreds of individual sequencing reactions. There are a total of 9750 sequences, which are highly unevenly distributed in the chloroplast genome (Figure 1). Only four protein coding genes exhibited high sequence densities across the entire RNA, two of them known to be highly expressed, namely psbA and rbcL (Figure 1). In addition to mRNAs, the abundant chloroplast rRNAs and tRNAs are covered well by sequences (Figure 1). For the present analysis, we excluded areas of continuous high sequence density. Instead, we focused on isolated peaks of multiple sequences that stood out over the otherwise low background of sequences in coding regions and intergenic spacers (Figure 1). We collected only clusters of minimally three sequences, which represent a combined number of at least 10 reads. We defined the core of each sequence cluster as the nucleotides that are found in 50% of all overlapping sequences. By these benchmarks, we found a total of 50 sRNAs with a median representation of 30 reads per sequence peak (Supplementary Table S1). In the following, we will refer to the core of a sequence cluster as chloroplast sRNA. A special class of sRNAs was identified in the 3′-regions of several chloroplast mRNAs. These RNAs are predicted to fold into a single stem-loop structure with a predicted free energy lower than −20 kcal/mol (Supplementary Table S1). Their connection with secondary structure elements distinguishes them from sRNAs and suggests a different biogenesis.
Figure 1.
Map of RNA sequencing data onto the chloroplast genome of A. thaliana. Individual RNA sequencing reads were extracted from the sRNA data set generated by Rajagopalan et al. (8) and aligned with the chloroplast chromosome of Arabidopsis (accession no. NC_000932) using the Geneious software package. Only sequences matching 100% with the genome were considered. The bars below the genome map indicate groups of identical reads from a single library. Thus, several bars can represent identical sequence stretches but are from different RNA seq libraries—e.g. two identical bars could indicate a cluster of identical reads from the leaf library and the silique library. Note that abundant RNA species are covered across their entire length by sequencing reads like the rRNAs, whereas reads are sparse for most protein coding genes, e.g. for the psbE operon. In several cases, e.g. for the rRNA operon, the number of reads piled up at certain genomic positions exceeds the space between two rows of the genome map and are thus cut off. Genes are shown as arrows, with arrow points indicating the 3′-end of the open reading frame/mature tRNA or rRNA. Filled arrows = genes for rRNAs or tRNAs; open arrows = protein-coding genes. Small horizontal arrows denote sRNAs and indicate their 5′-to-3′ direction. Small vertical arrows next to schematic stem-loops indicate sequencing reads that are likely accumulating due to the stability of secondary structure elements in the RNA. Gene names are given above the genome map. Numbers indicate the genomic position as given in GenBank entry NC_000932. The second version of the chloroplast inverted repeat (IRA) is not shown here.
sRNAs in intergenic regions are often found proximal to start codons
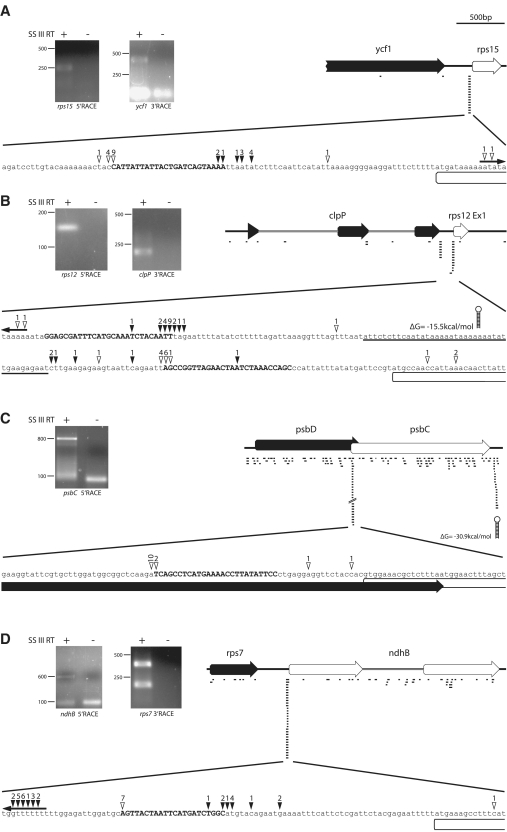
sRNAs are not randomly distributed in the chloroplast transcriptome: sRNAs are concentrated in non-coding regions (46 of 50, Supplementary Table S1). Few sRNAs were found in intronic sequences (6 of 50, Supplementary Table S1) and even less antisense to known chloroplast RNA species (3 of 50, Supplementary Table S1). Of the sRNAs in non-coding regions, 17 are found closer than 50 nt to the next downstream start codon (Supplementary Table S1). Their uneven distribution supports a functional significance for sRNAs and argues against them being the result of randomly stable RNA degradation products or spurious mini-transcription units. Their proximity to start codons might indicate a function related to translation. Four of the 17 sRNAs found in proximity of start codons are special in that they are located inside an upstream ORF: a sRNA 43 nt upstream of the ndhA start codon lies within ndhH, another is 19 nt away from the psbC start codon within psbD and a third 35 nt upstream from rpl2 within rpl23. The corresponding reading frames of psbC/psbD are overlapping by 17 nt, while ndhA and ndhH are spaced by 1 bp and rpl23/rpl2 by 18 bp. Thus, the peculiar location of these four sRNAs might indicate their role for the downstream ORF of these closely-knit di-cistronic transcripts. Noteworthy, psbC possesses an additional promoter serving exclusively its own transcription without an intact upstream psbC reading frame (12). This would locate the corresponding sRNAs in a true UTR region even though it encompasses a partial coding sequence. Whether mono-cistronic forms for the other two ORFs with sRNAs in 5′-adjacent reading frames (rpl2 and ndhA) can also be found is unknown. A fourth sRNA overlaps with the annotated start codon of ndhB—the only such case among all sRNAs identified. However, when comparing alignments of hundreds of ndhB genes using the NCBI protein cluster tool, we found a downstream ATG much more conserved than the one annotated as start codon in Arabidopsis (http://www.ncbi.nlm.nih.gov/sutils/prkview.cgi?result=align&cluster=CHL00049). This suggests that the ndhB 5′ sRNA is in fact at position −45 relative to the true start codon (see also Figure 5D). In sum, sRNAs are highly biased toward non-coding regions and many are found a short distance (∼30–70 nt) upstream of start codons.
Figure 5.
Mapping of transcript ends in the vicinity of selected sRNAs (A = rps15 5′; B = rps12 5′; C = psbD-psbC; D = rps7-ndhB). Total Arabidopsis RNA was ligated with RNA oligos selectively either at the 3′- or at the 5′-end, reverse-transcribed and amplified by PCR with combinations of gene-specific and oligo-specific primers.The amplification products were separated on an agarose gel (left side of each panel). PCR products were gel-purified and cloned. Clones were selected and sequenced. The last base before the sequence of the RNA oligo corresponds to the end of the original chloroplast RNA ligated. These ends are indicated by open arrowheads (for 5′-RACE experiments) or by filled arrowheads (for 3′-RACE experiments) above a blowup of a sequence stretch covering parts of the intergenic region containing the sRNAs (upper case) at the center. The numbers above the arrowheads point out numbers of clones that correspond to a particular transcript end. In case of 5′-RACE, the numbers refer to independent clones as evidenced by different bar-codes introduced via randomized nucleotides in the ligated 5′-RNA oligo. Number of clones indicating independent ends outside of the blowup region are indicated above outward-facing arrows at the ends of the sequence shown here. All further symbols and numbers are explained in Figure 1.
A subset of sRNAs is conserved across angiosperms
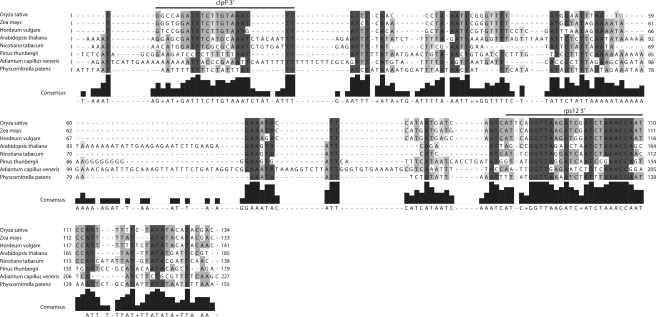
If sRNAs are functional and under selection, they should be evolutionarily conserved. We tested this for a number of sRNAs by comparing homologous intergenic regions containing sRNAs in Arabidopsis. An example is shown Figure 2 with an alignment of the clpP–rps12 intergenic spacer. Here, a sRNA 3′ of clpP and a sRNA 5′ of rps12 are found. Both regions exhibit conservation exceeding adjacent non-coding regions (Figure 2). However, the degree of conservation differs between the two sRNAs. The clpP 3′ sRNA is conserved only in angiosperms, including the GATTTC hexamer and a following triple-A stretch. Additional bases are found in most, but not all angiosperms. The sRNA 5′ to the rps12 reading frame is conserved even in the moss P. patens and in the fern A. capillus-veneris with 10 nt being identical between all species in the alignment. Such conservation down to Bryophytes was, however found only one more time among the eight sRNAs we prepared alignments for: the sequence of the psbH–petB intergenic spacers shows conservation of a sRNA in all species analyzed with the exception of A. capillus-veneris (Supplementary Figure S2). In contrast, conservation of sRNAs within angiosperms is more common. Like for the clpP 3′ sRNA, sRNAs in the 5′-region of atpH and petL also show higher identity values than adjacent sequences (Supplementary Figure S2). This suggests that at least some sRNAs are under selective pressure, i.e. are functional. A second group of sRNAs does not exhibit conservation in the set of species analyzed here. For example, sRNAs in the atpH–atpF intergenic spacer, and sRNAs upstream of rps15, ycf3 and rps14 do not stand out in conservation relative to the surrounding sequence (Supplementary Figure S2). Thus, there are highly conserved sRNAs, as well as lineage-specific sRNAs.
Figure 2.
Alignment of the clpP–rps12 intergenic regions in various embryophtes. The alignment was prepared using ClustalW2 (10) and the Jalview software package for graphical output (11). The two sRNAs in this intergenic spacer are indicated by solid bars above the alignment. A graphical and a sequence consensus are given at the bottom of the alignment. Shading refers to different levels of sequence conservation: dark gray = 7 or 8 of 8 sequences share the same base; intermediate gray = 5 or 6 of 8 share the same base; light gray = 4 of 8 share the same base.
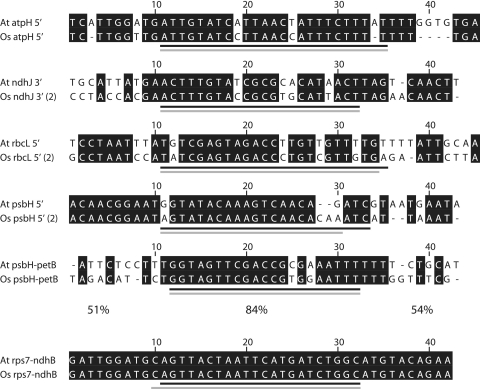
In order to gain further insights into the evolution of sRNAs, we turned to deep-sequencing datasets available online for rice (9) and compared them with our findings for Arabidopsis. Sequences pooled from different experiments were aligned with the chloroplast genome and evaluated in the same manner as outlined above for Arabidopsis (Supplementary Figure S3). Like for Arabidopsis, we found a bias of sRNAs towards intergenic regions (supplementary Table S2). In total, we found 18 rice sRNAs in regions for which we also identified sRNAs in Arabidopsis (supplementary Table S1). As an initial test for orthology, we aligned all rice and Arabidopsis sRNAs (with the exception of stem-loop sRNAs) and calculated a neighbor-joining tree based on percent identity between sRNAs. In this tree, we found 10 Arabidopsis–rice pairs of sRNAs that are situated in the same intergenic region (Supplementary Figure S4). The similarity between sRNAs with longer identical sequence stretches are favored by this method, whereas short or interrupted homologies are not found as sequence pairs. We therefore expect that the number of orthologous sRNAs between Arabidopsis and rice is higher. Among the pairs identified here, four are in highly conserved regions, namely in the chloroplast inverted repeat (rrn16, rrn23, rps7–ndhB) or within the psbC reading frame. These four pairs each show either perfect or >95% identity, which is however not different from adjacent sequences and is expected for the inverted repeat region. The remaining six sRNAs are found upstream or downstream (ndhJ 3′) of reading frames and show strong sequence conservation with an average of 84% identical bases (Figure 3). The 10 adjacent bases upstream and downstream of these sRNAs were on average only 51 and 54% identical, respectively. Evidently, these sRNAs are under selection and can be considered bona fide orthologs.
Figure 3.
Alignment of sRNAs conserved between rice and Arabidopsis. Pairs of sRNAs from Arabidopsis and rice with high sequence similarities were identified by phylogenetic analysis (Supplementary Figure S4). Here, only the six pairs situated immediately upstream or downstream of a reading frame are shown. The percentages refer to sequence identities found in sections of the alignments shown: 51 and 54% of all residues shown upstream or downstream of the sRNAs, respectively are identical, whereas 84% of the residues within the region spanned by the sRNAs are identical. Identical bases are shaded in black; At = A. thaliana; Os = O. sativa; black line = sRNA in Arabidopsis; gray line = sRNA in rice.
sRNAs from chloroplasts have been identified previously from tobacco (2). There are several overlaps with the dataset presented here (Supplementary Table S1). Tobacco Ntc-1, Ntc-2 and Ntc-8 correspond to the Arabidopsis sRNAs psbH–petB, rps7–ndhB and ndhE–psaC. Identity values are in all cases striking: Ntc-1/psbH–petB share 19 of 22 bases; Ntc-2/rps7–ndhB are 100% identical; Ntc-8/ndhE–psaC share a stretch starting from their 5′-end of 17 identical bases with only five mismatches in 28 bases total. Finally, we screened for sRNAs in a taxon only distantly related to the land plants in focus here, the green algae Chlamydomonas reinhardtii. Again, we identified a number of sRNAs in deep sequencing datasets that will be presented in detail elsewhere. Surprisingly, for one sRNA, we could find sequence conservation with angiosperms: the sRNA upstream of psbH is 65 % identical with the corresponding sRNA in Arabidopsis (Supplementary Figure S5). In conclusion, selected sRNAs accumulate not only in Arabidopsis, but also in tobacco and in rice, i.e. are conserved in dicot and monocot angiosperms and are bona fide orthologs. In rare cases, conservation extends even to green algae. This strongly speaks for a selective pressure behind the accumulation of a subset of sRNAs.
A comparison of all Arabidopsis sRNAs identifies sequence conservation for selected pairs of sRNAs
It is unclear how sRNAs are generated but if the underlying machinery has any sequence preferences, we might detect intraspecific similarities between sRNAs. To test such sRNA homologies, we aligned all Arabidopsis sRNAs (with the exception of those corresponding to stem-loops) using the ClustalW2 algorithm allowing for alignment gaps and calculated a neighbor-joining distance tree using percent identity (Supplementary Figure S6). The sequence with the least similarity to all other sRNAs was used as outgroup. We found several pairs of sRNAs with remarkable similarity. The four most similar pairs are shown in Figure 4 and are on average, 75% identical. We did, however not find more widespread similarities between sRNAs or any sort of consensus. This speaks for an individual origin of most sRNAs, whereas in a few cases, pairs of sRNAs could be produced by a common mechanism.
Figure 4.
Top four most similar pairs of Arabidopsis sRNAs. Similarity between sRNAs was identified by cross-comparing all sRNAs identified in Arabidopsis using a distance matrix (Supplementary Figure S6). The top four most similar sequence pairs are displayed here with identical bases shaded in black. The percent identity values for each alignment are given on the right.
sRNAs correlate with known and newly determined transcript termini
According to the footprint model, chloroplast sRNAs should co-localize with mRNA ends (4). Therefore, ends in the vicinity of Arabidopsis sRNAs were mapped by a modified rapid amplification of cDNA ends (RACE) protocol. This included sRNAs in seven randomly chosen intergenic regions, clpP–rps12, atpH–atpF, cemA–petA, rpl18–rps20, rps7–ndhB, rps15 5′ and psaI–ycf4. In addition we analyzed the sRNA in the coding region of psbD. As shown in Figure 5, single PCR-products were obtained for the 5′- and 3′-mapping of the rps15 5′ sRNA. Clones obtained for these PCR products do end close to the 5′- and 3′-end of the sRNA. Similarly, 5′-ends detected in the rps7–ndhB intergenic region are all found in the immediate vicinity of an sRNA's 5′-terminus. For the rps7 3′-RACE, two PCR products were obtained and cloned separately. The rps7/ndhB inter-cistronic cleavage has been mapped to this area previously by RNase protection assays (13). Clones from the longer PCR product correspond to RNA 3′-ends coinciding with the sRNA's 3′-end. The location of this sRNA and its conservation in rice and Arabidopsis makes it likely that it is caused by the action of CRR2 as has been pointed out previously (4). Clones from the shorter PCR product map to upstream sites at positions 100–112 nt relative to the rps7 stop codon. These latter transcript ends have been described previously as well (13), but do not match one of the sRNAs. For the psbD–psbC sRNA, a single 5′-RACE PCR product was obtained and again, this identified a transcript terminus in the vicinity of the sRNA's 5′-end. In contrast, we were unable to obtain a PCR product for the 3′-end of the psbD transcript in this region despite usage of two alternative primers for amplification. The same observation was made for psbD–psbC as described in the accompanying manuscript (14). This is in congruence with RNA accumulation data that did not detect a psbD transcript with a length suggestive of a terminus in the proximity of the psbD–psbC sRNA (15). Possibly, degradation of the di-cistronic psbD/psbC mRNA from the 3′-end could be an extremely rare event.
The congruence of ends of transcripts with ends of sRNAs was also found for sRNAs downstream of cemA, rps18 and psaI (Supplementary Figure S7).
An interesting case is the situation of transcript ends and sRNAs in the clpP–rps12 intergenic spacer. In barley and maize, only a single sRNA was identified that demarks both the 5′-end of the rps12 mRNA, as well as the 3′-end of the clpP mRNA, thus in perfect accordance with most sRNAs delineating overlapping transcript ends. As noted above, this sRNA is highly conserved in land plants including the moss P. patens. In Arabidopsis, two sRNAs are found in the same region, with the one closer to rps12 being orthologous to the conserved barley sRNA. When mapping 5′-and 3′-ends of transcripts in this region, we found a correlation of rps12 5′-ends with the downstream sRNA and a correlation of clpP 3′-ends with the upstream sRNA (Figure 5D). In contrast, no clpP 3′-ends correspond to the second sRNA nor are there 5′-ends of rps12 that would map to the upstream end of the sRNA closer to clpP. Thus, in Arabidopsis, the major ends of the clpP and rps12 transcripts within this intergenic region are generated by independent processes, and likely independent proteins. In any case, clpP–rps12 transcript ends correlate well with ends of sRNAs.
To support these findings statistically, we analyzed the number of mapped mRNA ends that are found in proximity to sRNA ends. For this, we counted how many of the mRNA ends fall into a window of 31 nt centered on the ends of sRNAs relevant in our RACE experiments. We repeated this analysis with a window of only 7 nt. Of the total 185 mRNA ends, 142 map within the 31-nt window and still 107 ends within the 7-nt window. Thus, we find a strong correlation of mRNA ends and ends of sRNAs.
In addition to our own data, there are some interesting overlaps with previously identified transcript ends and the position of sRNAs identified here. Most strikingly, the end of the mature, 5′-processed rbcL mRNA determined previously (16) matches exactly the 5′-end of the sRNA we find in this area. This end depends on the presence of the PPR protein MRL1 that is conserved not only in land plants, but even down to green algae (16). In line with conservation on the protein side, we find a highly similar sRNA in rice (Figure 3), although we could not detect a sRNA in the 5′-region of Chlamydomonas rbcL. Another PPR protein with a function in RNA stabilization is PGR3, which has recently been shown to bind the 5′-region of the petL and ndhA mRNAs (17). The sRNA in the petL 5′-region identified here, starts 1 nt downstream of the transcript's 5′-end at −59 (17) and this region is part of the RNA bound by PGR3 in vitro (17). Similarly, a sRNA found in the 5′-region of ndhA is located 4 nt downstream of the mono-cistronic messages’ 5′-end at −66 (17). Again, this region is included in a longer probe bound by PGR3 in vitro and has slight similarities with the corresponding 5′-end of the petL mRNA. More mapping experiments demonstrating correspondence between sRNA termini and mRNA termini are described in the accompanying manuscript in this same issue (14).
In sum, sRNAs co-localize with transcript ends and potential binding sites of PPR proteins. This is in line with and extends previous data suggesting they are remnants of PPR proteins protecting RNAs against exonucleolytic degradation (4).
hcf152 mutants fail to accumulate an sRNA corresponding to the HCF152 binding site
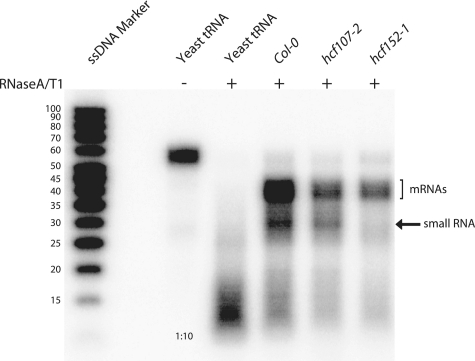
The PPR protein HCF152 binds to a conserved sequence represented by a sRNA in the psbH–petB intergenic region in maize, rice and Arabidopsis (14,18), and is required for the accumulation of processed 5′- and 3′ termini whose ends match those of the sRNA (4,18). To determine whether this sRNA accumulates due to protection by HCF152, we have analyzed the accumulation of this sRNA by RNase protection experiments in a null mutant of HCF152 (18) (Figure 6). As a control, we have also analyzed RNA from mutants of HCF107 (19,20). HCF152 and HCF107 are each required for the accumulation of different RNA segments from the psbB operon and display comparable phenotypic deviations from wild-type, i.e. seedling lethality and high chlorophyll fluorescence (HCF) (18,19,20). The probe used in our RNase Protection assay detects the unprocessed and processed RNAs, as well as the sRNA. The sRNA-precursors are reduced to comparative levels relative to wild-type in both hcf mutants analyzed. In contrast, the sRNA was only detected in hcf107 mutants, but not in hcf152 mutants. This demonstrates that a PPR protein is specifically responsible for the accumulation of the sRNA harboring its binding site.
Figure 6.
RNase protection experiments identify the absence of a sRNA in hcf152 mutants. Total leaf RNA (5 µg each sample) was analyzed from wild-type (Col-0), and from hcf152 and hcf172 mutant lines. The probe was designed to encompass the sRNA and adjacent sequences that are only detected if hybridization to mRNAs takes place. The probe itself is longer than the maximally protected fragment, because it contains a short, non-chloroplast-encoded sequence stretch. The full-length probe is visualized in the lane designated ‘yeast tRNA’ that was not subject to RNase degradation. The amount of probe loaded here is one-tenth of the amount used in protection experiments shown in the other lanes. When yeast tRNA is incubated together with the probe in the presence of RNase A and RNase T1, almost complete degradation of the radio-labeled probe is found. This demonstrates that signals obtained with protection assays using total plant RNA are specific. The lane designated ‘ssDNA marker’ contained radio-labeled single-stranded DNA of the indicated lengths. Note that ssDNA runs faster than ssRNA molecules and thus only gives a relative estimate of the sizes of bands detected in this assay.
DISCUSSION
We here identified 50 non-coding short chloroplast RNAs. These RNAs are distributed non-randomly in the chloroplast transcriptome and are in particular prevalent in intergenic regions.
Our data support the previously-proposed model on protection of short RNA segements by PPR proteins (4,21)
The positional bias of sRNAs towards intergenic regions of the chloroplast transcriptome is consistent with the position of RNA binding proteins (RBPs), including PPR proteins that use these regions as points of entry to stabilize and translate mRNAs (22,23).
The similarities we find for related sRNA pairs (on average 75%) is similar or higher than base identity values found for genetically identified pairs or triplets of binding sites of individual PPR proteins (Table 2 in 24). Also, the two target sites of PPR10 upstream of atpH and rpl33 share 75% of all their residues (4). Thus, similarities uncovered here for pairs of sRNAs are within the range of target sequence divergence tolerated by PPR proteins. As such, we hypothesize that members of such pairs are protected by the same protein.
A number of sRNAs show a striking conservation between Arabidopsis, tobacco, Chinese cabbage and rice. The conservation extends to sRNAs found in maize and barley (14). This is mirrored by the amino acid sequence conservation encountered between PPR proteins of land plants, that is exceptionally high when compared with other large plant protein families (25). This suggests that sRNAs are selectively constrained because they serve as target sequences for conserved RNA binding proteins like PPR proteins. PPR proteins are known to be conserved between angiosperms and some even down to bryophytes (25). Thus, ancient PPR proteins could be behind the conserved sequences identified here. Next to conserved sRNAs, a large number of lineage-specific sRNAs was found as well. A non-find of a sRNA in one species versus another could of course always be explained by experimental differences, e.g. choice of tissue, age of material, etc. Alternatively, they could be the result of lineage-specific PPR proteins. For example, despite the overall impressive conservation of PPR proteins between angiosperms, there is also a fraction of PPR proteins that seem to be specific to Arabidopsis and are not found in the rice genome (25). Finally, it is also possible that particular PPR proteins are capable of accommodating divergent RNA sequences and thus, would allow for a more freely evolving target sequence despite conservation of the PPR protein.
Support for sRNAs as footprints of PPR proteins comes also from the finding that overlapping chloroplast transcript ends co-localize with ends of sRNAs. Most of the mRNA ends we were able to map correspond to ends of sRNAs. Furthermore, ends of sRNAs map to a number of transcript ends reported previously. Importantly, for several of these ends, functionally-linked RNA binding proteins are known, among them the PPR proteins CRR2, PGR3, MRL1 and of course, PPR10 [and HCF152 as confirmed by Zhelyakova et al. (14)]. That this co-localization of PPR binding sites, transcript ends and sRNA ends were a chance event seems unlikely. Particularly striking are cases, where the sRNA's ends co-localize with the overlapping ends of upstream and downstream messages, i.e. in perfect agreement with the model for PPR proteins as bidirectional roadblocks against exonucleases (4,21). Data presented in the accompanying paper (14) on sRNAs in barley chloroplasts and the interaction of the PPR protein HCF152 with its cognate mRNA extend these previous findings and complement data presented here on Arabidopsis and rice.
Finally, we here show that the accumulation of a particular sRNA depends on the presence of its cognate PPR protein, HCF152. A similar link was made for the PPR proteins CRP1 and PPR10 in the accompanying paper (14).Most parsimoniously, loss of these PPR proteins eliminates the direct protection of the sRNAs against exonucleases and thus leads to complete sRNA degradation.
Footprints by RNA binding proteins other than PPR10-like PPR proteins
We expect that the number of links between PPR proteins and sRNAs will rise as more PPR proteins will be analyzed biochemically for their exact target location in the chloroplast transcriptome. Given the large number of PPR proteins in chloroplasts (5), and given the evidence provided here and by Zhelyakova et al. (14), we expect PPR proteins to be involved in the generation of most sRNAs found here. However, it cannot be excluded that in a minority of cases, other RNA binding proteins are generating sRNAs as well.
For instance, the related tetratricopeptide repeat protein HCF107 has been suggested to make contact with the 5′-area of psbH (26), where we found an sRNA conserved between Arabidopsis, rice and Chlamydomonas. The transcript's 5′-end was previously mapped to a position 2 nt upstream of this sRNA (20). Conservation of the sRNA is paralleled by conservation of HCF107, which is orthologous to the Chlamydomonas Mbb1 protein (20). From the class of RNA recognition motif (RRM) proteins, the chloroplast ribonucleoprotein CP31A has been implicated in stabilization of the ndhF message (27) and may be involved in generating the footprint in the 3′-region of ndhF identified here. Identification of minimal binding sites for more of the multitude of chloroplast RNA binding proteins, in particular for PPR proteins, is obviously urgently needed to investigate this.
Outlook: the use of sRNAs
If we accept that most sRNAs will be footprints of RBPs, we have to expect that their abundance will change with changes in the abundance of the corresponding proteins. In turn, a change of sRNA abundance will be telling of changes in the stability and translation status of the corresponding mRNA. Indeed, in the deep sequencing dataset on the chloroplast transcriptome of Chinese cabbage, the abundance of selected sRNAs changes in response to heat stress (3). It will be exciting to determine sRNA patterns in the future under different conditions and thus link outside signals with chloroplast RBPs and the chloroplast RNA pool. This has the potential to uncover regulatory roles for chloroplast RBPs—such regulatory functions have been notoriously difficult to nail down so far. Moreover, the question of whether sRNAs are really only footprints or serve a role by themselves is wide open. If sRNAs are really only useless remnants of a degraded 5′-non-coding region, their accumulation could be detrimental, because they would titrate their cognate RBP away from its real job on the mRNA. So, is each sRNA covered by its cognate RBP? If not, why are they not degraded down to nucleotides? A quantitative understanding of the relation between the abundance of the mRNA, the RBP and the sRNA will be necessary to answer these problems. In the future, overexpression of sRNAs or expression of antisense sRNAs could probe putative roles for sRNAs as modulators of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–7 and Supplementary References [9–11].
FUNDING
The Deutsche Forschungsgemeinschaft (DFG) (Emmy-Noether stipend SCHM 1698/2-1 to C.S.L.). Funding for open access charge: own funding, DFG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ian Small and Alice Barkan for critical discussion of the data and Alice Barkan for sharing data prior to publication and comments on this manuscript. We are in particular grateful to Peter Westhoff, Dagmar Lyska and Karin Meierhoff for providing seeds for hcf107 and hcf152 mutant Arabidopsis lines.
REFERENCES
- 1.Vera A, Sugiura M. A novel RNA gene in the tobacco plastid genome: its possible role in the maturation of 16S rRNA. EMBO J. 1994;13:2211–2217. doi: 10.1002/j.1460-2075.1994.tb06498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Yu X, Wang H, Lu Y, de Ruiter M, Prins M, He Y. A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa) BMC Genomics. 2011;12:289. doi: 10.1186/1471-2164-12-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–D833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 11.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao WB, Meng BY, Tanaka M, Sugiura M. An additional promoter within the protein-coding region of the psbD-psbC gene cluster in tobacco chloroplast DNA. Nucleic Acids Res. 1989;17:9583–9591. doi: 10.1093/nar/17.23.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas-Suarez M, Börner T, Barkan A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012;40:3092–3105. doi: 10.1093/nar/gkr1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meurer J, Berger A, Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai W, Okuda K, Peng L, Shikanai T. Proton Gradient Regulation 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011;67:318–327. doi: 10.1111/j.1365-313X.2011.04593.x. [DOI] [PubMed] [Google Scholar]
- 18.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sane AP, Stein B, Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- 20.Felder S, Meurer J, Meierhoff K, Klaff P, Bechtold N, Westhoff P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peled-Zehavi H, Danon A. Translation and translational regulation in chloroplasts. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol. 19. New York: Springer; 2007. pp. 249–282. [Google Scholar]
- 23.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. A Study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009;21:3686–3699. doi: 10.1105/tpc.109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008;25:1120–1128. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 26.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz-Linneweber C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl Acad. Sci. USA. 2009;106:6002–6007. doi: 10.1073/pnas.0808529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.